De Novo Assembly and Species-Specific Marker Development as a Useful Tool for the Identification of Scutellaria L. Species
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling and Genomic DNA Extraction
2.2. Polymerase Chain Reaction (PCR) Amplification
2.3. Sequel Library Construction
2.4. Preparation of Species-Specific Primers Using Barcode DNA
3. Results
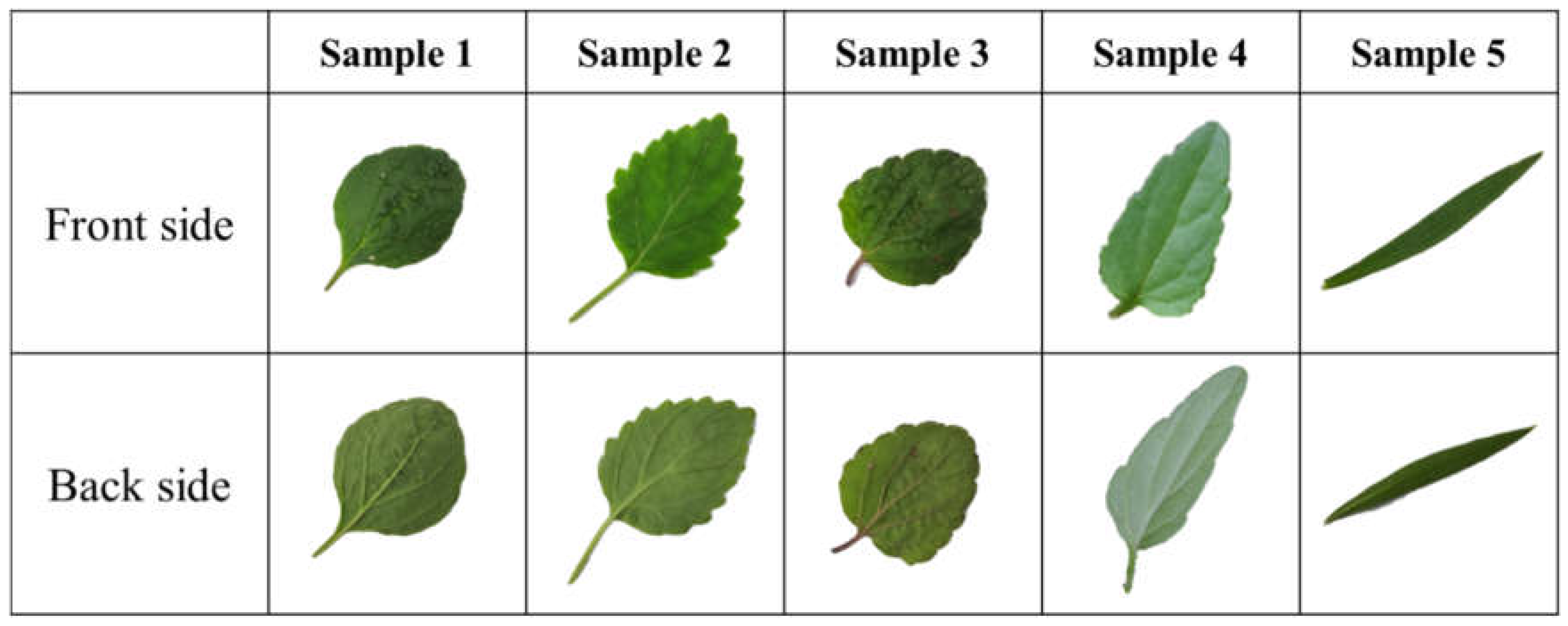
3.1. Leaf Profiles of Five Species of Scutellaria L.
3.2. RNA-Seq-Based De Novo Assembly of Sample 3
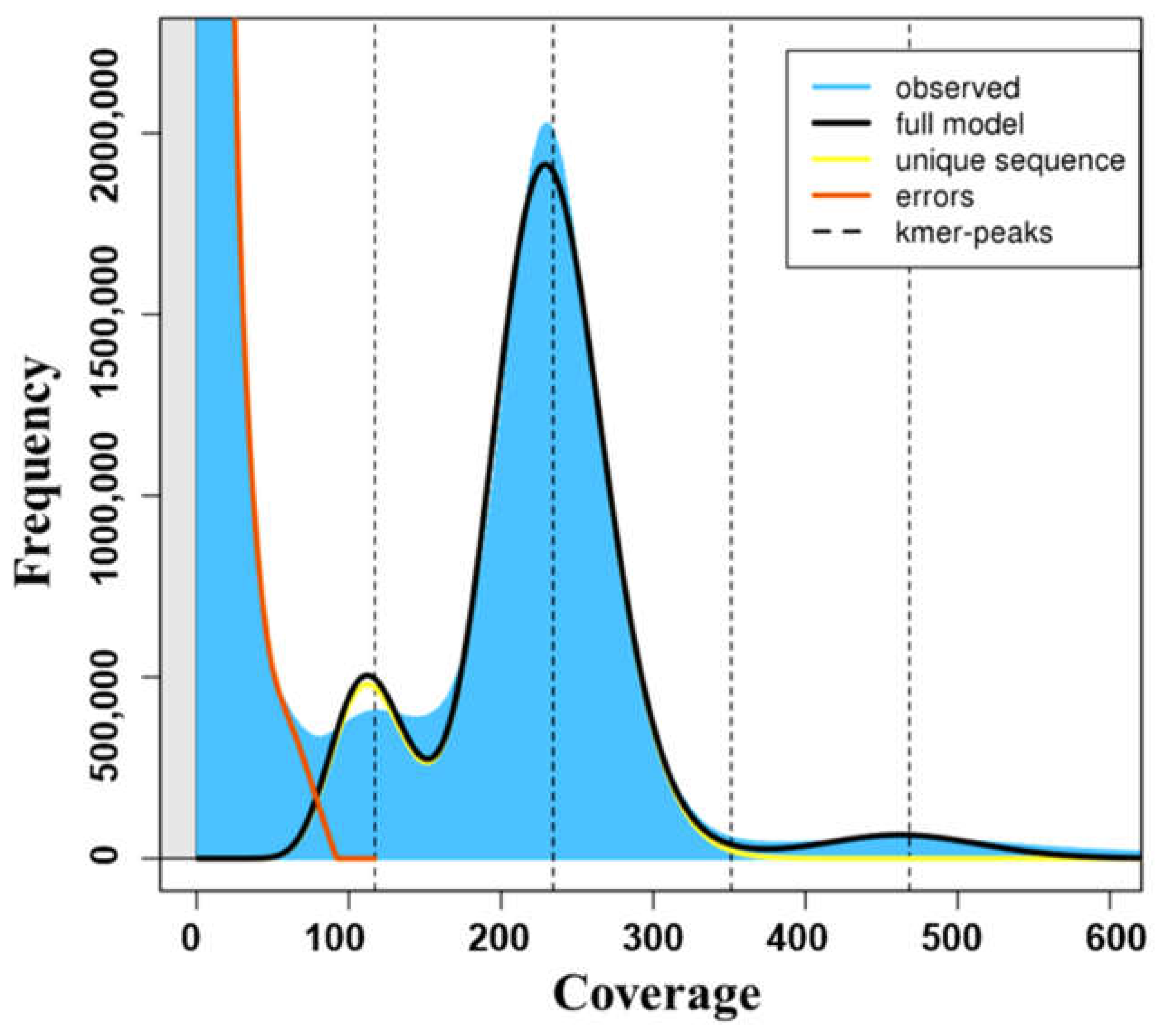
3.3. K-Mer Analysis
3.4. Assembly Results
3.5. Self-Mapping Results
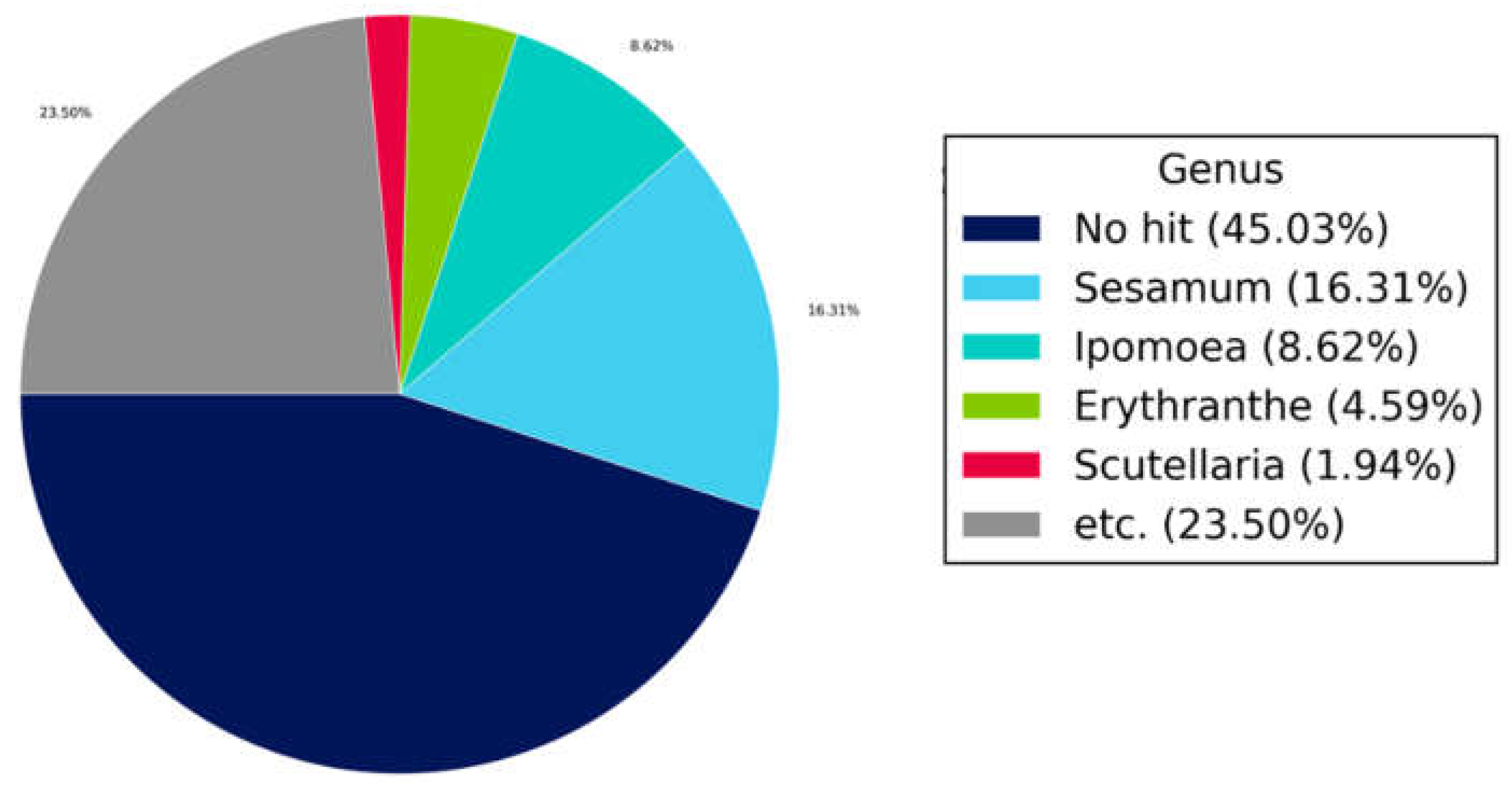
3.6. Blast Results
3.7. BUSCO Results
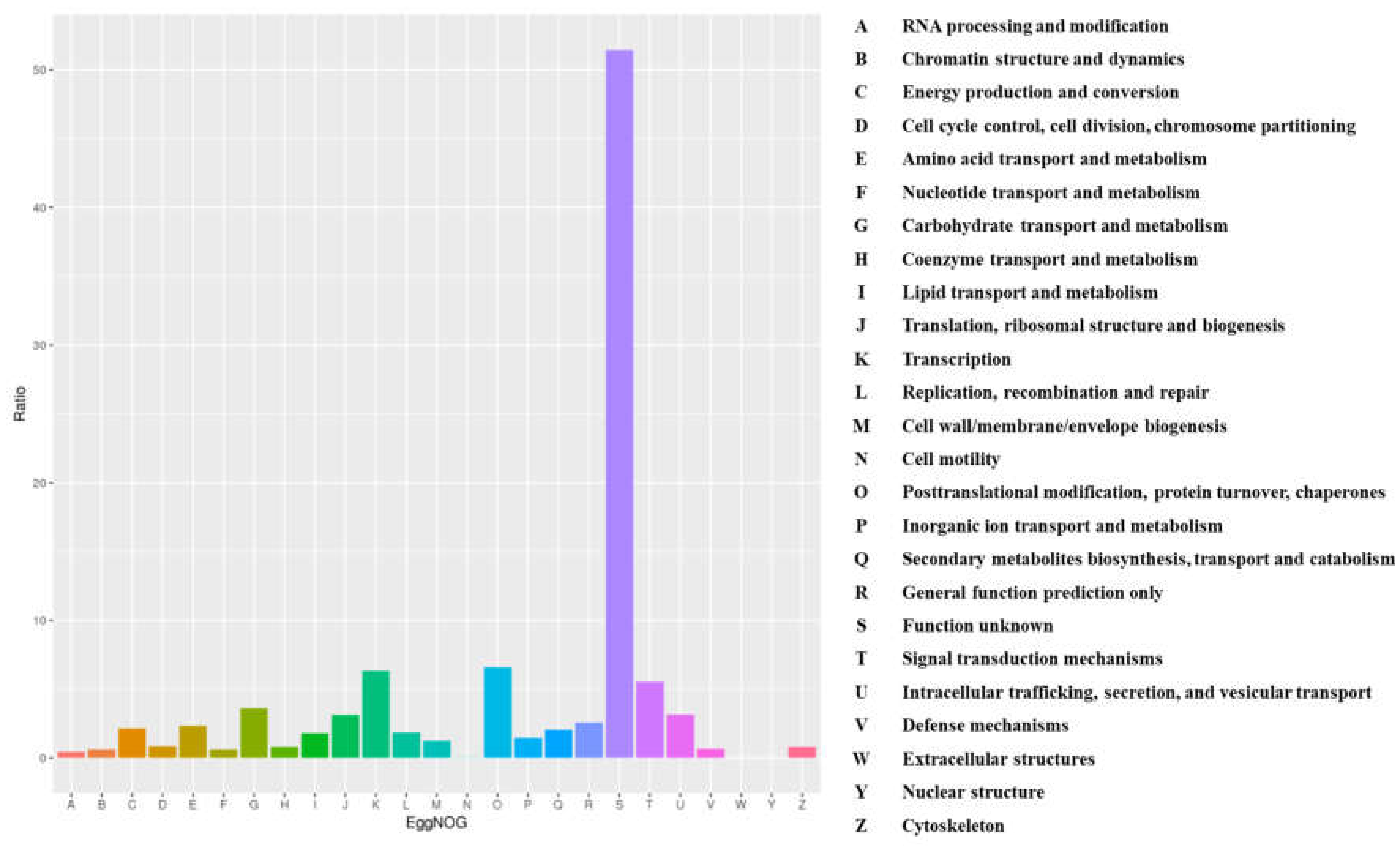
3.8. Gene Prediction and Annotation Results
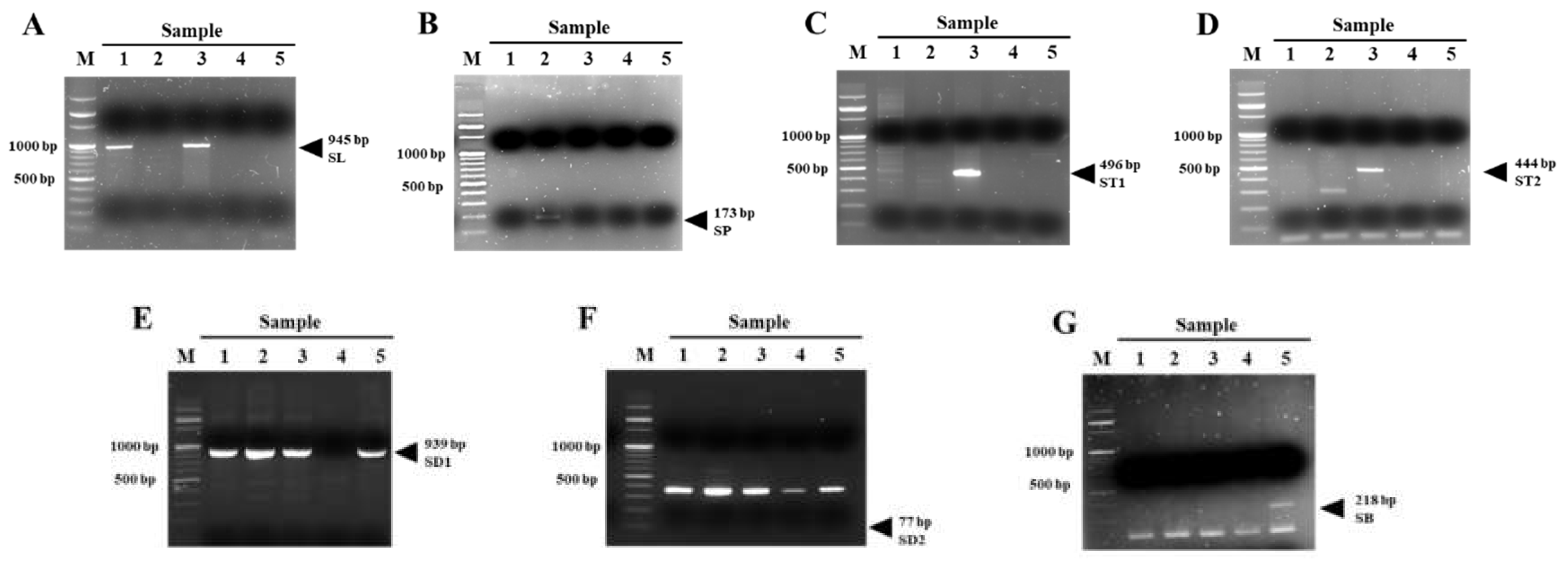
3.9. Validation of Species-Specific Primers Using Barcode DNA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, Y.; Kim, S. The complete chloroplast genome of Scutellaria indica var. coccinea (Lamiaceae), an endemic taxon in Korea. Mitochondrial DNA. Part B Resour. 2019, 4, 2539–2540. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Lee, S.T. Taxonomy of the genus Scutellaria (Lamiaceae) in Korea. Korean J. Plant Taxon. 1995, 25, 71–102. [Google Scholar] [CrossRef]
- Kim, C.-S.; Kim, S.-Y.; Byun, G.-O. A new record for Korean flora: Scutellaria tuberifera CY Wu & C. Chen (Lamiaceae). Korean J. Plant Taxon. 2011, 41, 249–252. [Google Scholar]
- Shang, X.; He, X.; Li, M.; Zhang, R.; Fan, P.; Zhang, Q.; Jia, Z. The genus Scutellaria an ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2010, 128, 279–313. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, J.-H.; Lee, B.Y.; Kim, J.-S.; Kim, S. A new distribution record of Scutellaria barbata D. Don (Lamiaceae) and an erroneously identified Scutellaria in Korea. Korean J. Plant Taxon. 2018, 48, 123–128. [Google Scholar] [CrossRef][Green Version]
- Kim, S.-T.; Lee, S.-T. Fruit surface morphology of Scutellaria (Lamiaceae) in Korea and its taxonomic implication. Korean J. Plant Taxon. 1995, 25, 165. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.K.; Singh, S.; Singh, B.D. Next-Generation Sequencing (NGS) Tools and Impact in Plant Breeding. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Springer: Berlin/Heidelberg, Germany, 2015; pp. 563–612. [Google Scholar]
- Punia, A.; Yadav, R.; Arora, P.; Chaudhury, A. Molecular and morphophysiological characterization of superior cluster bean (Cymopsis tetragonoloba) varieties. J. Crop Sci. Biotechnol. 2009, 12, 143. [Google Scholar] [CrossRef]
- Pathak, R.; Singh, S.; Singh, M.; Henry, A. Molecular assessment of genetic diversity in cluster bean (Cyamopsis tetragonoloba) genotypes. J. Genet. 2010, 89, 243–246. [Google Scholar] [CrossRef]
- Kuravadi, N.A.; Tiwari, P.B.; Tanwar, U.K.; Tripathi, S.K.; Dhugga, K.S.; Gill, K.S.; Randhawa, G.S. Identification and characterization of EST-SSR markers in cluster bean (Cyamopsis spp.). Crop Sci. 2014, 54, 1097–1102. [Google Scholar] [CrossRef]
- Kuravadi, N.A.; Yenagi, V.; Rangiah, K.; Mahesh, H.; Rajamani, A.; Shirke, M.D.; Russiachand, H.; Loganathan, R.M.; Lingu, C.S.; Siddappa, S. Comprehensive analyses of genomes, transcriptomes and metabolites of neem tree. PeerJ 2015, 3, e1066. [Google Scholar] [CrossRef]
- Pathak, R. Clusterbean: Physiology, Genetics and Cultivation; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Kumar, S.; Parekh, M.J.; Patel, C.B.; Zala, H.N.; Sharma, R.; Kulkarni, K.S.; Fougat, R.S.; Bhatt, R.K.; Sakure, A.A. Development and validation of EST-derived SSR markers and diversity analysis in cluster bean (Cyamopsis tetragonoloba). J. Plant Biochem. Biotechnol. 2016, 25, 263–269. [Google Scholar] [CrossRef]
- Taheri, S.; Lee Abdullah, T.; Yusop, M.R.; Hanafi, M.M.; Sahebi, M.; Azizi, P.; Shamshiri, R.R. Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules 2018, 23, 399. [Google Scholar] [CrossRef]
- Lee, J.; Kang, S.-J.; Shim, H.; Lee, S.-C.; Kim, N.-H.; Jang, W.; Park, J.Y.; Kang, J.H.; Lee, W.H.; Lee, T.J.; et al. Characterization of chloroplast genomes, nuclear ribosomal DNAs, and polymorphic SSR markers using whole genome sequences of two euonymus hamiltonianus phenotypes. Plant Breed. Biotechnol. 2019, 7, 50–61. [Google Scholar] [CrossRef]
- Lee, J.; Joh, H.J.; Kim, N.-H.; Lee, S.-C.; Jang, W.; Choi, B.S.; Yu, Y.; Yang, T.-J. High-throughput development of polymorphic simple sequence repeat markers using two whole genome sequence data in Peucedanum japonicum. Plant Breed. Biotechnol. 2017, 5, 134–142. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- Lenz, P.H.; Roncalli, V.; Hassett, R.P.; Wu, L.-S.; Cieslak, M.C.; Hartline, D.K.; Christie, A.E. De novo assembly of a transcriptome for Calanus finmarchicus (Crustacea, Copepoda)–the dominant zooplankter of the North Atlantic Ocean. PLoS ONE 2014, 9, e88589. [Google Scholar] [CrossRef]
- Singh, V.; Goel, R.; Pande, V.; Asif, M.H.; Mohanty, C.S. De novo sequencing and comparative analysis of leaf transcriptomes of diverse condensed tannin-containing lines of underutilized Psophocarpus tetragonolobus (L.) DC. Sci. Rep. 2017, 7, 44733. [Google Scholar] [CrossRef]
- Kajitani, R.; Yoshimura, D.; Okuno, M.; Minakuchi, Y.; Kagoshima, H.; Fujiyama, A.; Kubokawa, K.; Kohara, Y.; Toyoda, A.; Itoh, T. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat. Commun. 2019, 10, 1702. [Google Scholar] [CrossRef]
- English, A.C.; Richards, S.; Han, Y.; Wang, M.; Vee, V.; Qu, J.; Qin, X.; Muzny, D.M.; Reid, J.G.; Worley, K.C.; et al. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 2012, 7, e47768. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Sanchez Alvarado, A.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, A.Y.; Jo, A.; Choi, H.; Cho, S.S.; Choi, C. Development of user-friendly method to distinguish subspecies of the Korean medicinal herb perilla frutescens using multiplex-PCR. Molecules 2017, 22, 665. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, H.; Shin, J.; Jo, A.; Lee, K.E.; Cho, S.S.; Hwang, Y.P.; Choi, C. Molecular Discrimination of Cynanchum wilfordii and Cynanchum auriculatum by InDel Markers of Chloroplast DNA. Molecules 2018, 23, 1337. [Google Scholar] [CrossRef]
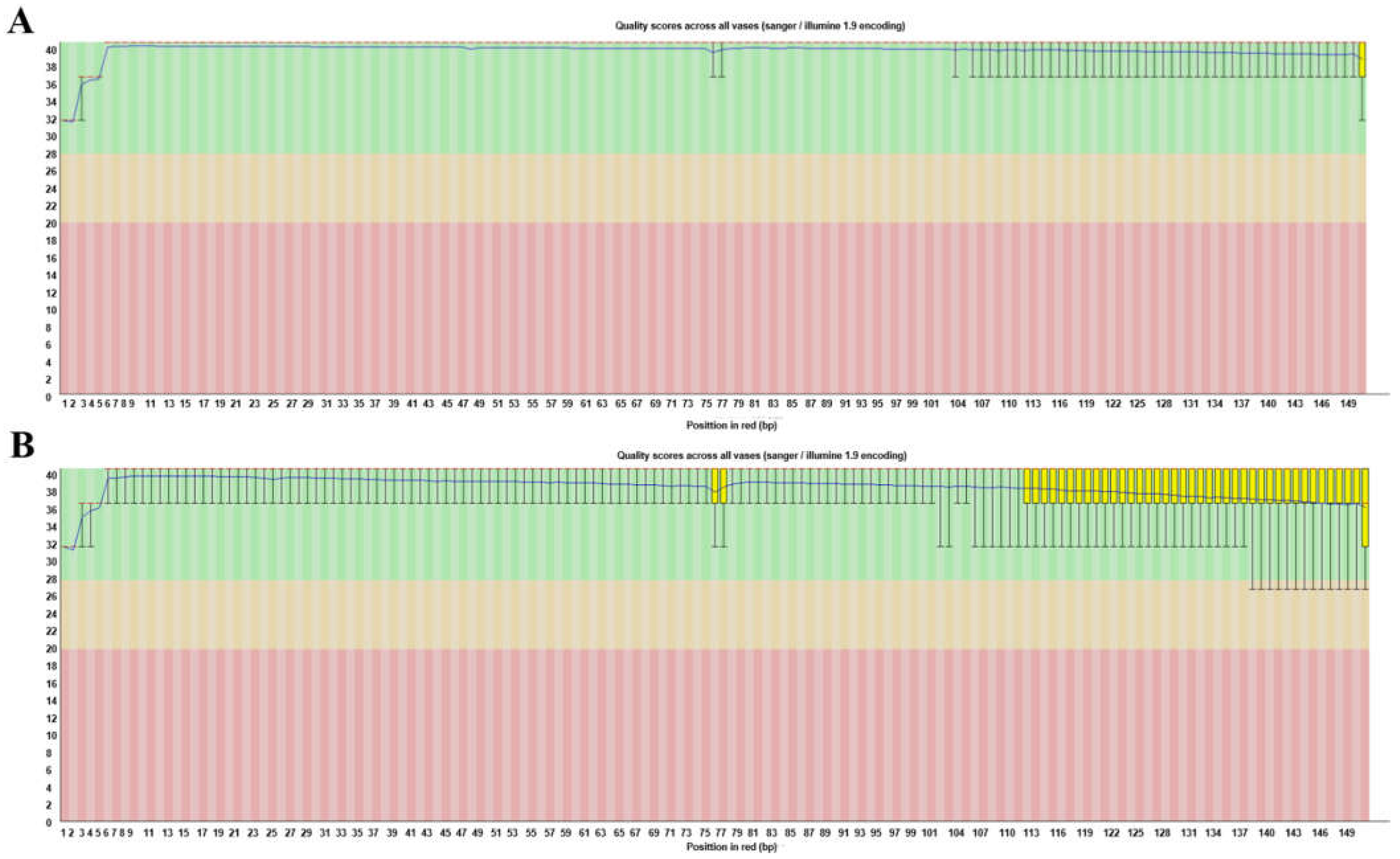
| Raw Data Stats | Filtered Data Stats | |
|---|---|---|
| Total read bases | 147,017,334,834 | 112,735,647,909 |
| Total reads | 973,24,734 | 749,527,438 |
| GC (%) | 37.4 | 36.69 |
| Q20 (%) | 95.51 | 98.91 |
| Q30 (%) | 90.45 | 96.49 |
| K-Mer Coverage | Heterozygosity | Genome Length | Genome Repeat Length | |
|---|---|---|---|---|
| 21mer | 234.2 | 0.404 | 352,670,975 | 166,169,989 |
| Contigs | Scaffolds | |
|---|---|---|
| No. | 19,561 | 14,625 |
| Sum | 298,515,080 | 318,741,328 |
| N50 | 42,020 | 78,430 |
| Longest | 411,840 | 803,009 |
| Shortest | 1000 | 1000 |
| Average length | 15,260 | 21,794 |
| Library Name | Total Reads | Mapped Reads | Coverage (%) | Depth | Ins. Size (Std.) |
|---|---|---|---|---|---|
| DNA | 749,527,438 | 655,896,731 (87.51%) | 94.65 | 276.51 | 475.88 (113.04) |
| Status | Of BUSCOs | Percentage |
|---|---|---|
| Complete BUSCOs (C) | ||
| Complete and single-copy BUSCOs (S) | 229 | 75.58% |
| Complete and duplicated BUSCOs (D) | 50 | 16.50% |
| Fragmented BUSCOs (F) | 4 | 1.32% |
| Missing BUSCOs (M) | 20 | 6.60% |
| Total BUSCO groups searched | 303 | 100.00% |
| Sample | Contigs | Bases | Genes | CDSs | tRNAs | rRNAs |
|---|---|---|---|---|---|---|
| DNA | 14,625 | 318,741,328 | 23,814 | 22,788 | 670 | 356 |
| Primer Name | Sequence (5′->3′) | Product Size | Tm (℃) | NCBI Accession of Target Genome | Locus | Target Species | |
|---|---|---|---|---|---|---|---|
| SL Primer | Forward | TGCTTACCTGCTTCCACAGG | 945 bp | 54 | KM526800.1 | CYC2B | Scutellaria indica L. |
| Reverse | TCGGTGGCGACGTTATATGG | ||||||
| SP Primer | Forward | GAAATTACTTTTAAATTCAT | 173 bp | 42 | KX060016.1 | trnH-PsbA | Scutellaria pekinensis var. transitra (Makino) H. Hara |
| Reverse | GTAGTCTTTCCTAGACTTTA | ||||||
| ST1 Primer | Forward | TTGTGGCATCACTAACCCCC | 491 bp | 54 | MZ714561 | AtpI_0 | Scutellaria indica var. tsusimensis (H. Hara) Ohwi |
| Reverse | AGGGGTAGGCTGAACGTACT | ||||||
| ST2 Primer | Forward | CGCATTCCTCCAGCCTATGT | 444 bp | 54 | MZ714562 | rbcL_3 | Scutellaria indica var. tsusimensis (H. Hara) Ohwi |
| Reverse | ATCACGGCAGTAGTGTGCAA | ||||||
| SD1 Primer | Forward | ATAACTTCCCTCTAGACTTA | 939 bp | 43 | KT750009.1 | - | Scutellaria barbata D. Don |
| Reverse | TGAATTTCAATTATTTTTTC | ||||||
| SD2 Primer | Forward | ACCCTTGATTCGCACACTGA | 77 bp | 55 | KX059898.1 | PsbK-PsbI | Scutellaria barbata D. Don |
| Reverse | TGAGGAAACGGACGTAAGCC | ||||||
| SB Primer | Forward | TCCCCAAAAAGTGGATCCCG | 218 bp | 55 | MF521633.1 | - | Scutellaria baicalensis Georgi |
| Reverse | GGGCCTCATTGGTAAGTGCT | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.; Kang, W.S.; Kim, J.S.; Na, C.-S.; Kim, S. De Novo Assembly and Species-Specific Marker Development as a Useful Tool for the Identification of Scutellaria L. Species. Curr. Issues Mol. Biol. 2021, 43, 2177-2188. https://doi.org/10.3390/cimb43030152
Choi H, Kang WS, Kim JS, Na C-S, Kim S. De Novo Assembly and Species-Specific Marker Development as a Useful Tool for the Identification of Scutellaria L. Species. Current Issues in Molecular Biology. 2021; 43(3):2177-2188. https://doi.org/10.3390/cimb43030152
Chicago/Turabian StyleChoi, Hakjoon, Wan Seok Kang, Jin Seok Kim, Chang-Su Na, and Sunoh Kim. 2021. "De Novo Assembly and Species-Specific Marker Development as a Useful Tool for the Identification of Scutellaria L. Species" Current Issues in Molecular Biology 43, no. 3: 2177-2188. https://doi.org/10.3390/cimb43030152
APA StyleChoi, H., Kang, W. S., Kim, J. S., Na, C.-S., & Kim, S. (2021). De Novo Assembly and Species-Specific Marker Development as a Useful Tool for the Identification of Scutellaria L. Species. Current Issues in Molecular Biology, 43(3), 2177-2188. https://doi.org/10.3390/cimb43030152







