MIFNN: Molecular Information Feature Extraction and Fusion Deep Neural Network for Screening Potential Drugs
Abstract
1. Introduction
2. Materials and Methods
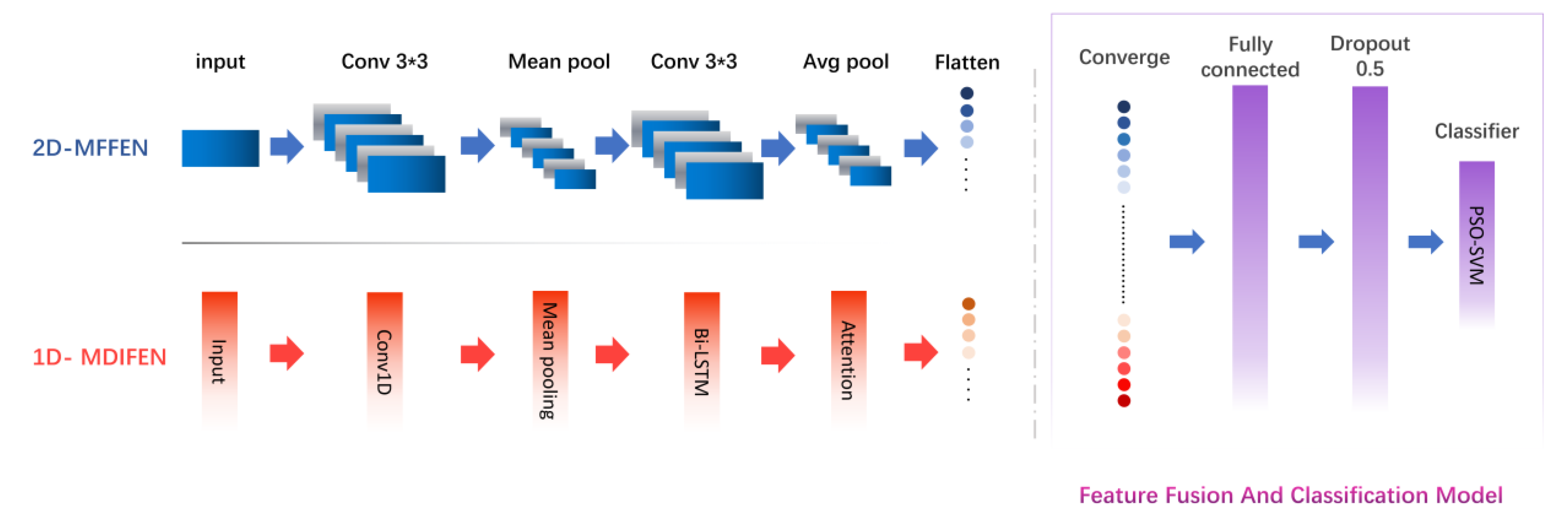
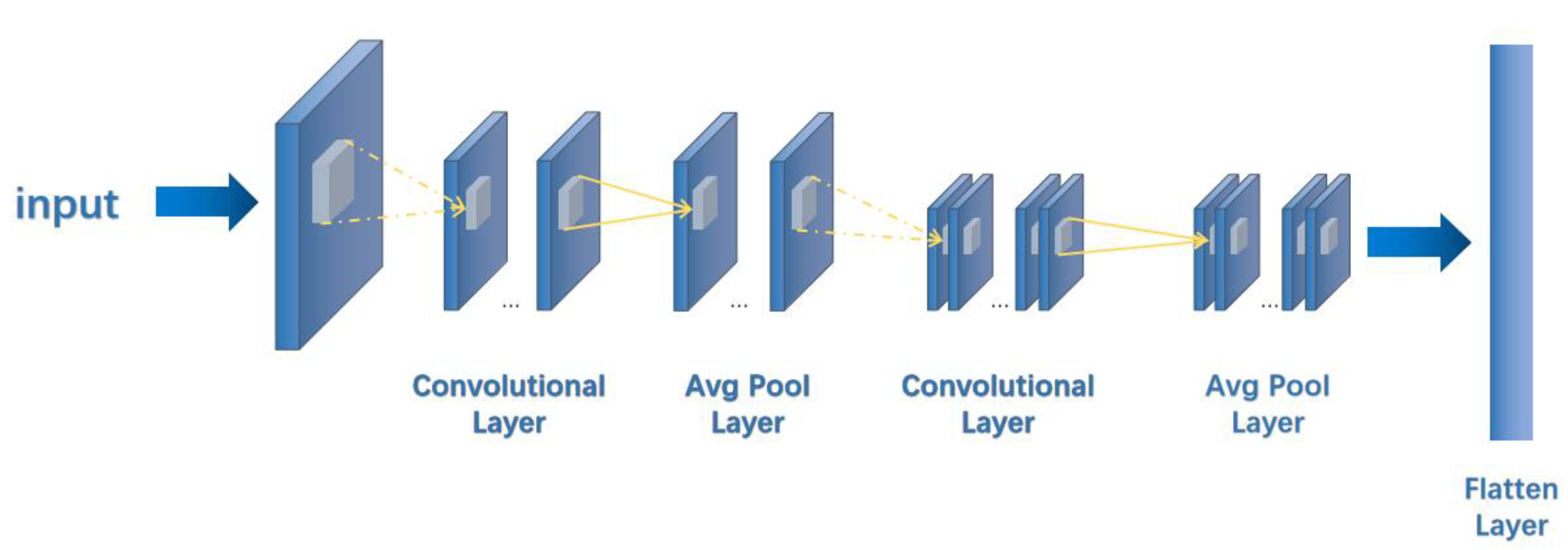
2.1. Network Model
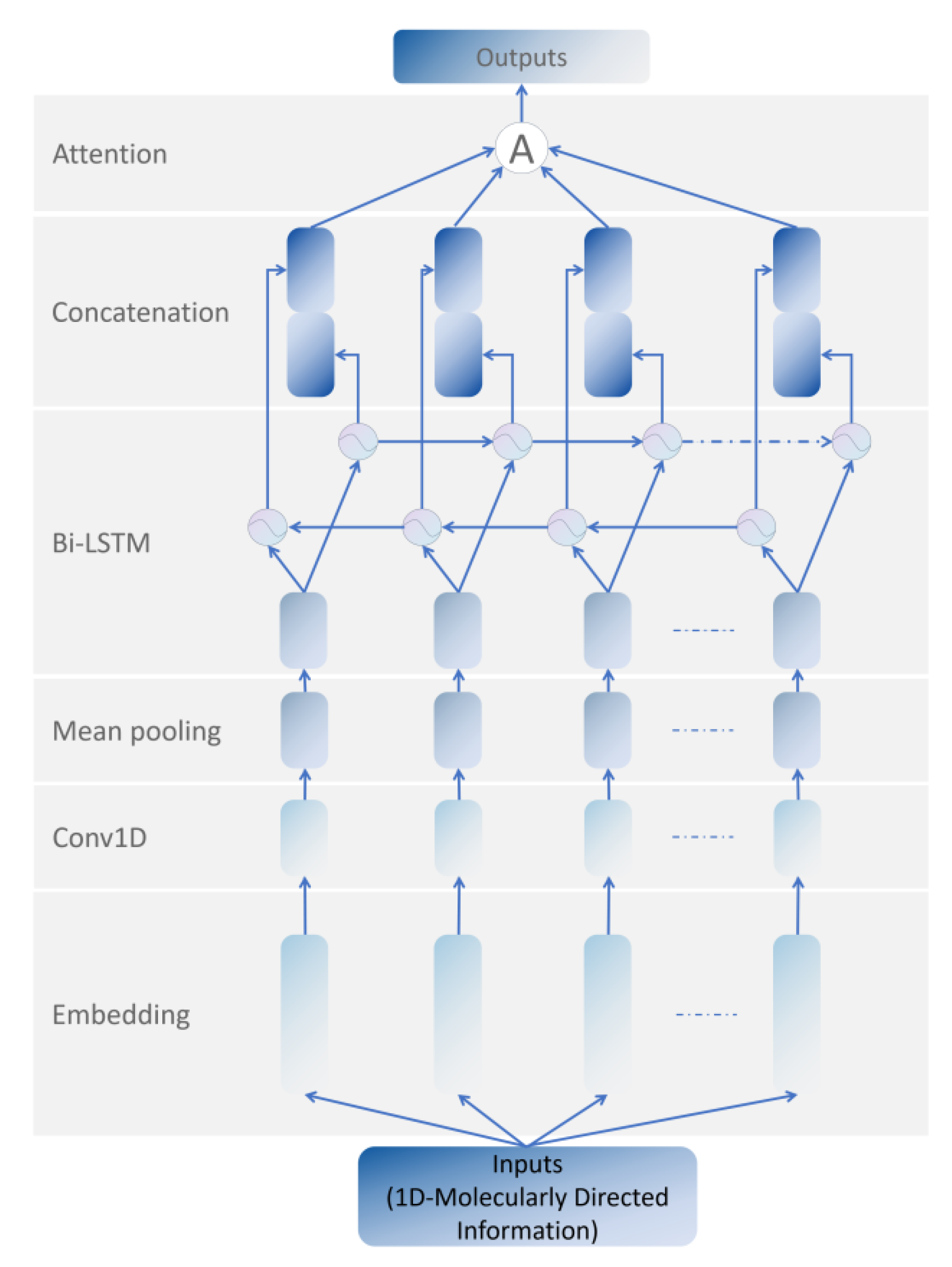
2.1.1. Molecular Feature Extraction Module based on bi-LSTM
2.1.2. Feature Fusion and Classification Model
2.2. Directed Message Passing Information
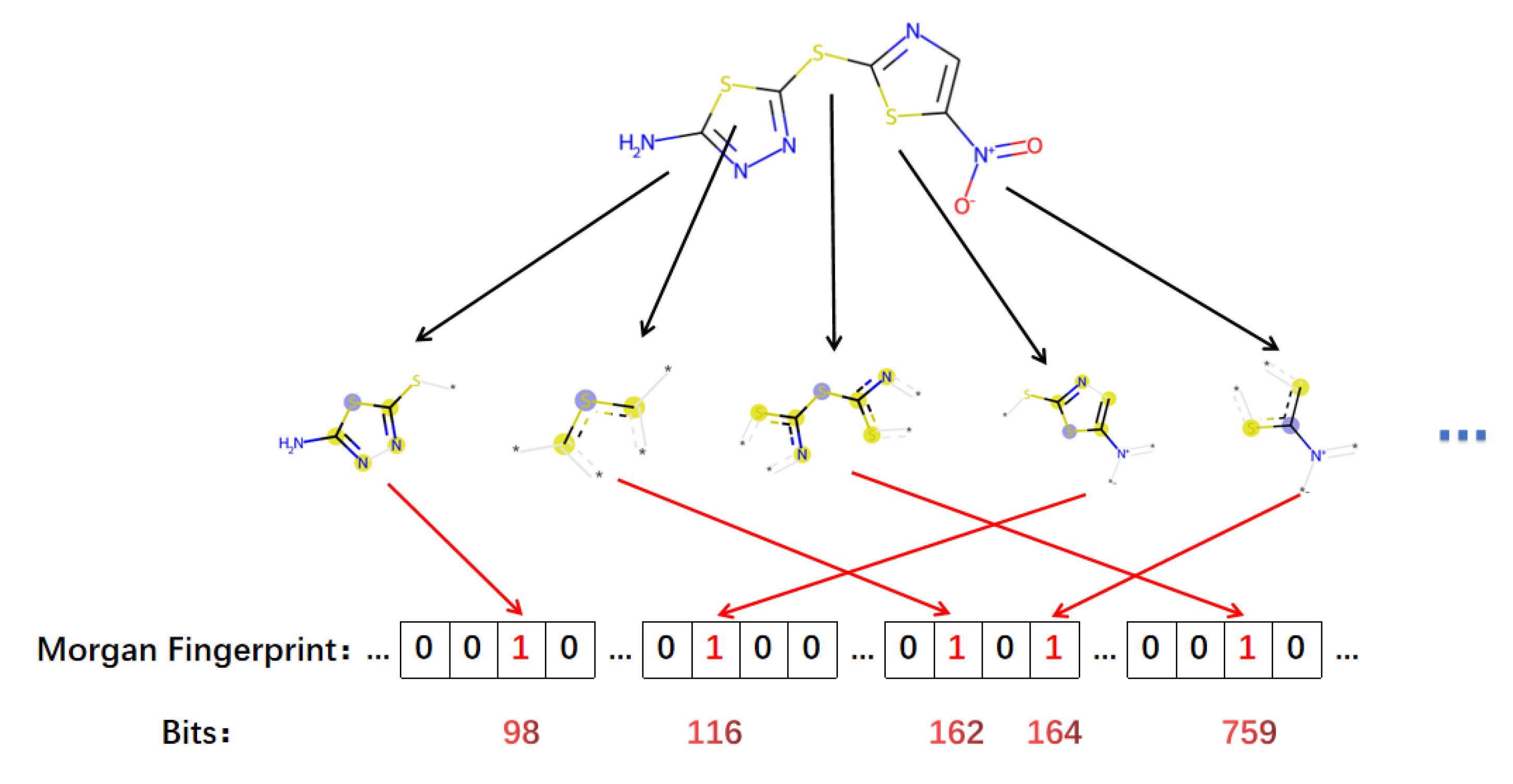
2.3. Morgan Fingerprint
3. Experiment
3.1. Model Experiment
3.1.1. Data
3.1.2. Model Performance Evaluation
3.2. Experimental Results
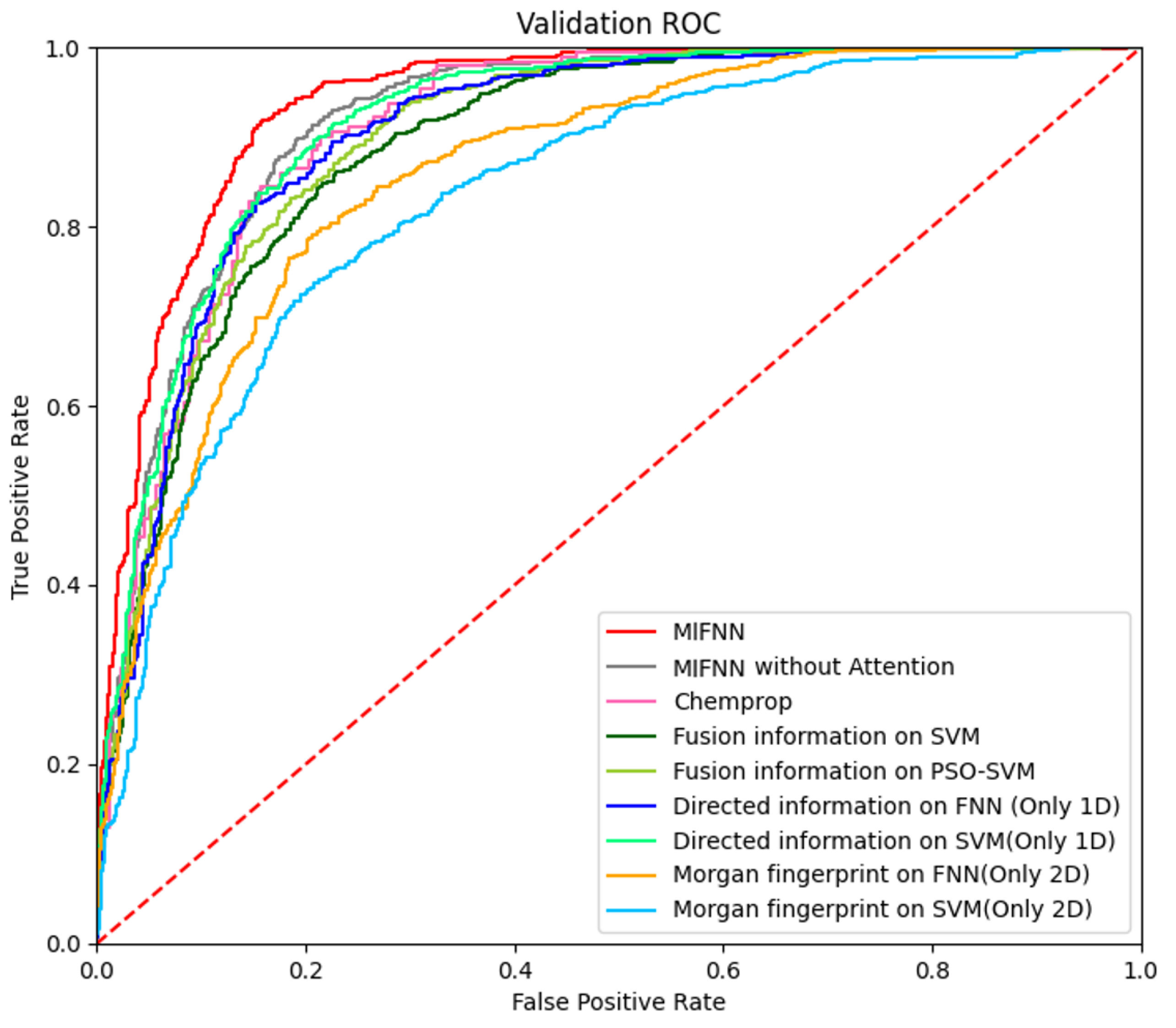
3.2.1. Improvements to Other Baseline Models
3.2.2. Improvement under Different Conditions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Ding, J.; Wang, Z.; Cao, D.; Ding, X.; Hou, T. From machine learning to deep learning: Advances in scoring functions for protein–ligand docking. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2020, 10, 1429. [Google Scholar] [CrossRef]
- Kearnes, S.; McCloskey, K.; Berndl, M.; Pande, V.; Riley, P. Molecular graph convolutions: Moving beyond fingerprints. J. Comput.-Aided Mol. Des. 2016, 30, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Kadurin, A.; Nikolenko, S.; Khrabrov, K.; Aliper, A.; Zhavoronkov, A. druGAN: An Advanced Generative Adversarial Autoencoder Model for de Novo Generation of New Molecules with Desired Molecular Properties in Silico. Mol. Pharm. 2017, 14, 3098–3104. [Google Scholar] [CrossRef]
- Schneider, G. Virtual screening: An endless staircase? Nat. Rev. Drug Discov. 2010, 9, 273–276. [Google Scholar] [CrossRef]
- Weininger, D. Smiles. 3. Depict. Graphical Depiction of Chemical Structures. J. Chem. Inf. Comput. Sci. 1990, 30, 237–243. [Google Scholar] [CrossRef]
- Yang, K.; Swanson, K.; Jin, W.; Coley, C.; Eiden, P.; Gao, H.; Guzman-Perez, A.; Hopper, T.; Kelley, B.; Mathea, M.; et al. Analyzing Learned Molecular Representations for Property Prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. [Google Scholar] [CrossRef]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef]
- Li, C.; Wei, W.; Li, J.; Yao, J.; Zeng, X.; Lv, Z. 3DMol-Net: Learn 3D Molecular Representation using Adaptive Graph Convolutional Network Based on Rotation Invariance. IEEE J. Biomed. Health Inform. 2021, 14, 2168–2194. [Google Scholar] [CrossRef]
- Senese, C.L.; Duca, J.; Pan, D.; Hopfinger, A.J.; Tseng, Y.J. 4D-fingerprints, universal QSAR and QSPR descriptors. J. Chem. Inf. Comput. Sci. 2004, 44, 1526–1539. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Brooks, B.R. A deep learning approach for the blind logP prediction in SAMPL6 challenge. J. Comput.-Aided Mol. Des. 2020, 34, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Steijaert, M.; Wegner, J.K.; Ceulemans, H.; Clevert, D.-A. Hochreiter, S.. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Ence 2018, 9, 5441–5451. [Google Scholar]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2017, 9, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.J.; Hopfinger, A.J.; Esposito, E.X. The great descriptor melting pot: Mixing descriptors for the common good of QSAR models. J. Comput.-Aided Mol. Des. 2012, 26, 39–43. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Chang, Q. Feature selection methods for big data bioinformatics: A survey from the search perspective. Methods 2016, 111, 21–31. [Google Scholar] [CrossRef]
- Pérez-Castillo, Y.; Lazar, C.; Taminau, J.; Froeyen, M.; Cabrera-Pérez, M.; Nowé, A. VGA(M)E-QSAR: A novel, fully automatic genetic-algorithm-(meta)-ensembles approach for binary classification in ligand-based drug design. J. Chem. Inf. Model. 2012, 52, 2366–2386. [Google Scholar] [CrossRef]
- Su, B.-H.; Tu, Y.-S.; Esposito, E.X.; Tseng, Y.J. Predictive toxicology modeling: Protocols for exploring hERG classification and Tetrahymena pyriformis end point predictions. J. Chem. Inf. Model. 2012, 52, 1660–1673. [Google Scholar] [CrossRef]
- Ye, H.; Lounkine, E.; Bajorath, J. Filtering and counting of extended connectivity fingerprint features maximizes compound recall and the structural diversity of hits. Chem. Biol. Drug Des. 2009, 74, 92–98. [Google Scholar]
- Fu, L.; Liu, L.; Yang, Z.; Li, P.; Ding, J.J.; Yun, Y.H.; Lu, A.P.; Hou, T.J.; Cao, D.S. Systematic Modeling of logD 7.4 Based on Ensemble Machine Learning, Group Contribution and Matched Molecular Pair Analysis. J. Chem. Inf. Model. 2020, 60, 63–76. [Google Scholar] [CrossRef]
- Jiang, D.; Lei, T.; Wang, Z.; Shen, C.; Cao, D.; Hou, T. ADMET evaluation in drug discovery. 20. Prediction of breast cancer resistance protein inhibition through machine learning. J. Cheminform. 2020, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Taherkhani, A.; Cosma, G.; McGinnity, T.M. Deep-FS: A feature selection algorithm for Deep Boltzmann Machines. Neurocomputing 2018, 322, 22–37. [Google Scholar] [CrossRef]
- Tetko, I.V.; Tropsha, A.; Zhu, H.; Papa, E.; Gramatica, P.; Öberg, T.; Fourches, D.; Varnek, A. Comparison of applicability domains of QSAR models: Application to the modelling of the environmental toxicity against Tetrahymena pyriformis. Chem. Cent. J. 2008, 2, 14. [Google Scholar] [CrossRef][Green Version]
- Tetko, I.V.; Livingstone, D.J.; Luik, A.I. Neural network studies. 1. Comparison of overfitting and overtraining. J. Chem. Inf. Comput. Sci. 1995, 35, 826–833. [Google Scholar] [CrossRef]
- Xie, L.; Xu, L.; Kong, R.; Chang, S.; Xu, X. Improvement of Prediction Performance With Conjoint Molecular Fingerprint in Deep Learning. Front. Pharmacol. 2020, 11, 606668. [Google Scholar] [CrossRef]
- Jiang, D.; Wu, Z.; Hsieh, C.-Y.; Chen, G.; Ben Liao, B.; Wang, Z.; Shen, C.; Cao, D.; Wu, J.; Hou, T. Could graph neural networks learn better molecular representation for drug discovery? A comparison study of descriptor-based and graph-based models. J. Cheminform. 2021, 13, 12. [Google Scholar] [CrossRef]
- Chen, D.; Gao, K.; Nguyen, D.D.; Chen, X.; Jiang, Y.; Wei, G.-W.; Pan, F. Algebraic graph-assisted bidirectional transformers for molecular property prediction. Nat. Commun. 2021, 12, 3521. [Google Scholar] [CrossRef]
- Lenselink, E.B.; Ten Dijke, N.; Bongers, B.; Papadatos, G.; van Vlijmen, H.W.T.; Kowalczyk, W.; Ijzerman, A.P.; van Westen, G.J.P. Beyond the hype: Deep neural networks outperform established methods using a ChEMBL bioactivity benchmark set. J. Cheminform 2017, 9, 45. [Google Scholar] [CrossRef]
- Öztürk, H.; Özgür, A.; Ozkirimli, E. DeepDTA: Deep drug-target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Deng, W.; Zhao, H.; Yang, X.; Xiong, J.; Sun, M.; Li, B. Study on an improved adaptive PSO algorithm for solving multi-objective gate assignment. Appl. Soft Comput. 2017, 59, 288–302. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Gao, L.; Li, P. A new subset based deep feature learning method for intelligent fault diagnosis of bearing. Expert Syst. Appl. 2018, 100, 125–142. [Google Scholar] [CrossRef]
- Duan, B. Research on Application of Support Vector Machine in Machine Learning. J. Electron. Res. Appl. 2019, 3, 11–14. [Google Scholar] [CrossRef]
- Mustaqeem; Kwon, S. MLT-DNet: Speech emotion recognition using 1D dilated CNN based on multi-learning trick approach. Expert Syst. Appl. 2020, 167, 114117. [Google Scholar]
- Ji, Y.; Zhang, H.; Zhang, Z.; Liu, M. CNN-based encoder-decoder networks for salient object detection: A comprehensive review and recent advances. Inf. Sci. Int. J. 2021, 546, 835–857. [Google Scholar] [CrossRef]
- Shi, Q.; Zhang, H. Fault Diagnosis of an Autonomous Vehicle With an Improved SVM Algorithm Subject to Unbalanced Datasets. IEEE Trans. Ind. Electron. 2021, 68, 6248–6256. [Google Scholar] [CrossRef]
- Wang, F.; Sahana, M.; Pahlevanzadeh, B.; Pal, S.C.; Shit, P.K.; Piran, J.; Janizadeh, S.; Band, S.S.; Mosavi, A. Applying different resampling strategies in machine learning models to predict head-cut gully erosion susceptibility. Alex. Eng. J. 2021, 60, 5813–5829. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural Message Passing for Quantum Chemistry. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 11 August 2017. [Google Scholar]
- Dai, H.; Dai, B.; Song, L. Discriminative Embeddings of Latent Variable Models for Structured Data. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 19 June 2016. [Google Scholar]
- Mahé, P.; Ueda, N.; Akutsu, T.; Perret, J.L.; Vert, J.P. Extensions of marginalized graph kernels. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, Canada, 4 July 2004. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified Linear Units Improve Restricted Boltzmann Machines. In Proceedings of the 27th International Conference on Machine Learning, Haifa, Israel, 21 June 2010. [Google Scholar]
- Cao, D.S.; Xu, Q.S.; Hu, Q.N.; Liang, Y.Z. ChemoPy: Freely available python package for computational biology and chemoinformatics. Bioinformatics 2013, 29, 1092–1094. [Google Scholar] [CrossRef]
- Huang, T.; Mi, H.; Lin, C.Y.; Zhao, L.; Zhong, L.L.; Liu, F.B.; Zhang, G.; Lu, A.P.; Bian, Z.X. MOST: Most-similar ligand based approach to target prediction. BMC Bioinform. 2017, 18, 165. [Google Scholar] [CrossRef]
- Cortes-Ciriano, I. Bioalerts: A python library for the derivation of structural alerts from bioactivity and toxicity data sets. J. Cheminform. 2016, 8, 13. [Google Scholar] [CrossRef]



| Data Set | Category | Description | Size |
|---|---|---|---|
| HIV | Biophysics | Inhibition of HIV replication | 41,127 |
| BACE | Biophysics | Inhibition of human β-secretase 1 | 1513 |
| BBBP | Physiology | Ability to penetrate the blood–brain barrier | 2039 |
| Tox21 | Physiology | Toxicity | 7831 |
| ToxCast | Physiology | Toxicity | 8576 |
| SIDER | Physiology | Side-effects of drugs | 1427 |
| ClinTox | Physiology | Toxicity | 1478 |
| ChEMBL | Physiology | Biological assays | 456,331 |
| Dataset | MIFNN | Mayr’s | MolNet | Chemprop |
|---|---|---|---|---|
| HIV | 0.867 | 0.81 | 0.798 | 0.81 |
| BACE | 0.922 | 0.834 | 0.715 | 0.821 |
| BBBP | 0.909 | 0.891 | 0.736 | 0.893 |
| Tox21 | 0.876 | 0.791 | 0.809 | 0.823 |
| ToxCast | 0.849 | 0.698 | 0.605 | 0.741 |
| SIDER | 0.59 | 0.586 | 0.605 | 0.625 |
| ClinTox | 0.842 | 0.817 | 0.82 | 0.85 |
| ChEMBL | 0.895 | 0.784 | 0.754 | 0.775 |
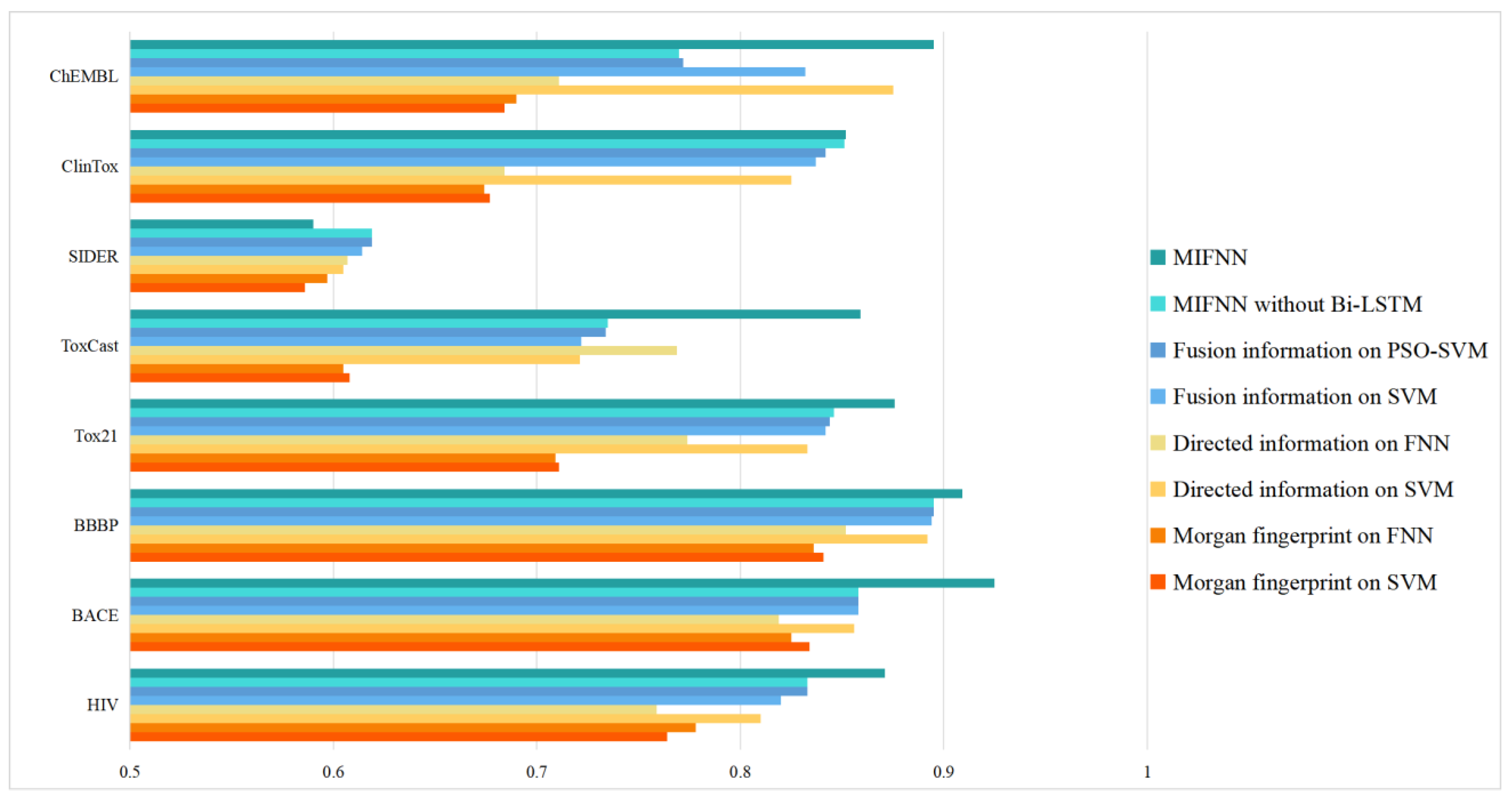
| Dataset | Morgan Fingerprint on SVM | Morgan Fingerprint on FNN | Directed Information on SVM | Directed Information on FNN | Fusion Information on SVM | Fusion Information on PSO-SVM | MIFNN without Bi-LSTM | MIFNN |
|---|---|---|---|---|---|---|---|---|
| HIV | 0.764 | 0.778 | 0.81 | 0.759 | 0.811 | 0.816 | 0.833 | 0.871 |
| BACE | 0.834 | 0.825 | 0.856 | 0.819 | 0.837 | 0.862 | 0.868 | 0.925 |
| BBBP | 0.841 | 0.836 | 0.89 | 0.852 | 0.870 | 0.877 | 0.895 | 0.909 |
| Tox21 | 0.711 | 0.709 | 0.833 | 0.774 | 0.817 | 0.839 | 0.846 | 0.876 |
| ToxCast | 0.608 | 0.605 | 0.721 | 0.769 | 0.724 | 0.731 | 0.735 | 0.859 |
| SIDER | 0.586 | 0.597 | 0.605 | 0.607 | 0.601 | 0.615 | 0.619 | 0.59 |
| ClinTox | 0.677 | 0.674 | 0.825 | 0.684 | 0.696 | 0.815 | 0.851 | 0.852 |
| ChEMBL | 0.684 | 0.69 | 0.875 | 0.711 | 0.841 | 0.786 | 0.77 | 0.895 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, H.; Zhao, W.; Pang, T.; Sun, Z.; Zhang, B.; Xu, H. MIFNN: Molecular Information Feature Extraction and Fusion Deep Neural Network for Screening Potential Drugs. Curr. Issues Mol. Biol. 2022, 44, 5638-5654. https://doi.org/10.3390/cimb44110382
Wang J, Li H, Zhao W, Pang T, Sun Z, Zhang B, Xu H. MIFNN: Molecular Information Feature Extraction and Fusion Deep Neural Network for Screening Potential Drugs. Current Issues in Molecular Biology. 2022; 44(11):5638-5654. https://doi.org/10.3390/cimb44110382
Chicago/Turabian StyleWang, Jingjing, Hongzhen Li, Wenhan Zhao, Tinglin Pang, Zengzhao Sun, Bo Zhang, and Huaqiang Xu. 2022. "MIFNN: Molecular Information Feature Extraction and Fusion Deep Neural Network for Screening Potential Drugs" Current Issues in Molecular Biology 44, no. 11: 5638-5654. https://doi.org/10.3390/cimb44110382
APA StyleWang, J., Li, H., Zhao, W., Pang, T., Sun, Z., Zhang, B., & Xu, H. (2022). MIFNN: Molecular Information Feature Extraction and Fusion Deep Neural Network for Screening Potential Drugs. Current Issues in Molecular Biology, 44(11), 5638-5654. https://doi.org/10.3390/cimb44110382






