Status of Fungicide Resistance and Physiological Characterization of Tebuconazole Resistance in Rhizocotonia solani in Sichuan Province, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of R. solani
2.2. Fungicides and Chemicals
2.3. Determination of Resistance Frequency of R. solani
2.4. Sensitivity Baseline or Resistance Ratio of R. solani to the Tested Fungicides
2.5. Fitness Determination of Tebuconazole-Resistant Isolates
2.6. Determination of Cell Membrane Permeability
2.7. Clone of CYP51 Gene from R. solani Isolates
2.8. Functional Domain and Structural Analysis of CYP51 Gene
2.9. Statistical Analysis
3. Results
3.1. Resistance Frequency of R. solani in Sichuan Province
3.2. Sensitivity Baseline of R. solani to Four Test Fungicides
3.3. Resistance Levels of R. solani Isolates to Four Fungicides
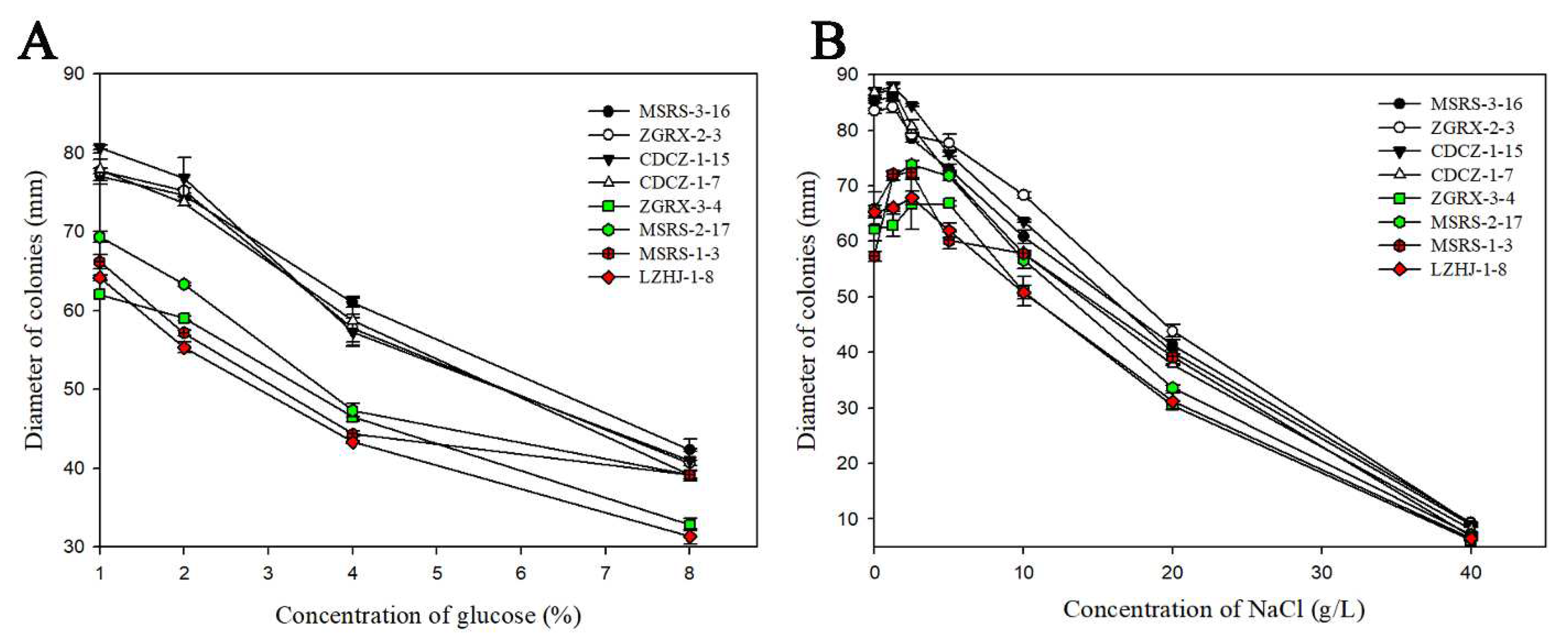
3.4. Fitness of Tebuconazole-Resistant Isolates
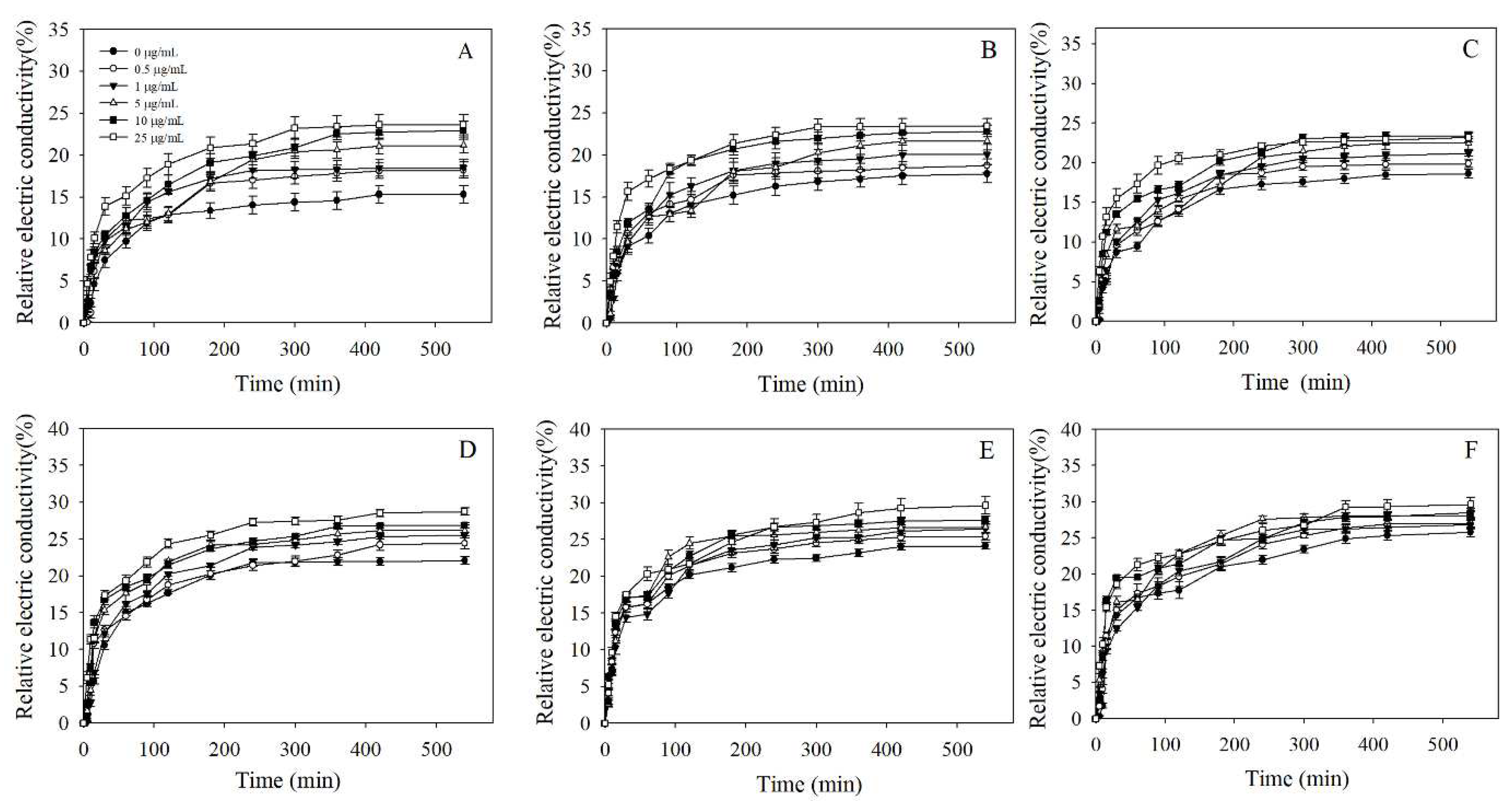
3.5. Cell Membrane Permeability
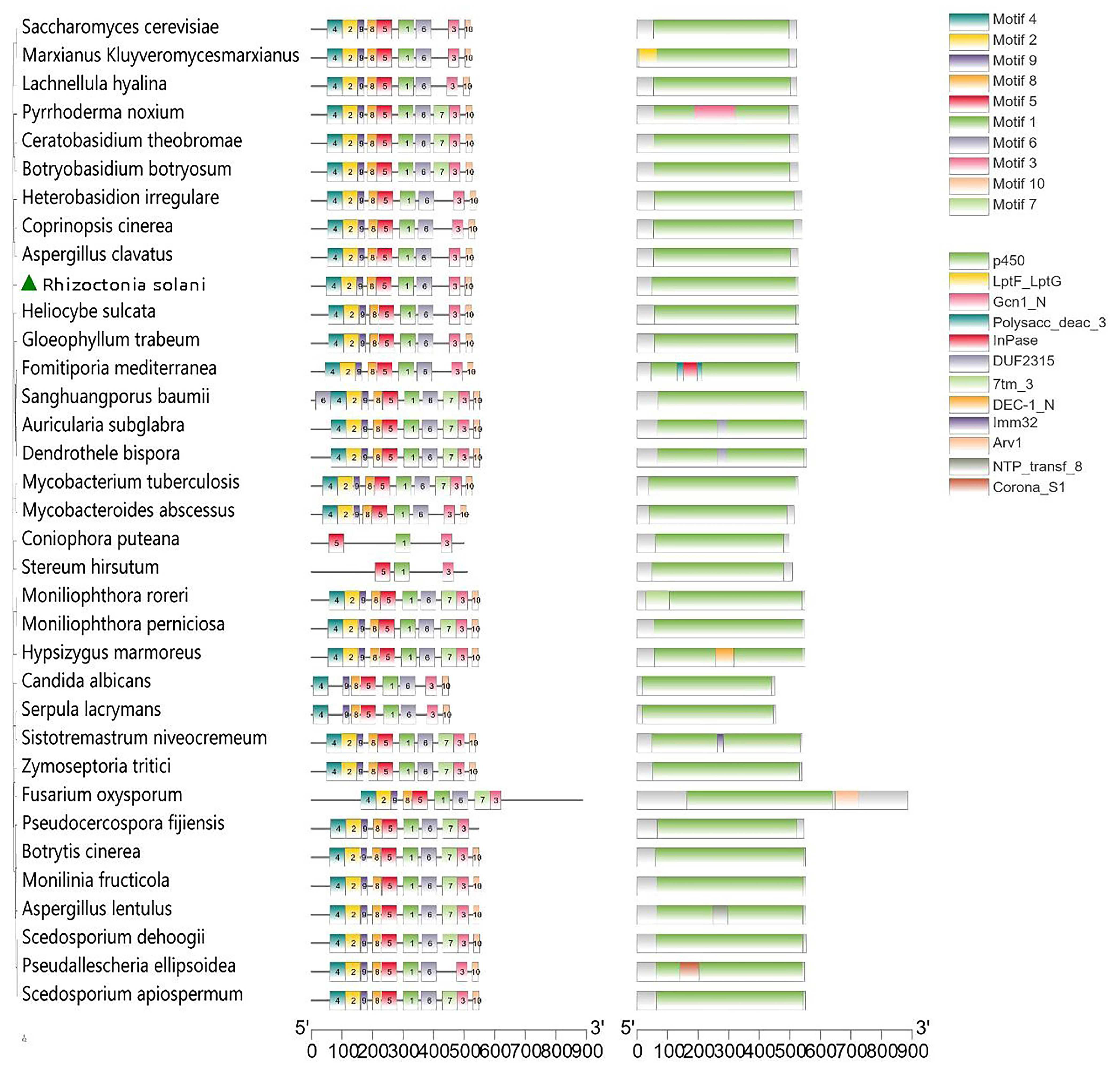
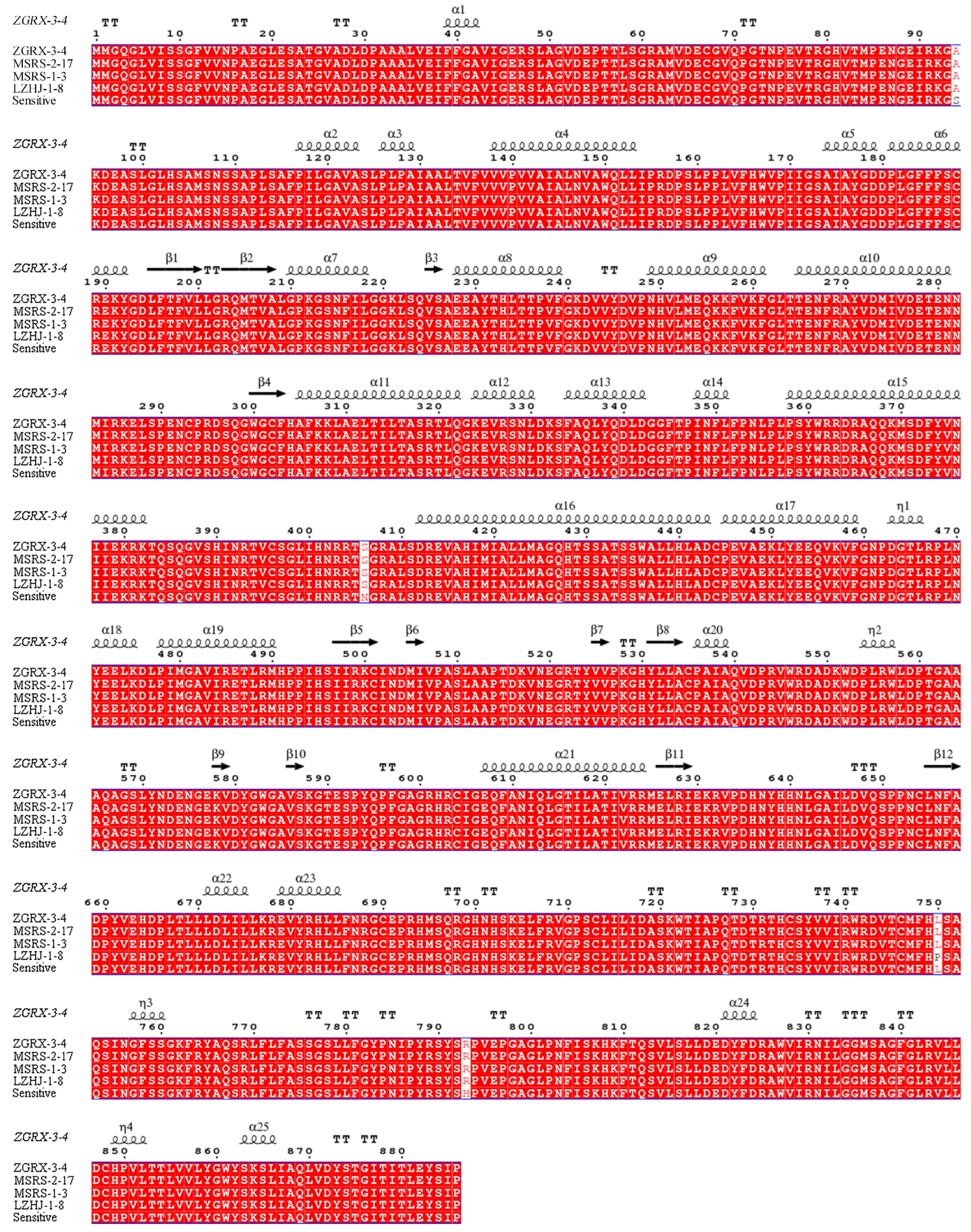
3.6. Functional Domain Analysis of Sterol 14α-Demethylase (CYP51)
3.7. Detection of CYP51 Mutation of Tebuconazole-Susceptible and -Resistant Isolates
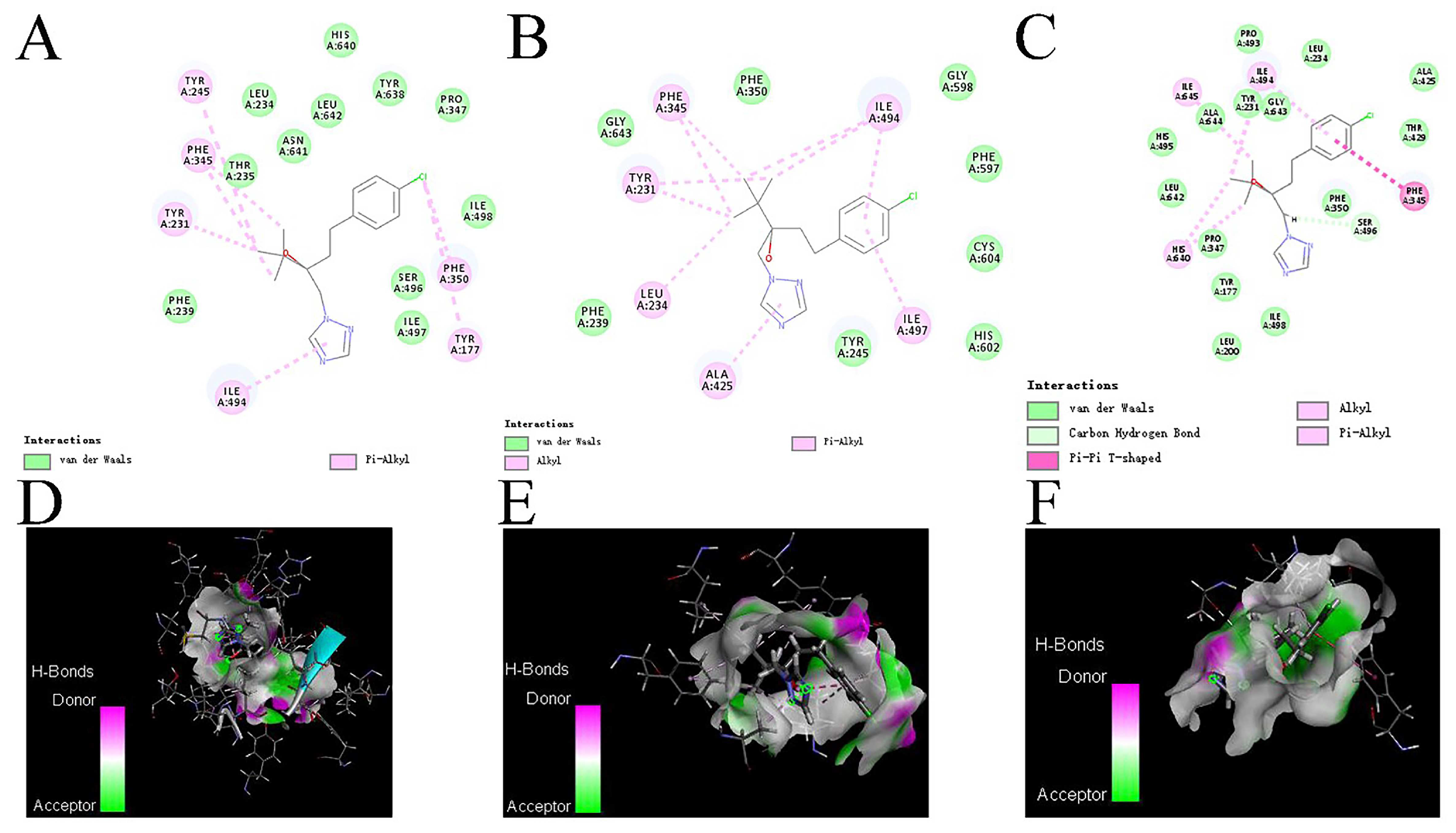
3.8. Effects of Mutations on the Affinity of CYP51 Protein and Tebuconazole
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naeimi, S.; Okhovvat, S.M.; Javan-Nikkhah, M.; Vágvölgyi, C.; Khosravi, V.; Kredics, L. Biological control of Rhizoctonia solani AG1-1A, the causal agent of rice sheath blight with Trichoderma strains. Phytopathol. Mediterr. 2010, 49, 287–300. [Google Scholar]
- Prasad, B.; Eizenga, G.C. Rice sheath blight disease resistance identified in Oryza spp. accessions. Plant Dis. 2008, 92, 1503–1509. [Google Scholar] [CrossRef]
- Klikocka, H.; Haneklaus, S.; Bloem, E.; Schnug, E. Influence of sulfur fertilization on infection of potato tubers with Rhizoctonia solani and Streptomyces scabies. J. Plant Nutr. 2005, 28, 819–833. [Google Scholar] [CrossRef]
- Ajayi-Oyetunde, O.O.; Bradley, C.A. Rhizoctonia solani: Taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant pathol. 2018, 67, 3–17. [Google Scholar] [CrossRef]
- Moore, W.F. Density of sclerotia of Rhizoctonia solani and incidence of sheath blight in rice fields in Mississippi. Plant Dis. 1993, 77, 257–260. [Google Scholar]
- Zhao, C.; Zhang, X.; Hua, H.; Han, C.; Wu, X. Sensitivity of Rhizoctonia spp. to flutolanil and characterization of the point mutation in succinate dehydrogenase conferring fungicide resistance. Eur. J. Plant Pathol. 2019, 155, 13–23. [Google Scholar] [CrossRef]
- Chen, Y.A.; Zhang, W.; Wang, W.X.; Zhang, Y.; Gao, T.C. Baseline sensitivity and efficacy of thifluzamide in Rhizoctonia solani. Ann. Appl. Biol. 2012, 161, 247–254. [Google Scholar] [CrossRef]
- Mu, W.; Wang, Z.; Bi, Y.; Ni, X.; Hou, Y.; Zhang, S.; Liu, X. Sensitivity determination and resistance risk assessment of Rhizoctonia solani to SDHI fungicide thifluzamide. Ann. Appl. Biol. 2017, 170, 240–250. [Google Scholar] [CrossRef]
- Ajayi-Oyetunde, O.O.; Butts-Wilmsmeyer, C.J.; Bradley, C.A. Sensitivity of Rhizoctonia solani to succinate dehydrogenase inhibitor and demethylation inhibitor fungicides. Plant Dis. 2017, 101, 487–495. [Google Scholar] [CrossRef]
- Suemoto, H.; Matsuzaki, Y.; Iwahashi, F. Metyltetraprole, a novel putative complex III inhibitor, targets known QoI-resistant strains of Zymoseptoria tritici and Pyrenophora teres. Pest Manag. Sci. 2019, 75, 1181–1189. [Google Scholar] [CrossRef]
- Spanner, R.; Taliadoros, D.; Richards, J.; Rivera-Varas, V.; Neubauer, J.; Natwick, M.; Hamilton, O.; Vaghefi, N.; Pethybridge, S.; Secor, G.; et al. Genome-wide association and selective sweep studies reveal the complex genetic architecture of DMI fungicide resistance in Cercospora beticola. Genome Biol. Evol. 2021, 13, evab209. [Google Scholar] [CrossRef]
- Chen, S.; Schnabel, G.; Yuan, H.; Luo, C. LAMP detection of the genetic element ‘Mona’ associated with DMI resistance in Monilinia fructicola. Pest Manag. Sci. 2019, 75, 779–786. [Google Scholar] [CrossRef]
- Wei, L.L.; Chen, W.C.; Zhao, W.C.; Wang, J.; Wang, B.R.; Li, F.J.; Wei, M.D.; Guo, J.; Chen, C.J.; Zheng, J.Q. Mutations and overexpression of CYP51 associated with DMI-resistance in Colletotrichum gloeosporioides from chili. Plant Dis. 2020, 104, 668–676. [Google Scholar] [CrossRef]
- Avenot, H.F.; Solorio, C.; Morgan, D.P.; Michailides, T.J. Sensitivity and cross-resistance patterns to demethylation-inhibiting fungicides in California populations of Alternaria alternata pathogenic on pistachio. Crop Prot. 2016, 88, 72. [Google Scholar] [CrossRef]
- Sun, X.; Wang, J.; Feng, D.; Ma, Z.; Li, H. PdCYP51B, a new putative sterol 14α-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl. Microbiol. Biotechnol. 2011, 91, 1107–1119. [Google Scholar] [CrossRef]
- Cools, H.J.; Bayon, C.; Atkins, S.; Lucas, J.A.; Fraaije, B.A. Overexpression of the sterol 14α-demethylase gene (MgCYP51) in Mycosphaerella graminicola isolates confers a novel azole fungicide sensitivity phenotype. Pest Manag. Sci. 2012, 68, 1034–1040. [Google Scholar] [CrossRef]
- Zhang, T.; Cao, Q.; Li, N.; Liu, D.; Yuan, Y. Transcriptome analysis of fungicide-responsive gene expression profiles in two Penicillium italicum strains with different response to the sterol demethylation inhibitor (DMI) fungicide prochloraz. BMC Genom. 2020, 21, 156. [Google Scholar] [CrossRef]
- Collinge, D.B.; Jørgensen, H.J.; Lund, O.S.; Lyngkjær, M.F. Engineering pathogen resistance in crop plants: Current trends and future prospects. Annu. Rev. Phytopathol. 2010, 48, 269–291. [Google Scholar] [CrossRef]
- Yuan, N.N.; Chen, S.N.; Zhai, L.X.; Schnabel, G.; Yin, L.F.; Luo, C.X. Baseline sensitivity of Monilia yunnanensis to the DMI fungicides tebuconazole and triadimefon. Eur. J. Plant Pathol. 2013, 136, 651–655. [Google Scholar] [CrossRef]
- Ma, Z.H.; Proffer, T.J.; Jacobs, J.L.; Sundin, G.W. Overexpression of the 14 α-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microb. 2006, 72, 2581–2585. [Google Scholar] [CrossRef]
- Tucker, M.A.; Lopez-Ruiz, F.; Cools, H.J. Analysis of mutations in West Australian populations of Blumeria graminis f. sp. hordei CYP51 conferring resistance to DMI fungicides. Pest Manag. Sci. 2020, 76, 1265–1272. [Google Scholar] [CrossRef]
- Yan, Y.F.; Yang, C.J.; Shang, X.F.; Zhao, Z.M.; Liu, Y.Q.; Zhou, R.; Liu, H.; Wu, T.L.; Zhao, W.B.; Wang, Y.L.; et al. Bioassay-guided isolation of two antifungal compounds from Magnolia officinalis, and the mechanism of action of honokiol. Pestic. Biochem. Physiol. 2020, 170, 104705. [Google Scholar] [CrossRef]
- Shao, W.; Zhang, Y.; Ren, W.; Chen, C. Physiological and biochemical characteristics of laboratory induced mutants of Botrytis cinerea with resistance to fluazinam. Pestic. Biochem. Physiol. 2015, 117, 19–23. [Google Scholar] [CrossRef]
- Karaoglanidis, G.S.; Markoglou, A.N.; Bardas, G.A.; Doukas, E.G.; Konstantinou, S.; Kalampokis, J.F. Sensitivity of Penicillium expansum field isolates to tebuconazole, iprodione, fludioxonil and cyprodinil and characterization of fitness parameters and patulin production. Int. J. Food Microbiol. 2011, 145, 195–204. [Google Scholar] [CrossRef]
- Lichtemberg, P.S.; Michailides, T.J.; Puckett, R.D.; Zeviani, W.M.; May-De-Mio, L.L. Fitness costs associated with G461S mutants of Monilinia fructicola could favor the management of tebuconazole resistance in Brazil. Trop. Plant Pathol. 2019, 44, 140–150. [Google Scholar] [CrossRef]
- Fillinger, S.; Leroux, P.; Auclair, C.; Barreau, C.; Al-Hajj, C.; Debieu, D. Genetic analysis of fenhexamid-resistant field isolates of the phytopathogenic fungus Botrytis cinerea. Antimicrob. Agents Chemother. 2008, 52, 3933–3940. [Google Scholar] [CrossRef][Green Version]
- Hellin, P.; King, R.; Urban, M.; Hammond-Kosack, K.E.; Legrève, A. The adaptation of Fusarium culmorum to DMI fungicides is mediated by major transcriptome modifications in response to azole fungicide, including the overexpression of a PDR transporter (FcABC1, Front. Microbiol. 2018, 9, 1385. [Google Scholar] [CrossRef]
- Pereira, W.V.; Morales, R.G.F.; Bauer, A.I.G.; Kudlawiec, K.; May-De-Mio, L.L. Discontinuance of tebuconazole in the field restores sensitivity of Monilinia fructicola in stone fruit orchards. Plant Pathol. 2020, 69, 68–76. [Google Scholar] [CrossRef]
- Stammler, G.; Taher, K.; Koch, A.; Haber, J.; Liebmann, B.; Bouagila, A.; Yahyaoui, A.; Nasraoui, B. Sensitivity of Mycosphaerella graminicola isolates from Tunisia to epoxiconazole and pyraclostrobin. Crop Prot. 2012, 34, 32–36. [Google Scholar] [CrossRef]
- Stammler, G.; Carstensen, M.; Koch, A.; Semar, M.; Strobel, D.; Schlehuber, S. Frequency of different CYP51-haplotypes of Mycosphaerella graminicola and their impact on epoxiconazole-sensitivity and-field efficacy. Crop Prot. 2008, 27, 1448–1456. [Google Scholar] [CrossRef]
- Stammler, G.; Cordero, J.; Koch, A.; Semar, M.; Schlehuber, S. Role of the Y134F mutation in cyp51 and overexpression of cyp51 in the sensitivity response of Puccinia triticina to epoxiconazole. Crop Prot. 2009, 28, 891–897. [Google Scholar] [CrossRef]
- Taher, K.; Graf, S.; Fakhfakh, M.M.; Salah, H.B.; Yahyaoui, A.; Rezgui, S.; Nasraoui, B.; Stammler, G. Sensitivity of Zymoseptoria tritici isolates from Tunisia to pyraclostrobin, fluxapyroxad, epoxiconazole, metconazole, prochloraz and tebuconazole. J. Phytopathol. 2014, 162, 442–448. [Google Scholar] [CrossRef]
- Qian, H.; Du, J.; Chi, M.; Sun, X.; Liang, W.; Huang, J.; Li, B.D. The Y137H mutation in the cytochrome P450 FgCYP51B protein confers reduced sensitivity to tebuconazole in Fusarium graminearum. Pest Manag. Sci. 2018, 74, 1472–1477. [Google Scholar] [CrossRef]
- Al-Fadhal, F.A.; AL-Abedy, A.N.; Alkhafije, D.A. Isolation and molecular identification of Rhizoctonia solani and Fusarium solani isolated from cucumber (Cucumis sativus L.) and their control feasibility by Pseudomonas fluorescens and Bacillus subtilis. Egypt. J. Biol. Pest Control 2019, 29, 47. [Google Scholar] [CrossRef]
- Kano, R.; Sobukawa, H.; Murayama, S.Y.; Hirose, D.; Tanaka, Y.; Kosuge, Y.; Hasegawa, A.; Kamata, H. In vitro resistance of Aspergillus fumigatus to azole farm fungicide. J. Infect. Chemother. 2016, 22, 133–136. [Google Scholar] [CrossRef]
- Shih, H.D.; Liu, Y.C.; Hsu, F.L.; Mulabagal, V.; Dodda, R.; Huang, J.W. Fungichromin: A substance from Streptomyces padanus with inhibitory effects on Rhizoctonia solani. J. Agric. Food Chem. 2003, 51, 95–99. [Google Scholar] [CrossRef]
- Halifu, S.; Deng, X.; Song, X.; Song, R.; Liang, X. Inhibitory mechanism of Trichoderma virens ZT05 on Rhizoctonia solani. Plants 2020, 9, 912. [Google Scholar] [CrossRef]
- Zhong, W.; Chang, Y.; Lin, Y.; Zhang, A. Synthesis and antifungal activities of hydrophilic cationic polymers against Rhizoctonia solani. Fungal Biol. 2020, 124, 735–741. [Google Scholar] [CrossRef]
- Das, M.M.; Haridas, M.; Sabu, A. Biological control of black pepper and ginger pathogens, Fusarium oxysporum, Rhizoctonia solani and Phytophthora capsici, using Trichoderma spp. Biocatal. Agric. Biotechnol. 2019, 17, 177–183. [Google Scholar]
- Dolores, F.O.; Chen, F.; Schnabel, G. Resistance to cyprodinil and lack of fludioxonil resistance in Botrytis cinerea isolates from strawberry in North and South Carolina. Plant Dis. 2013, 97, 81–85. [Google Scholar]
- Wang, B.; Liu, F.; Li, Q.; Xu, S.; Zhao, X.; Xue, P.; Feng, X. Antifungal activity of zedoary turmeric oil against Phytophthora capsici through damaging cell membrane. Pestic. Biochem. Physiol. 2019, 159, 59–67. [Google Scholar] [CrossRef]
- Malandrakis, A.A.; Vattis, K.N.; Markoglou, A.N.; Karaoglanidis, G.S. Characterization of boscalid-resistance conferring mutations in the SdhB subunit of respiratory complex II and impact on fitness and mycotoxin production in Penicillium expansum laboratory strains. Pestic. Biochem. Physiol. 2017, 138, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Majid, A.; Hossain, M.; Ahmed, S.K.; Ashid, M.; Bhojiya, A.A.; Upadhyay, S.K.; Vishvakarma, N.K.; Alam, M. Identification of 1, 2, 4-triazine and its derivatives against Lanosterol 14-demethylase (CYP51) property of Candida albicans: Influence on the development of new antifungal therapeutic strategies. Front. Med. Technol. 2022, 4, 845322. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.T.; Bass, C.; Williamson, M.S.; Kaussmann, M.; Wölfel, K.; Gutbrod, O.; Nauen, R. Molecular and functional characterization of CYP6BQ23, a cytochrome P450 conferring resistance to pyrethroids in European populations of pollen beetle, Meligethes aeneus. Insect Biochem. Mol. 2014, 45, 18–29. [Google Scholar] [CrossRef]
- Dueholm, B.; Krieger, C.; Drew, D.; Olry, A.; Kamo, T.; Taboureau, O.; Weitzel, C.; Bourgaud, F.; Simonsen, H.T. Evolution of substrate recognition sites (SRSs) in cytochromes P450 from Apiaceae exemplified by the CYP71AJ subfamily. BMC Evol. Biol. 2015, 15, 122. [Google Scholar] [CrossRef]
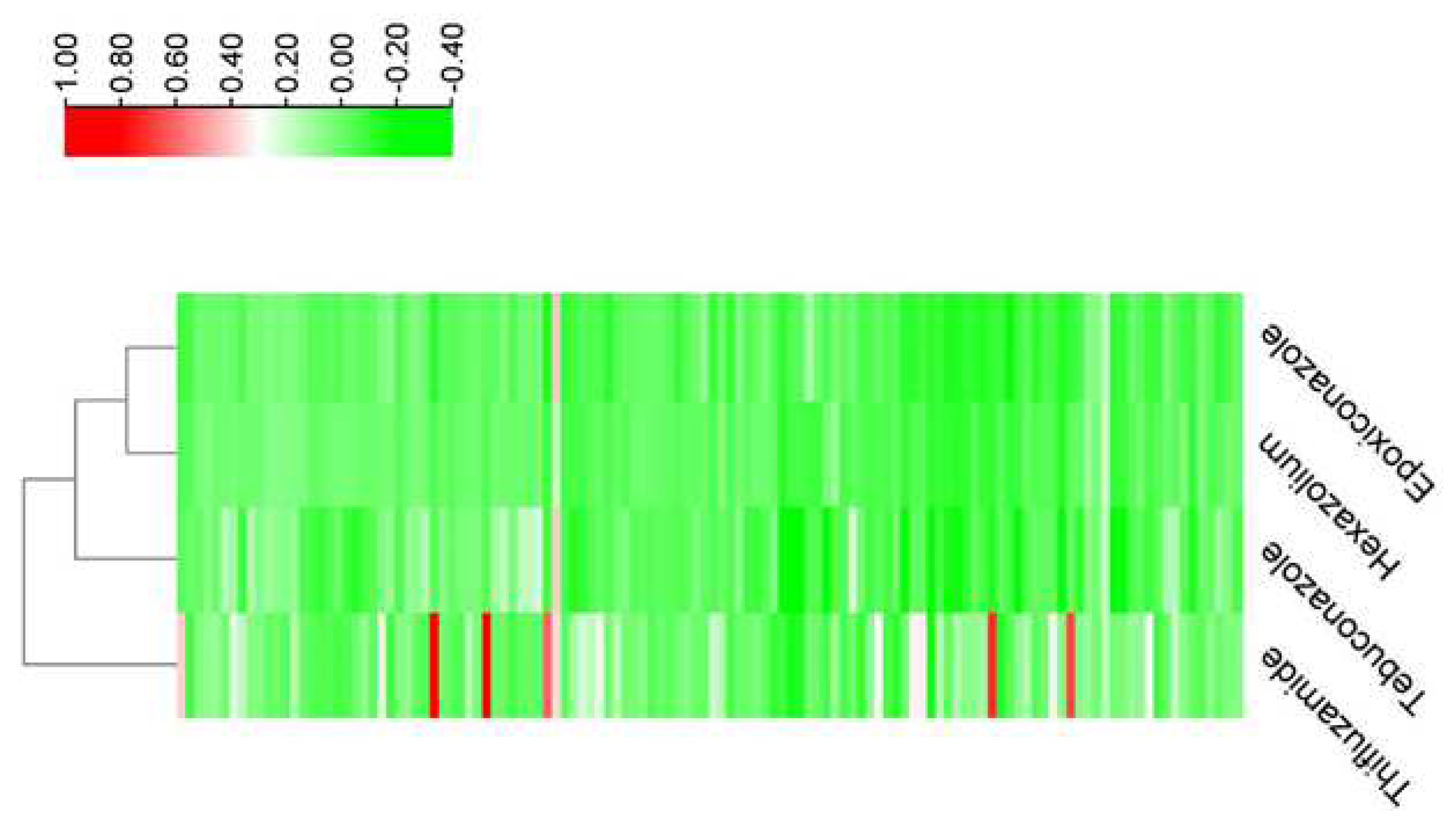
| Sampling Sites | Thifluzamide | Hexaconazole | Tebuconazole | Epoxiconazole | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | S | Res-Frequency | R | S | Res-Frequency | R | S | Res-Frequency | R | S | Res-Frequency | |
| CDCZ | 11 | 8 | 57.89% | 18 | 1 | 94.74% | 11 | 8 | 57.89% | 13 | 6 | 68.42% |
| CDDY | 2 | 1 | 66.67% | 3 | 0 | 100.00% | 1 | 2 | 33.33% | 1 | 2 | 33.33% |
| CDPX | 3 | 5 | 37.50% | 2 | 6 | 25.00% | 0 | 8 | 0.00% | 3 | 5 | 37.50% |
| MSRS | 30 | 10 | 75.00% | 8 | 32 | 20.00% | 7 | 33 | 17.50% | 7 | 33 | 17.50% |
| ZGRX | 28 | 11 | 71.79% | 18 | 21 | 46.15% | 28 | 11 | 71.79% | 14 | 25 | 35.90% |
| LZHJ | 4 | 4 | 50.00% | 6 | 2 | 75.00% | 7 | 1 | 87.50% | 5 | 3 | 20.83% |
| NJLC | 2 | 3 | 40.00% | 0 | 5 | 0.00% | 4 | 1 | 80.00% | 1 | 4 | 20.00% |
| Total | 80 | 42 | 65.57% | 55 | 67 | 45.08% | 58 | 64 | 47.54% | 44 | 78 | 36.07% |
| Test Fungicides | No. of Isolates | Regression Equation | EC50 (μg/mL) | 95% Confidence Interval | Correlation Index |
|---|---|---|---|---|---|
| Thifluzamide | CDPX-1-7 | Y = 5.8483 + 0.6012 X | 0.04 | (0.02–0.09) | 0.9656 |
| ZGRX-2-13 | Y = 6.0421 + 0.7504 X | 0.04 | (0.02–0.07) | 0.9877 | |
| MSRS-2-17 | Y = 5.8319 + 0.6303 X | 0.05 | (0.03–0.09) | 0.9625 | |
| CDPX-1-3 | Y = 5.9502 + 0.6050 X | 0.03 | (0.01–0.07) | 0.9583 | |
| ZGRX-2-19 | Y = 6.2788 + 0.8061 X | 0.05 | (0.01–0.06) | 0.9665 | |
| CDPX-1-7 | Y = 6.4815 + 0.9369 X | 0.03 | (0.01–0.05) | 0.9715 | |
| CDDY1-2 | Y = 6.2350 + 0.9628 X | 0.05 | (0.03–0.08) | 0.9844 | |
| MSRS-2-3 | Y = 5.8892 + 0.6085 X | 0.04 | (0.02–0.09) | 0.9825 | |
| ZGRX-3-1 | Y = 5.8052 + 0.6085 X | 0.05 | (0.02–0.09) | 0.9706 | |
| ZGRX-1-3 | Y = 6.5188 + 1.0734 X | 0.04 | (0.03–0.06) | 0.9370 | |
| Hexaconazole | ZGRX-3-1 | Y = 5.9980 + 0.8225 X | 0.06 | (0.04–0.09) | 0.9998 |
| ZGRX-2-3 | Y = 6.7999 + 1.4116 X | 0.05 | (0.04–0.07) | 0.9882 | |
| MSRS-3-13 | Y = 6.6624 + 1.0217 X | 0.02 | (0.01–0.04) | 0.9876 | |
| MSRS-2-12 | Y = 6.7086 + 1.1962 X | 0.04 | (0.03–0.06) | 0.9988 | |
| MSRS-3-6 | Y = 6.6118 + 1.1475 X | 0.04 | (0.03–0.06) | 0.9977 | |
| MSRS-3-16 | Y = 6.7126 + 1.3524 X | 0.05 | (0.04–0.08) | 0.9517 | |
| MSRS-2-6 | Y = 6.9664 + 1.7276 X | 0.07 | (0.05–0.10) | 0.9672 | |
| MSRS-3-17 | Y = 6.7536 + 1.4386 X | 0.06 | (0.04–0.09) | 0.9947 | |
| ZGRX-3-2 | Y = 5.9080 + 0.7256 X | 0.06 | (0.03–0.12) | 0.9886 | |
| ZGRX-2-19 | Y = 6.1238 + 0.9151 X | 0.06 | (0.03–0.11) | 0.9926 | |
| Tebuconazole | CDCZ-1-15 | Y = 5.5328 + 0.6969 X | 0.17 | (0.12–0.26) | 0.9912 |
| ZGRX-2-3 | Y = 5.6929 + 0.7746 X | 0.13 | (0.09–0.19) | 0.9878 | |
| CDCZ-1-10 | Y = 5.5518 + 0.6825 X | 0.16 | (0.10–0.24) | 0.9852 | |
| MSRS-3-16 | Y = 7.2183 + 1.3364 X | 0.02 | (0.01–0.06) | 0.9739 | |
| MSRS-3-10 | Y = 7.2672 + 1.8764 X | 0.06 | (0.05–0.08) | 0.9963 | |
| CDPX-1-7 | Y = 6.5625 + 1.3526 X | 0.07 | (0.05–0.10) | 0.9850 | |
| MSRS-3-13 | Y = 7.3102 + 1.8224 X | 0.05 | (0.04–0.08) | 0.9684 | |
| CDPX-1-6 | Y = 6.4646 + 1.4190 X | 0.09 | (0.07–0.12) | 0.9947 | |
| CDCZ-1-6 | Y = 5.9860 + 0.7552 X | 0.05 | (0.03–0.09) | 0.9683 | |
| MSRS-2-13 | Y = 5.5530 + 0.8550 X | 0.23 | (0.16–0.31) | 0.9893 | |
| Epoxiconazole | MSRS-2-13 | Y = 7.0486 + 1.3978 X | 0.03 | (0.02–0.05) | 0.9839 |
| MSRS-2-12 | Y = 6.0911 + 0.8262 X | 0.05 | (0.03–0.08) | 0.9725 | |
| MSRS-2-6 | Y = 5.9836 + 0.8701 X | 0.07 | (0.05–0.11) | 0.9132 | |
| MSRS-2-17 | Y = 5.9911 + 1.1652 X | 0.14 | (0.10–0.19) | 0.9826 | |
| MSRS-3-10 | Y = 6.0264 + 0.9587 X | 0.08 | (0.05–0.13) | 0.9878 | |
| MSRS-3-13 | Y = 6.1238 + 0.8946 X | 0.06 | (0.04–0.08) | 0.9768 | |
| ZGRX-1-3 | Y = 6.0133 + 1.1836 X | 0.14 | (0.11–0.18) | 0.9971 | |
| ZGRX-1-1 | Y = 5.9971 + 1.2023 X | 0.15 | (0.09–0.24) | 0.9951 | |
| CDCZ-1-9 | Y = 5.8796 + 1.0372 X | 0.14 | (0.09–0.23) | 0.9829 | |
| CDDY1-2 | Y = 5.7995 + 0.9206 X | 0.14 | (0.08–0.24) | 0.9826 |
| Test Fungicides | No. of Isolates | Regression Equation | EC50 (μg/mL) | 95% Confidence Interval | Correlation Index | Resistance Fold |
|---|---|---|---|---|---|---|
| Thifluzamide | MSRS-2-7 | Y = 5.1309 + 0.8174 X | 0.69 | (0.30–1.59) | 0.9825 | 16.43 |
| CDCZ-1-7 | Y = 5.5432 + 1.0143 X | 0.29 | (0.16–0.53) | 0.9783 | 6.90 | |
| CDPX-1-3 | Y = 5.6393 + 0.7784 X | 0.15 | (0.09–0.25) | 0.9493 | 3.57 | |
| ZGRX-2-1 | Y = 6.1624 + 1.1256 X | 0.09 | (0.07–0.12) | 0.9557 | 2.14 | |
| MSRS-3-6 | Y = 5.6066 + 0.5882 X | 0.09 | (0.05–0.18) | 0.8505 | 2.14 | |
| MSRS-3-13 | Y = 6.7033 + 1.5839 X | 0.08 | (0.05–0.13) | 0.9441 | 1.90 | |
| ZGRX-2-20 | Y = 6.3143 + 1.1751 X | 0.08 | (0.05–0.12) | 0.9937 | 1.90 | |
| ZGRX-2-4 | Y = 5.7924 + 0.6502 X | 0.06 | (0.03–0.11) | 0.9330 | 1.43 | |
| ZGRX-3-4 | Y = 5.1309 + 0.8714 X | 0.07 | (0.30–1.59) | 0.9825 | 1.67 | |
| LZHJ-1-3 | Y = 7.9279 + 2.4365 X | 0.06 | (0.05–0.08) | 0.8846 | 1.43 | |
| Hexaconazole | CDPX-1-5 | Y = 5.9542 + 1.2631 X | 0.18 | (0.11–0.28) | 0.9333 | 4.50 |
| MSRS-2-8 | Y = 6.0407 + 1.1371 X | 0.12 | (0.08–0.18) | 0.9880 | 3.00 | |
| CDCZ-1-16 | Y = 6.2837 + 1.7042 X | 0.18 | (0.14–0.22) | 0.9693 | 4.50 | |
| CDPX-1-3 | Y = 6.0707 + 1.2022 X | 0.13 | (0.09–0.18) | 0.9847 | 3.25 | |
| MSRS-2-20 | Y = 5.7696 + 0.9057 X | 0.14 | (0.09–0.22) | 0.9877 | 3.50 | |
| CDCZ-1-19 | Y = 6.2636 + 1.6420 X | 0.17 | (0.13–0.22) | 0.9783 | 4.25 | |
| ZGRX-1-1 | Y = 6.2531 + 1.5260 X | 0.15 | (0.11–0.21) | 0.9709 | 3.75 | |
| CDCZ-1-18 | Y = 6.5664 + 1.8577 X | 0.14 | (0.11–0.19) | 0.9893 | 3.50 | |
| CDPX-1-4 | Y = 5.8777 + 0.8436 X | 0.09 | (0.05–0.16) | 0.9277 | 2.25 | |
| MSRS-2-11 | Y = 5.8841 + 0.8176 X | 0.08 | (0.04–0.16) | 0.9984 | 2.00 | |
| Tebuconazole | LZHJ-1-4 | Y = 5.1587 + 1.1307 X | 0.72 | (0.54–0.97) | 0.9715 | 14.12 |
| ZGRX-3-4 | Y = 4.8774 + 0.9424 X | 1.35 | (0.96–1.90) | 0.9563 | 26.47 | |
| MSRS-1-3 | Y = 4.5709 + 1.0107 X | 2.66 | (1.64–4.30) | 0.9679 | 52.16 | |
| LZHJ-1-2 | Y = 5.2758 + 1.1849 X | 0.59 | (0.43–0.80) | 0.9887 | 11.57 | |
| MSRS-2-11 | Y = 4.9065 + 1.1902 X | 1.20 | (0.89–1.61) | 0.9717 | 23.53 | |
| CDCZ-1-19 | Y = 5.2303 + 0.9686 X | 0.58 | (0.38–0.87) | 0.9811 | 11.37 | |
| CDCZ-1-9 | Y = 4.9063 + 0.9036 X | 1.27 | (0.88–1.83) | 0.9795 | 24.90 | |
| CDCZ-1-7 | Y = 5.1560 + 0.5903 X | 0.54 | (0.28–1.05) | 0.9798 | 10.59 | |
| MSRS-2-17 | Y = 4.7848 + 1.3253 X | 1.45 | (1.09–1.94) | 0.9878 | 28.43 | |
| LZHJ-1-8 | Y = 4.2535 + 1.5181 X | 3.10 | (1.99–4.84) | 0.9996 | 60.78 | |
| Epoxiconazole | MSRS-2-11 | Y = 6.1163 + 1.7336 X | 0.23 | (0.15–0.34) | 0.9971 | 2.30 |
| MSRS-2-17 | Y = 5.9897 + 1.6367 X | 0.25 | (0.19–0.33) | 0.9982 | 2.50 | |
| ZGRX-2-20 | Y = 5.6710 + 1.0176 X | 0.22 | (0.14–0.33) | 0.9866 | 2.20 | |
| MSRS-2-8 | Y = 5.6199 + 1.4031 X | 0.36 | (0.28–0.46) | 0.9839 | 3.60 | |
| CDPX-1-3 | Y = 5.8153 + 1.4081 X | 0.26 | (0.20–0.34) | 0.9954 | 2.60 | |
| LZHJ-1-3 | Y = 5.9860 + 1.5160 X | 0.22 | (0.17–0.30) | 0.9821 | 2.20 | |
| ZGRX-3-4 | Y = 5.3441 + 0.8510 X | 0.39 | (0.22–0.72) | 0.9431 | 3.90 | |
| MSRS-2-7 | Y = 5.5624 + 0.9933 X | 0.27 | (0.17–0.43) | 0.9923 | 2.70 | |
| LZHJ-1-8 | Y = 4.4986 + 0.9143 X | 3.54 | (1.33–9.38) | 0.9804 | 35.40 | |
| MSRS-3-16 | Y = 5.6143 + 0.8581 X | 0.19 | (0.12–0.30) | 0.9619 | 1.90 |
| Isolate Type | Code | Resistant Type | Mutational Type |
|---|---|---|---|
| Susceptible isolate | MSRS-3-16 | - | - |
| ZGRX-2-3 | - | - | |
| CDCZ-1-15 | - | - | |
| CDCZ-1-7 | - | - | |
| Resistant isolate | ZGRX-3-4 | MR | S94A, N406S, H793R |
| MSRS-2-17 | MR | S94A, N406S, H793R | |
| MSRS-1-3 | HR | S94A, N406S, H793R | |
| LZHJ-1-8 | HR | S94A, N406S, L750P, H793R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, C.; Liu, M.; Liu, D.; Wang, Q.; Hasnain, A.; Zhan, X.; Pu, J.; Liang, Y.; Liu, X.; Wang, X. Status of Fungicide Resistance and Physiological Characterization of Tebuconazole Resistance in Rhizocotonia solani in Sichuan Province, China. Curr. Issues Mol. Biol. 2022, 44, 4859-4876. https://doi.org/10.3390/cimb44100330
Gong C, Liu M, Liu D, Wang Q, Hasnain A, Zhan X, Pu J, Liang Y, Liu X, Wang X. Status of Fungicide Resistance and Physiological Characterization of Tebuconazole Resistance in Rhizocotonia solani in Sichuan Province, China. Current Issues in Molecular Biology. 2022; 44(10):4859-4876. https://doi.org/10.3390/cimb44100330
Chicago/Turabian StyleGong, Changwei, Min Liu, Dan Liu, Qiulin Wang, Ali Hasnain, Xiaoxu Zhan, Jian Pu, Yueyang Liang, Xuemei Liu, and Xuegui Wang. 2022. "Status of Fungicide Resistance and Physiological Characterization of Tebuconazole Resistance in Rhizocotonia solani in Sichuan Province, China" Current Issues in Molecular Biology 44, no. 10: 4859-4876. https://doi.org/10.3390/cimb44100330
APA StyleGong, C., Liu, M., Liu, D., Wang, Q., Hasnain, A., Zhan, X., Pu, J., Liang, Y., Liu, X., & Wang, X. (2022). Status of Fungicide Resistance and Physiological Characterization of Tebuconazole Resistance in Rhizocotonia solani in Sichuan Province, China. Current Issues in Molecular Biology, 44(10), 4859-4876. https://doi.org/10.3390/cimb44100330







