Virtual Lead Identification of Farnesyltransferase Inhibitors Based on Ligand and Structure-Based Pharmacophore Techniques
Abstract
:1. Introduction
2. Experimental Section
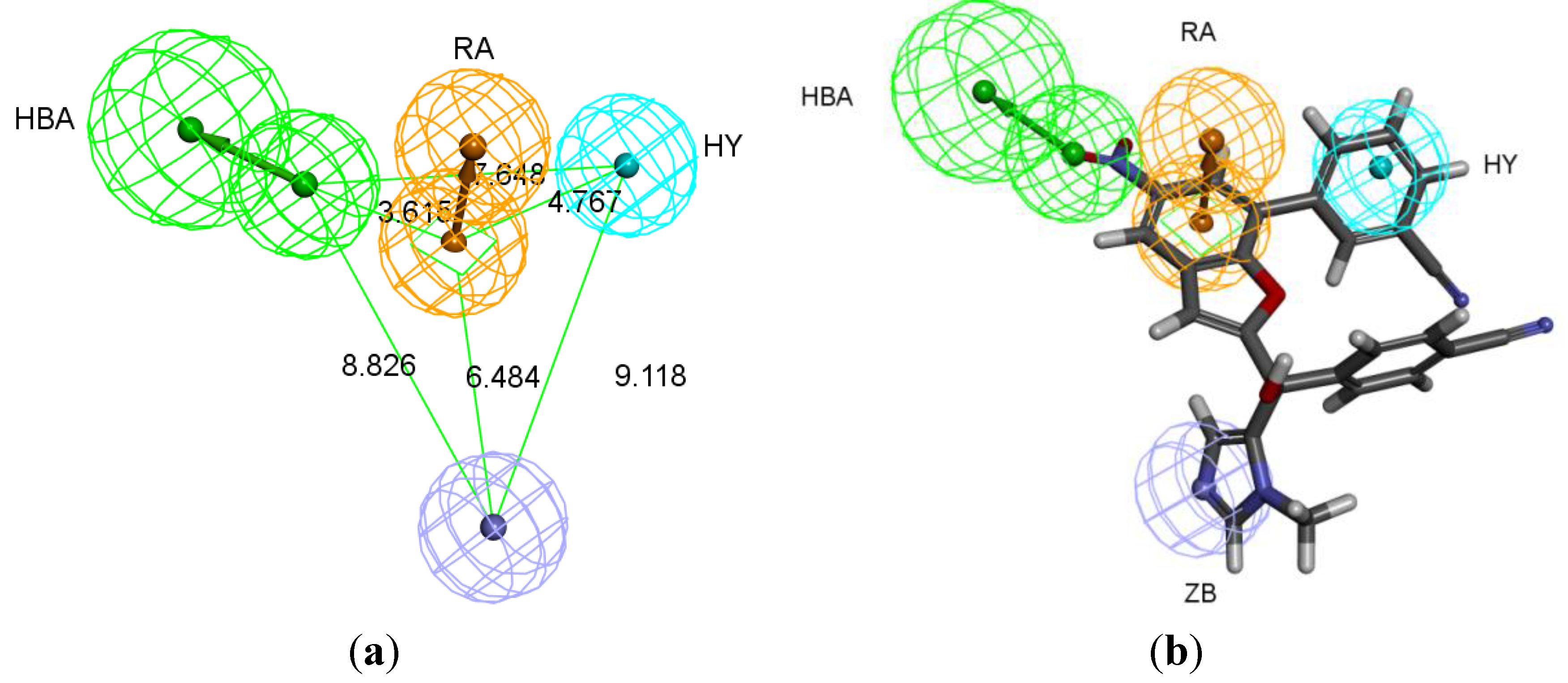
2.1. Generation of Pharmacophore Hypotheses: Common Feature Based Approach
| Pharmacophore Hypotheses a | Features b | Rank c |
|---|---|---|
| 1A | RHAA | 89.866 |
| 2A | RHAA | 89.741 |
| 3A | ZRHA | 88.981 |
| 4A | HHAA | 88.370 |
| 5A | HHAA | 88.132 |
| 6A | ZRHA | 87.756 |
| 7A | RHAA | 87.643 |
| 8A | RHAA | 86.766 |
| 9A | HHAA | 86.448 |
| 10A | RHAA | 86.374 |
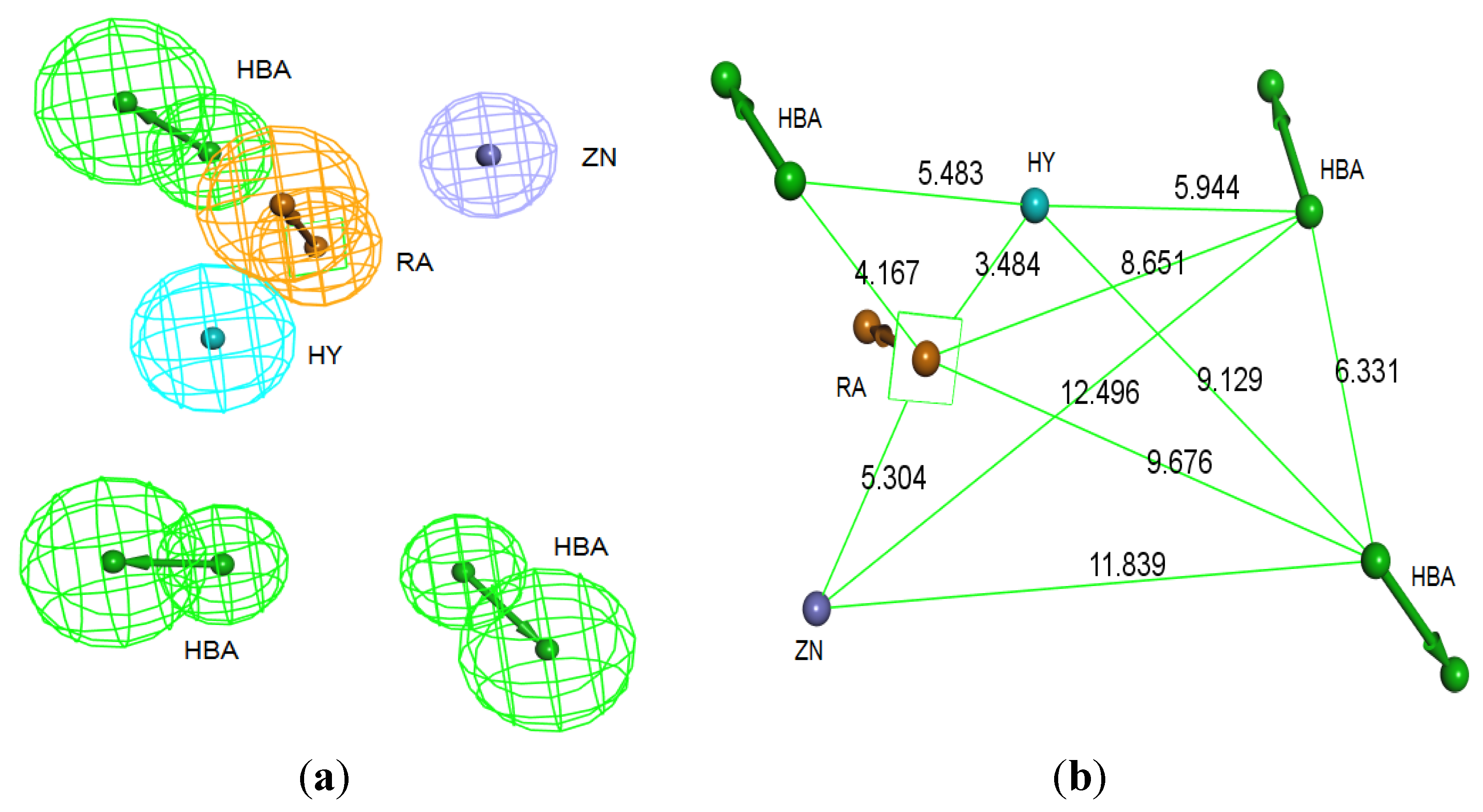
2.2. Generation of Pharmacophore Hypotheses: Structure-Based Approach
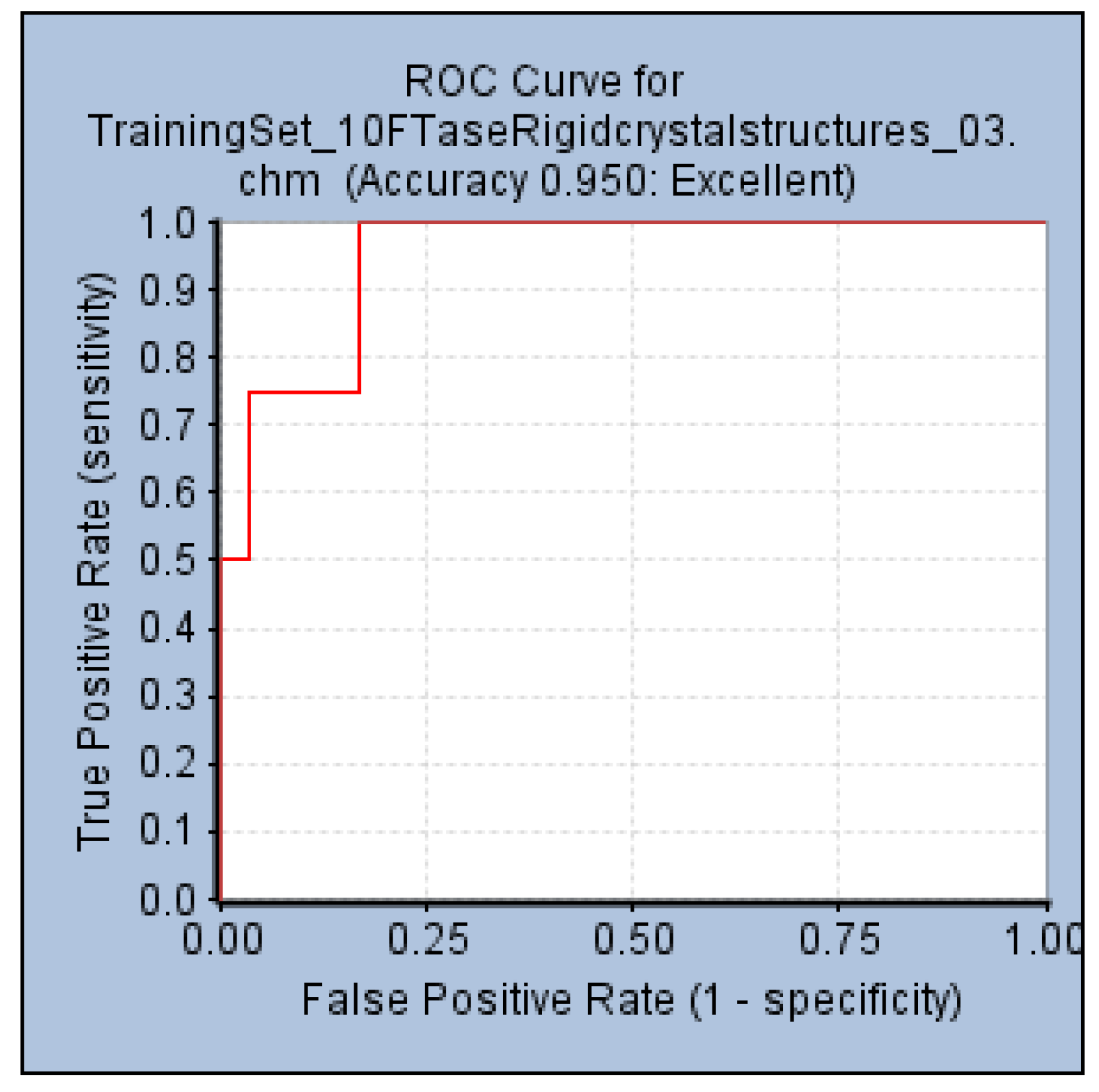
2.3. Validation of the Pharmacophore Hypotheses
2.4. Database Screening
2.5. Molecular Docking
3. Results and Discussion
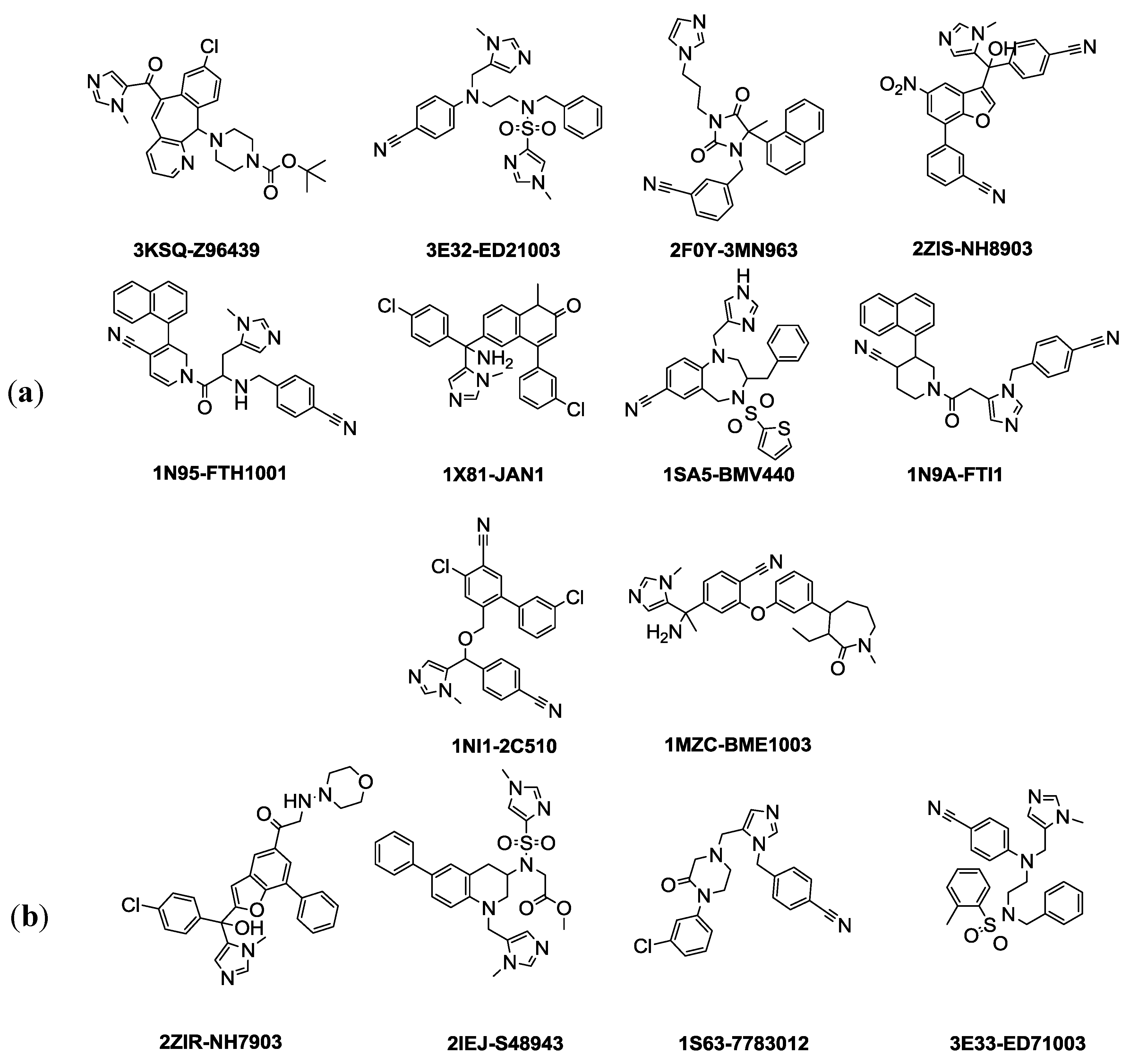
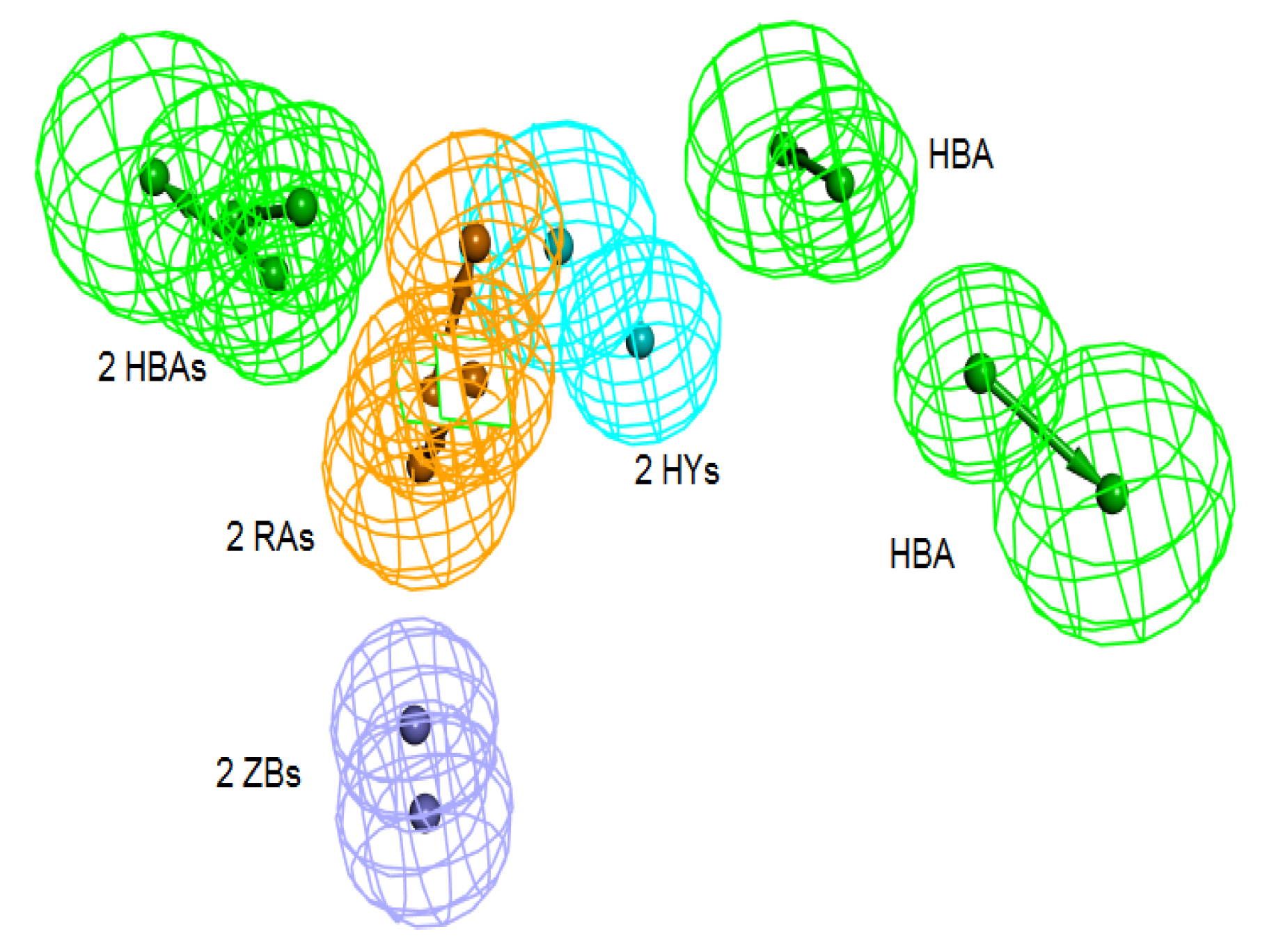
3.1. Common Feature Pharmacophore Hypothesis
| PDB ID | Inhibitor Name | Fit Value |
|---|---|---|
| Pharm-3A | ||
| 2ZIS | NH8903 | 4.000 |
| 1N95 | FTH1001 | 3.643 |
| 3E32 | ED21003 | 3.191 |
| 1NI1 | 2C510 | 3.178 |
| 3KSQ | Z96439 | 3.018 |
| 2F0Y | 3MN963 | 2.564 |
| 1SA5 | BMV440 | 2.112 |
| 1X81 | JAN1 | 1.826 |
| 1MZC | BME1003 | 1.164 |
| 1N9A | FTI1 | 0.788 |
| 2ZIR | NH7903 * | 3.585 |
| 3E33 | ED71003 * | 3.096 |
| 2IEJ | S48943 * | 2.748 |
| 1S63 | 7783012 * | 1.784 |
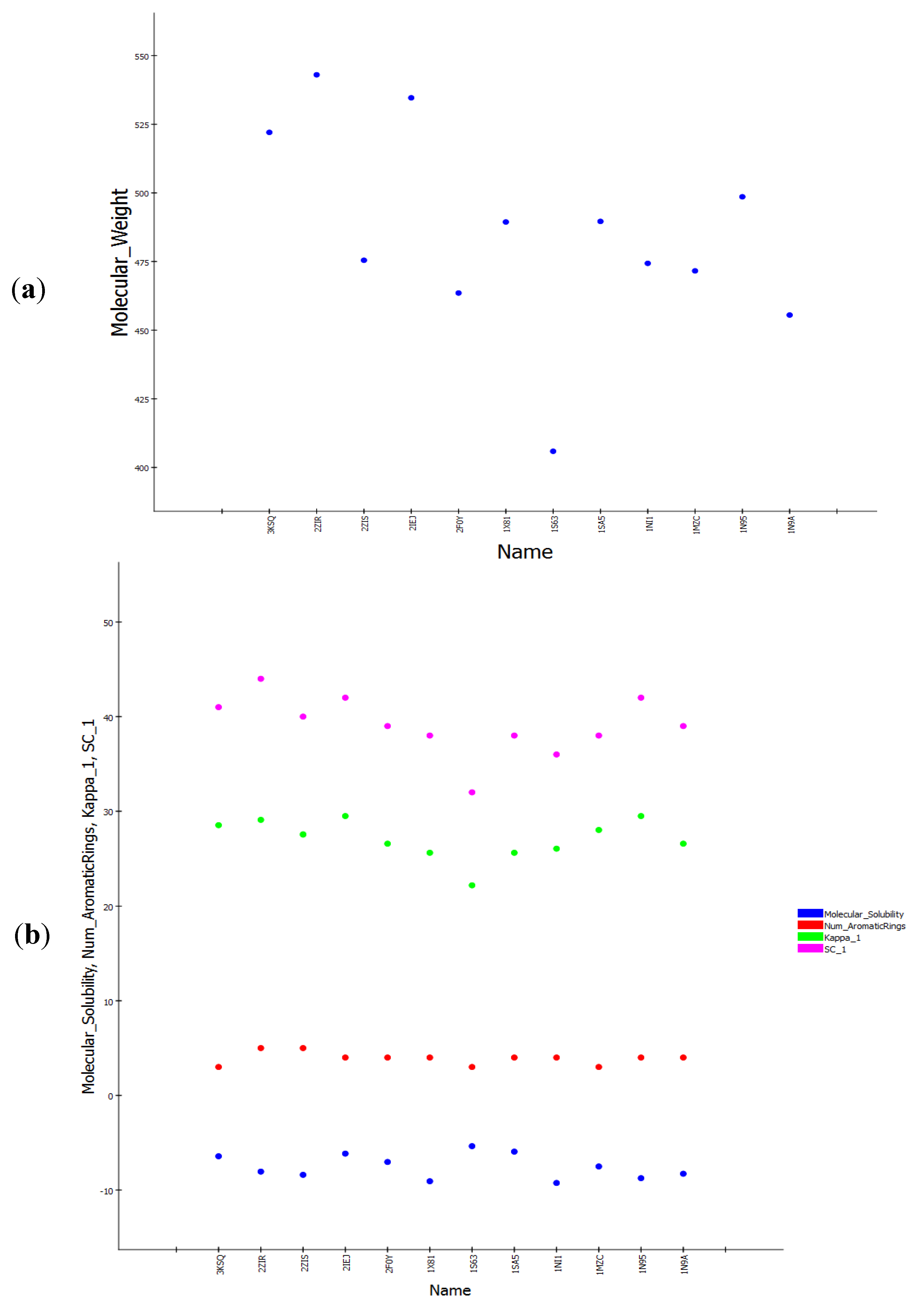
3.2. Structure-Based Pharmacophore Hypothesis
| PDB ID | Inhibitor Name | Fit Value |
|---|---|---|
| Pharm-B | ||
| 2ZIS | NH8903 | 5.088 |
| 1N95 | FTH1001 | 4.979 |
| 3E32 | ED21003 | 5.666 |
| 1NI1 | 2C510 | 4.713 |
| 3KSQ | Z96439 | 4.324 |
| 2F0Y | 3MN963 | 5.457 |
| 1SA5 | BMV440 | 4.956 |
| 1X81 | JAN1 | 4.627 |
| 1MZC | BME1003 | 4.778 |
| 1N9A | FTI1 | 4.962 |
| 2ZIR | NH7903 * | 4.933 |
| 3E33 | ED71003 * | 5.266 |
| 2IEJ | S48943 * | 5.579 |
| 1S63 | 7783012 * | 4.167 |
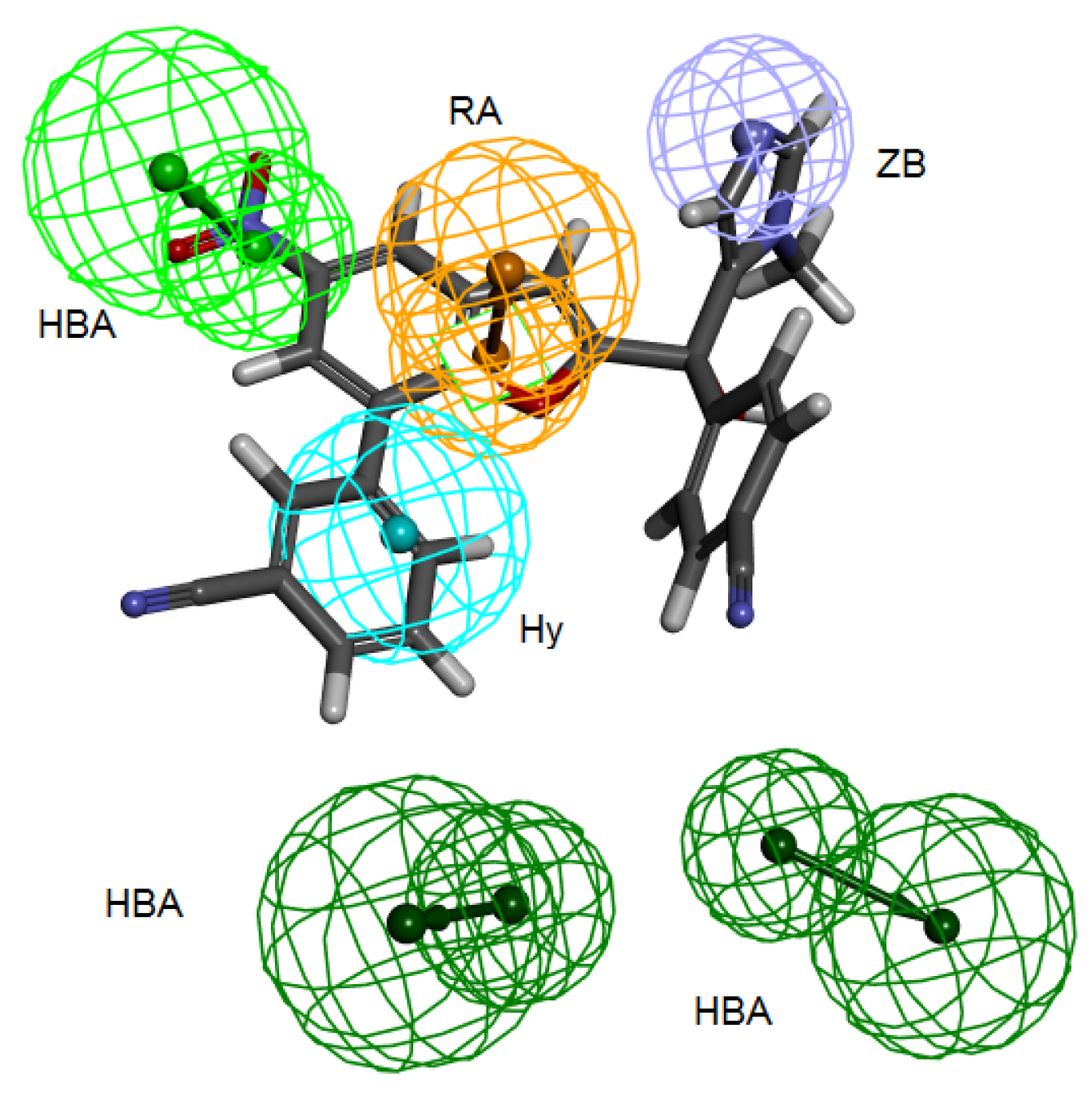
3.3. Validation of Pharmacophore Hypotheses
3.4. Database Screening
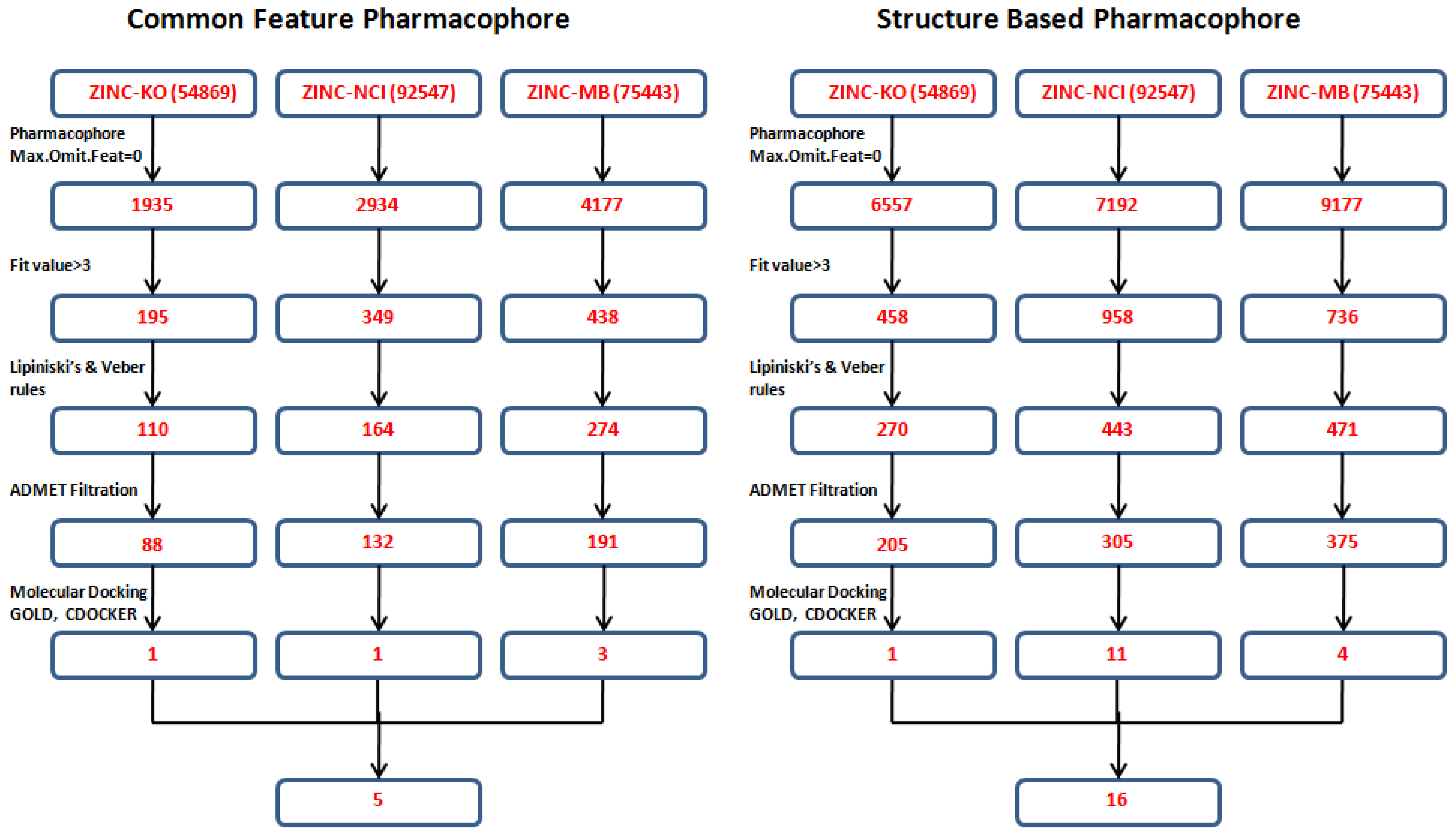
3.5. Molecular Docking
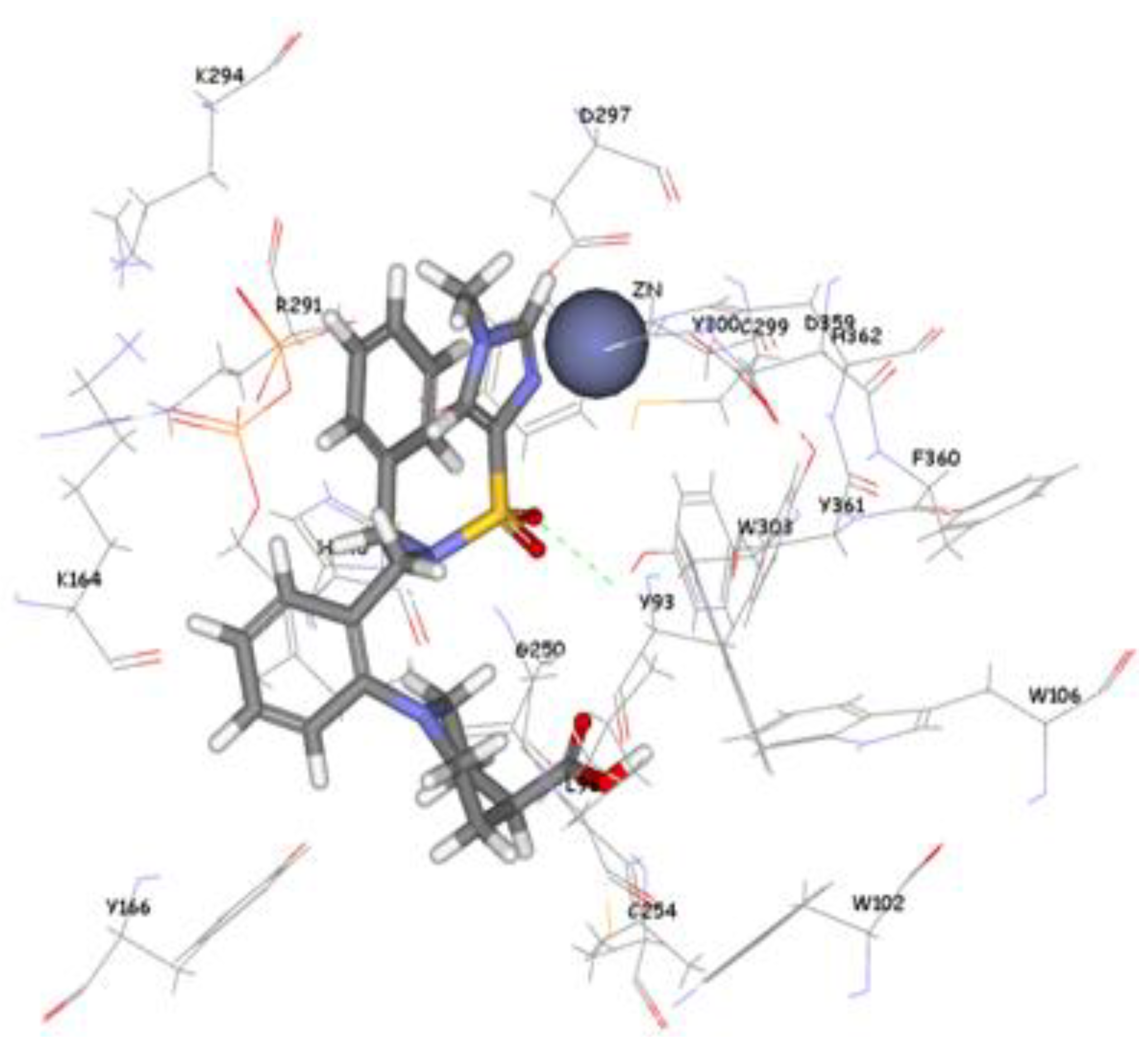
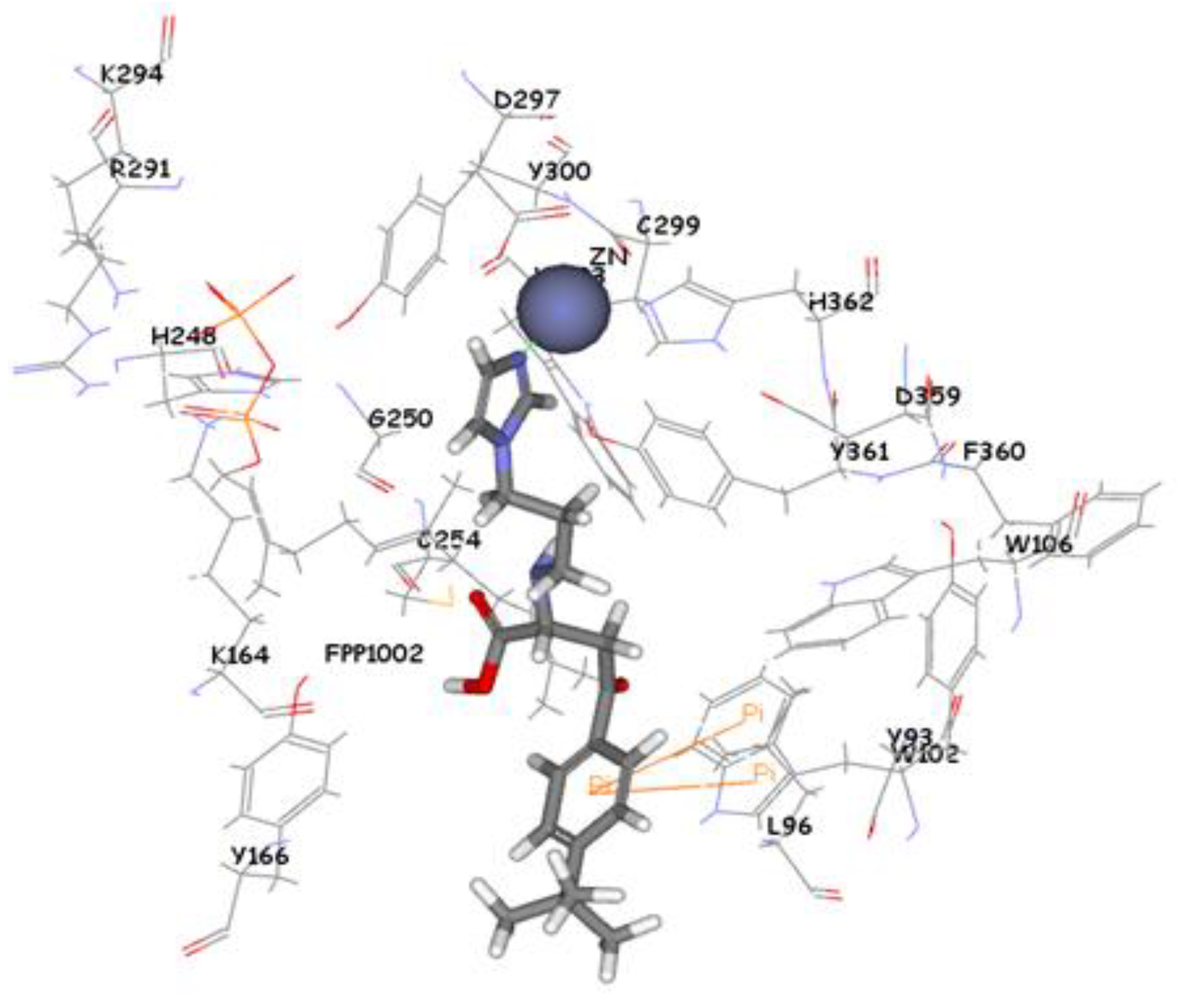
4. Conclusions
Conflict of Interest
References
- Sinensky, M.; Beck, L.A.; Leonard, S.; Evans, R. Differential inhibitory effects of lovastatin on protein isoprenylation and sterol synthesis. J. Biol. Chem. 1990, 265, 19937–19941. [Google Scholar]
- Habenicht, A.J.; Glomset, J.A.; Ross, R. Relation of cholesterol and mevalonic acid to the cell cycle in smooth muscle and Swiss 3T3 cells stimulated to divide by platelet-derived growth factor. J. Biol. Chem. 1980, 255, 5134–5140. [Google Scholar]
- Reiss, Y.; Brown, M.S.; Goldstein, J.L. Divalent cation and prenyl pyrophosphate specificities of the protein farnesyltransferase from rat brain, a zinc metalloenzyme. J. Biol. Chem. 1992, 267, 6403–6408. [Google Scholar]
- Reiss, Y.; Goldstein, J.L.; Seabra, M.C.; Casey, P.J.; Brown, M.S. Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell 1990, 62, 81–88. [Google Scholar] [CrossRef]
- Maltese, W.A.; Erdman, R.A. Characterization of isoprenoid involved in the post-translational modification of mammalian cell proteins. J. Biol. Chem. 1989, 264, 18168–18172. [Google Scholar]
- Zhang, F.L.; Casey, P.J. Protein Prenylation: Molecular Mechanisms and Functional Consequences. Annu. Rev. Biochem. 1996, 1996, 241–269. [Google Scholar] [CrossRef]
- Qian, Y.; Vogt, A.; Sebti, S.M.; Hamilton, A.D. Design and Synthesis of Non-Peptide Ras CAAX Mimetics as Potent Farnesyltransferase Inhibitors. J. Med. Chem. 1996, 39, 217–223. [Google Scholar] [CrossRef]
- Strickland, C.L.; Windsor, W.T.; Syto, R.; Wang, L.; Bond, R.; Wu, Z.; Schwartz, J.; Le, H.V.; Beese, L.S.; Weber, P.C. Cystal Structure of Farnesyl Protein Transferase Complexed with a CaaX Peptide and Farnesyl Diphosphate Analogue. Biochemistry 1998, 37, 16601–16611. [Google Scholar] [CrossRef]
- Waldron, K.J.; Robinson, N.J. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Micro. 2009, 7, 25–35. [Google Scholar] [CrossRef]
- Andreini, C.; Bertini, I.; Rosato, A. A hint to search for metalloproteins in gene banks. Bioinformatics 2004, 2004, 1373–1380. [Google Scholar]
- Roe, R.R.; Pang, Y.P. Zinc’s Exclusive Tetrahedral Coordination Governed by Its Electronic Structure. J. Mol. Model. 1999, 134–140. [Google Scholar] [CrossRef]
- Huang, C.C.; Casey, P.J.; Fierke, C.A. Evidence for a catalytic role of zinc in protein farnesyltransferase. Spectroscopy of Co2+-farnesyltransferase indicates metal coordination of the substrate thiolate. J. Biol. Chem. 1997, 272, 20–23. [Google Scholar]
- Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Farnesyltransferase New Insights into the Zinc-Coordination Sphere Paradigm: Evidence for a Carboxylate-Shift Mechanism. Biophys. J. 2005, 88, 483–494. [Google Scholar] [CrossRef]
- Equbal, T.; Silakari, O.; Rambabu, G.; Ravikumar, M. Pharmacophore mapping of diverse classes of farnesyltransferase inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 1594–1600. [Google Scholar] [CrossRef]
- Lu, A.; Zhang, J.; Yin, X.; Luo, X.; Jiang, H. Farnesyltransferase pharmacophore model derived from diverse classes of inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 243–249. [Google Scholar] [CrossRef]
- Vaidya, M.; Weigt, M.; Wiese, M. 3D-QSAR with the aid of pharmacophore search and docking-based alignments for farnesyltransferase inhibitors. Eur. J. Med. Chem. 2009, 44, 4070–4082. [Google Scholar] [CrossRef]
- Wu, G.; Robertson, D.H.; Brooks, C.L., 3rd; Vieth, M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER—A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003, 24, 1549–1562. [Google Scholar] [CrossRef]
- Nissink, J.W.; Murray, C.; Hartshorn, M.; Verdonk, M.L.; Cole, J.C.; Taylor, R. A new test set for validating predictions of protein-ligand interaction. Proteins 2002, 49, 457–471. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Hast, M.A.; Fletcher, S.; Cummings, C.G.; Pusateri, E.E.; Blaskovich, M.A.; Rivas, K.; Gelb, M.H.; van Voorhis, W.C.; Sebti, S.M.; Hamilton, A.D.; et al. Sructural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase. Chem. Biol. 2009, 16, 181–192. [Google Scholar] [CrossRef]
- Irwin, J.; Shoichet, B. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Al-Balas, Q.A.; Amawi, H.A.; Hassan, M.A.; Qandil, A.M.; Almaaytah, A.M.; Mhaidat, N.M. Virtual Lead Identification of Farnesyltransferase Inhibitors Based on Ligand and Structure-Based Pharmacophore Techniques. Pharmaceuticals 2013, 6, 700-715. https://doi.org/10.3390/ph6060700
Al-Balas QA, Amawi HA, Hassan MA, Qandil AM, Almaaytah AM, Mhaidat NM. Virtual Lead Identification of Farnesyltransferase Inhibitors Based on Ligand and Structure-Based Pharmacophore Techniques. Pharmaceuticals. 2013; 6(6):700-715. https://doi.org/10.3390/ph6060700
Chicago/Turabian StyleAl-Balas, Qosay A., Haneen A. Amawi, Mohammad A. Hassan, Amjad M. Qandil, Ammar M. Almaaytah, and Nizar M. Mhaidat. 2013. "Virtual Lead Identification of Farnesyltransferase Inhibitors Based on Ligand and Structure-Based Pharmacophore Techniques" Pharmaceuticals 6, no. 6: 700-715. https://doi.org/10.3390/ph6060700
APA StyleAl-Balas, Q. A., Amawi, H. A., Hassan, M. A., Qandil, A. M., Almaaytah, A. M., & Mhaidat, N. M. (2013). Virtual Lead Identification of Farnesyltransferase Inhibitors Based on Ligand and Structure-Based Pharmacophore Techniques. Pharmaceuticals, 6(6), 700-715. https://doi.org/10.3390/ph6060700





