Down-Regulation of the Longevity-Associated Protein SIRT1 in Peripheral Blood Mononuclear Cells of Treated HIV Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Subjects and Ethics Statements
2.2. Isolation of PBMCs
2.3. Quantitative RT-PCR
2.4. Protein Expression Analysis by Western Blot
2.5. Statistical Analysis and Presentation of Data
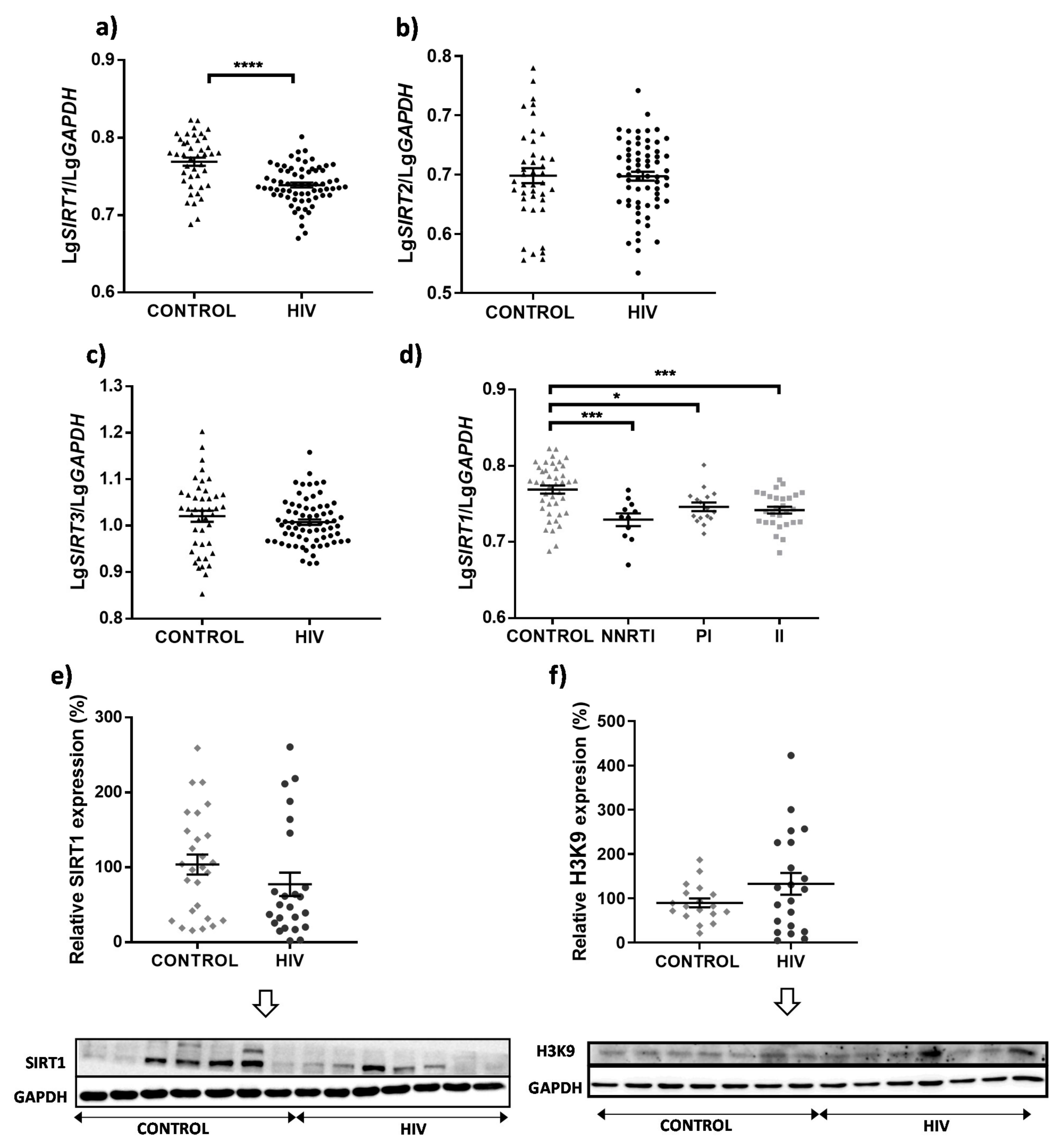
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Legarth, R.A.; Ahlström, M.G.; Kronborg, G.; Larsen, C.S.; Pedersen, C.; Pedersen, G.; Mohey, R.; Gerstoft, J.; Obel, N. Long-term mortality in HIV-Infected individuals 50 years or older: A nationwide, population-based cohort study. J. Acquir. Immune Defic. Syndr. 2016, 71, 213–218. [Google Scholar] [CrossRef]
- Hsu, D.C.; Sereti, I.; Ananworanich, J. Serious non-AIDS events: Immunopathogenesis and interventional strategies. AIDS Res. Ther. 2013, 10, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- High, K.P.; Brennan-Ing, M.; Clifford, D.B.; Cohen, M.H.; Currier, J.; Deeks, S.G.; Deren, S.; Effros, R.B.; Gebo, K.; Goronzy, J.J.; et al. HIV and aging: State of knowledge and areas of critical need for research. A report to the NIH Office of AIDS Research by the HIV and Aging Working Group. J. Acquir. Immune Defic. Syndr. 2012, 60, S1–S18. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, D.; Roy, D.; Cassol, E. Examining Relationships between Metabolism and Persistent Inflammation in HIV Patients on Antiretroviral Therapy. Mediat. Inflamm. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wing, E.J. HIV and aging. Int. J. Infect. Dis. 2016, 53, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Buler, M.; Andersson, U.; Hakkola, J. Who watches the watchmen? Regulation of the expression and activity of sirtuins. Faseb J. 2016, 30, 3942–3960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, B.J. Seven sirtuins for seven deadly diseases of aging. Free Radic. Biol. Med. 2013, 56, 133–171. [Google Scholar] [CrossRef]
- Grabowska, W.; Sikora, E.; Bielak-Zmijewska, A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology 2017, 18, 447–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeng, M.Y.; Hull, P.A.; Fei, M.; Kwon, H.S.; Tsou, C.L.; Kasler, H.; Ng, C.P.; Gordon, D.E.; Johnson, J.; Krogan, N.; et al. Metabolic reprogramming of human CD8+ memory T cells through loss of SIRT1. J. Exp. Med. 2018, 215, 51–62. [Google Scholar] [CrossRef]
- Pagans, S.; Pedal, A.; North, B.J.; Kaehlcke, K.; Marshall, B.L.; Dorr, A.; Hetzer-Egger, C.; Henklein, P.; Frye, R.; McBurney, M.W.; et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005, 3, e41. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.S.; Brent, M.M.; Getachew, R.; Jayakumar, P.; Chen, L.F.; Schnolzer, M.; McBurney, M.W.; Marmorstein, R.; Greene, W.C.; Ott, M. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell Host Microbe 2008, 3, 158–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruevska, A.; Moragrega, A.B.; Galindo, M.J.; Esplugues, J.V.; Blas-Garcia, A.; Apostolova, N. p53 and p53-related mediators PAI-1 and IGFBP-3 are downregulated in peripheral blood mononuclear cells of HIV-patients exposed to non-nucleoside reverse transcriptase inhibitors. Antivir. Res. 2020, 178, 104784. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Pinzone, M.R.; Cacopardo, B.; Condorelli, F.; di Rosa, M.; Nunnari, G. Sirtuin-1 and HIV-1: An overview. Curr. Drug Targets 2013, 14, 648–652. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, C.; Shi, C.; Fan, T.; Chu, K.; Ma, Y. Role of miR-124a in T cell activation and immunity in AIDS patients. Exp. Med. 2017, 14, 4807–4812. [Google Scholar] [CrossRef] [PubMed]
- Bogoi, R.N.; de Pablo, A.; Valencia, E.; Martín-Carbonero, L.; Moreno, V.; Vilchez-Rueda, H.H.; Asensi, V.; Rodriguez, R.; Toledano, V.; Rodés, B. Expression profiling of chromatin-modifying enzymes and global DNA methylation in CD4+ T cells from patients with chronic HIV infection at different HIV control and progression states. Clin. Epigenetics 2018, 10, 20. [Google Scholar] [CrossRef]
- Kwon, Y.; Kim, J.; Lee, C.Y.; Kim, H. Expression of SIRT1 and SIRT3 varies according to age in mice. Anat. Cell Biol. 2015, 48, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Kilic, U.; Gok, O.; Erenberk, U.; Dundaroz, M.R.; Torun, E.; Kucukardali, Y.; Elibol-Can, B.; Uysal, O.; Dundar, T. A remarkable age-related increase in SIRT1 protein expression against oxidative stress in elderly: SIRT1 gene variants and longevity in human. PLoS ONE 2015, 10, e0117954. [Google Scholar] [CrossRef]
- Hu, G.; Liao, K.; Yang, L.; Pendyala, G.; Kook, Y.; Fox, H.S.; Buch, S. Tat-Mediated Induction of miRs-34a & -138 Promotes Astrocytic Activation via Downregulation of SIRT1: Implications for Aging in HAND. J. Neuroimmune Pharm. 2017, 12, 420–432. [Google Scholar] [CrossRef]
- Feng, J.; Bao, L.; Wang, X.; Li, H.; Chen, Y.; Xiao, W.; Li, Z.; Xie, L.; Lu, W.; Jiang, H.; et al. Low expression of HIV genes in podocytes accelerates the progression of diabetic kidney disease in mice. Kidney Int. 2021, 99, 914–925. [Google Scholar] [CrossRef]
- Jurkowska, K.; Szymanska, B.; Knysz, B.; Kuzniarski, A.; Piwowar, A. Sirtuins as Interesting Players in the Course of HIV Infection and Comorbidities. Cells 2021, 10, 2739. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.; Kumar, V.; Lackner, A.A.; Alvarez, X. Dysregulated miR-34a-SIRT1-acetyl p65 axis is a potential mediator of immune activation in the colon during chronic simian immunodeficiency virus infection of rhesus macaques. J. Immunol. 2015, 194, 291–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, Y.; Li, R.; Rezk, A.; Misirliyan, H.; Moore, C.; Farooqi, N.; Solis, M.; Goiry, L.G.; de Faria Junior, O.; Dang, V.D.; et al. A novel microRNA-132-surtuin-1 axis underlies aberrant B-cell cytokine regulation in patients with relapsing-remitting multiple sclerosis. PLoS ONE 2014, 9, e105421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Yan, C.; Xin, M.; Han, L.; Zhang, Y.; Sun, M. Sirtuin 1 (Sirt1) Overexpression in BaF3 cells contributes to cell proliferation promotion, apoptosis resistance and pro-inflammatory cytokine production. Med. Sci Monit. 2017, 23, 1477–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Genes | Gene Symbol | Forward (F) and Reverse (R) Primers 5′-3′ | Size (bp) |
|---|---|---|---|
| Sirtuins | SIRT1 | F: TGGGTACCGAGATAACCTTCT R: TGTTCGAGGATCTGTGCCAA | 181 |
| SIRT2 | F: CATCCCCGACTTTCGCTCTC R: ATGGTTGGCTTGAACTGCCC | 165 | |
| SIRT3 | F: TCTGCCACCTGCACAGTCTGC R: CAGCGGCTCCCCAAAGAACAC | 138 | |
| Lymphocyte markers | CD4 | F: TTCCCAGAAGAAGAGCATACAA R: TGGCAGTCAATCCGAACAC | 254 |
| CD8A | F: CCCTTTACTGCAACCACAGG R: GGAAGGACTTGCTCCCTCAA | 167 | |
| CD19 | F: AGCGAATGACTGACCCCACC R: AGCCCTCCCCTTCCTCTTCT | 255 | |
| CD14 | F: CCCGAGTCAACAGGGCATT R: GATGTTTCAGGGAGGGGGAC | 121 | |
| Housekeeping gene | GAPDH | F: CTTCTTTTGCGTCGCCAGCC R: TTCTCAGCCTTGACGGTGCC | 232 |
| General Characteristics of the HIV Patients | ||
|---|---|---|
| gender | male | 56 (80%) |
| female | 14 (20%) | |
| age (years) median (IQR) | 52.00 (45.75–56.00) | |
| BMI (kg/m2) median (IQR) | 24.90 (23.33–27.87) | |
| smoking | yes | 25 (35.72%) |
| no | 33 (47.14%) | |
| previous smoker | 12 (17.14%) | |
| alcohol use | yes | 1 (1%) |
| no | 69 (99%) | |
| Comorbidity-Related Parameters | ||
| HCV coinfection | yes | 20 (28.57%) |
| no | 48 (68.57%) | |
| no data | 2 (2.86%) | |
| HBV coinfection | yes | 2 (2.86%) |
| no | 63 (90.00%) | |
| no data | 5 (7.14%) | |
| diabetes mellitus | yes | 15 (21.43%) |
| no | 55 (78.57%) | |
| dyslipidemia | yes | 22 (31.43%) |
| no | 48 (68.57%) | |
| cardiovascular risk | none | 16 (22.86%) |
| low | 16 (22.86%) | |
| medium | 20 (28.57%) | |
| high | 16 (22.86%) | |
| no data | 2 (2.86%) | |
| Charlson index median (IQR) | 2.00 (0.00–6.00) | |
| HIV-Related Parameters | ||
| years with HIV median (IQR) | 19 (8.50–27.00) | |
| years with antiretroviral therapy median (IQR) | 15 (4.00–21.25) | |
| mode of transmission | HO/BI | 25 (35.71%) |
| HTSX | 17 (24.29%) | |
| IDU | 21 (30.00%) | |
| other | 7 (10.00%) | |
| AIDS | yes | 18 (25.71%) |
| no | 52 (74.29%) | |
| antiretroviral therapy (in addition to NRTI backbone) | NNRTIs | 11 (15.71%) |
| PIs | 16 (22.86%) | |
| IIs | 28 (40%) | |
| combination | 15 (21.43%) | |
| antiretroviral therapy line median (IQR) | 5 (2.00–8.25) | |
| max viral load (copies/mL) median (IQR) | 79,500 (5925–361,000) | |
| CD4 nadir count (cell/mL) median (IQR) | 207.5 (71.50–339.00) | |
| CD4:CD8 median (IQR) | 0.730 (0.478–1.080) | |
| Characteristics of the Uninfected Control Population | ||
|---|---|---|
| gender | male | female |
| 33 (76.74%) | 10 (23.26%) | |
| age (years) median (IQR) | 47 (42.00–54.25) | |
| BMI (kg/m2) median (IQR) | 26.10 (24.01–28.32) | |
| smoking | yes | no |
| 10 (23.26%) | 33 (76.74%) | |
| alcohol consumption | yes | no |
| 33 (76.74%) | 10 (23.26%) | |
| Markers of PBMC Subpopulations | ||
|---|---|---|
| Control | HIV+ | |
| CD4 | rs = 0.186 | rs = 0.295 |
| p = 0.231 | p = 0.017 | |
| CD8A | rp = 0.634 | rp = 0.342 |
| p < 0.0001 | p = 0.005 | |
| CD14 | rs = 0.131 | rp = −0.018 |
| p = 0.408 | p = 0.888 | |
| CD19 | rp = 0.599 | rp = 0.340 |
| p < 0.0001 | p = 0.006 | |
| Drug Family | SIRT1 vs. CD4 | SIRT1 vs. CD8A | SIRT1 vs. CD14 | SIRT1 vs. CD19 |
|---|---|---|---|---|
| NNRTIs | rs = 0.527 | rp = 0.168 | rp = −0.342 | rp = 0.329 |
| p = 0.100 | p = 0.622 | p = 0.303 | p = 0.323 | |
| PIs | rs = 0.024 | rs = 0.475 | rp = −0.416 | rp = −0.018 |
| p = 0.940 | p = 0.076 | p = 0.139 | p = 0.951 | |
| IIs | rs = 0.157 | rp = 0.336 | rp = −0.276 | rp = 0.515 |
| p = 0.474 | p = 0.117 | p = 0.203 | p = 0.012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruevska, A.; Moragrega, Á.B.; Galindo, M.J.; Esplugues, J.V.; Blas-García, A.; Apostolova, N. Down-Regulation of the Longevity-Associated Protein SIRT1 in Peripheral Blood Mononuclear Cells of Treated HIV Patients. Cells 2022, 11, 348. https://doi.org/10.3390/cells11030348
Gruevska A, Moragrega ÁB, Galindo MJ, Esplugues JV, Blas-García A, Apostolova N. Down-Regulation of the Longevity-Associated Protein SIRT1 in Peripheral Blood Mononuclear Cells of Treated HIV Patients. Cells. 2022; 11(3):348. https://doi.org/10.3390/cells11030348
Chicago/Turabian StyleGruevska, Aleksandra, Ángela B. Moragrega, María J. Galindo, Juan V. Esplugues, Ana Blas-García, and Nadezda Apostolova. 2022. "Down-Regulation of the Longevity-Associated Protein SIRT1 in Peripheral Blood Mononuclear Cells of Treated HIV Patients" Cells 11, no. 3: 348. https://doi.org/10.3390/cells11030348