The Role of Non-Coding Regions in Breast Cancer: From Gene Regulation to Therapeutic Implications
Abstract
1. Introduction
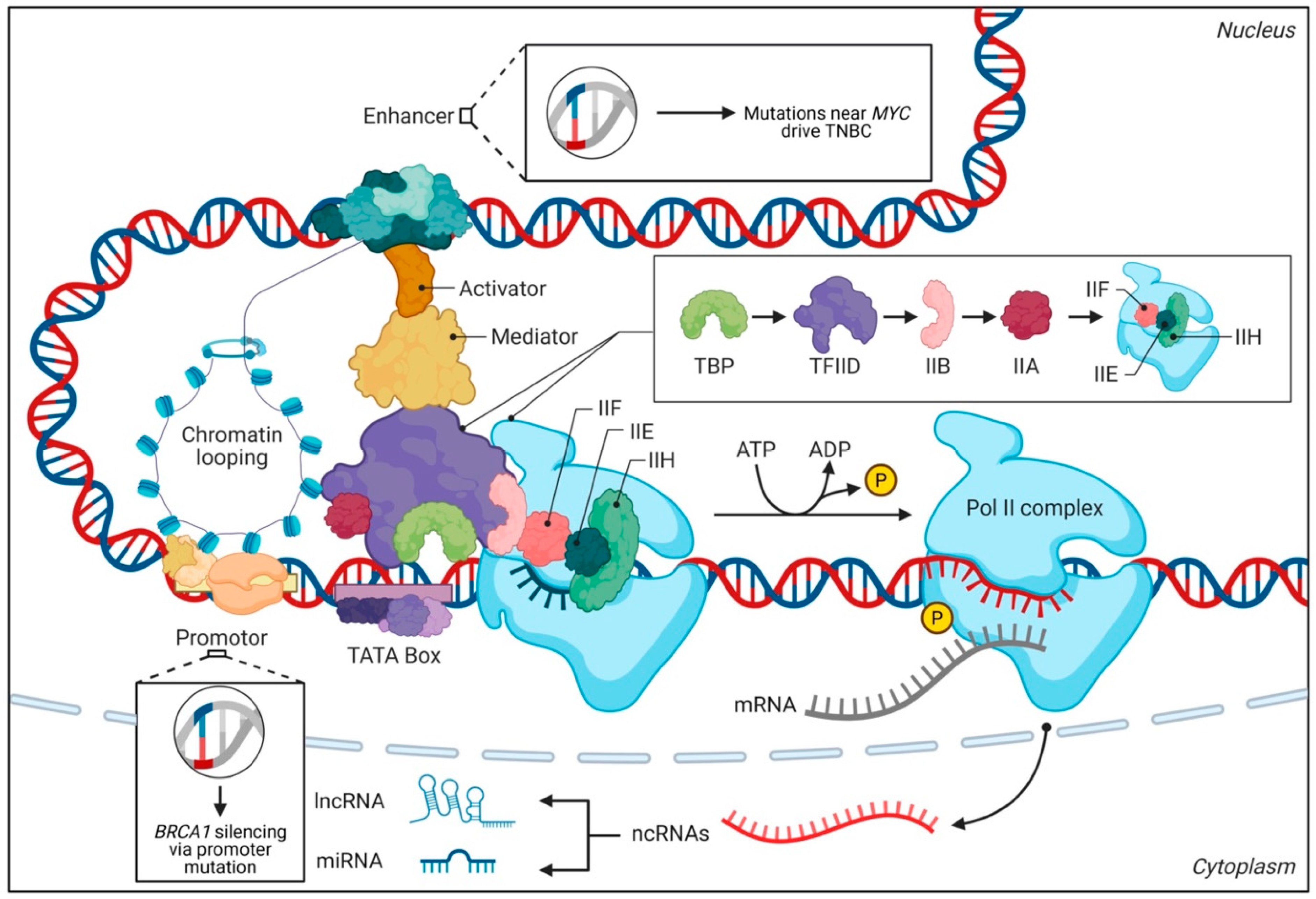
2. Key Non-Coding Elements and Their Regulatory Functions
2.1. Enhancers and Super-Enhancers
2.2. Introns
2.3. Long Non-Coding RNAs (lncRNAs)
2.4. MicroRNAs (miRNAs)
| Type of ncRNA | Example | Functional Role in BC | Implications | References |
|---|---|---|---|---|
| Long Non-Coding RNAs (lncRNAs) | HOTAIR | Promotes chromatin remodeling via interaction with PRC2; silences tumor suppressor genes. | Drives metastasis and therapy resistance in luminal and TNBC subtypes. | [43] |
| MALAT1 | Regulates alternative splicing and gene expression; enhances chemoresistance. | Associated with aggressive tumor behavior and poor prognosis in TNBC. | [48] | |
| NEAT1 | Facilitates nuclear paraspeckle formation; regulates DNA repair and cell cycle progression. | Promotes tumor growth and chemoresistance in TNBC. | [49] | |
| PVT1 | Interacts with MYC to upregulate PD-L1; promotes immune evasion. | Enhances immune evasion and resistance to immunotherapy in basal-like/TNBC. | [50] | |
| XIST | Regulates X-chromosome inactivation; modulates chromatin structure. | Implicated in tumor progression and therapy resistance in luminal and TNBC subtypes. | [51] | |
| MicroRNAs (miRNAs) | miR-21 | Targets tumor suppressors (PTEN, PDCD4); promotes oncogenic signaling. | Drives tumor growth and chemoresistance in TNBC and HER2-enriched subtypes. | [52] |
| miR-200 family | Inhibits EMT by targeting ZEB1/ZEB2; suppresses metastasis. | Downregulated in TNBC; restoration inhibits EMT and reduces tumor aggressiveness. | [53] | |
| miR-125a/b | Targets HER2; inhibits oncogenic signaling. | Downregulated in HER2-positive BCs; restoration reduces HER2 levels and tumor growth. | [54] | |
| miR-34a | Regulates cell cycle and apoptosis; targets MYC and BCL2. | Downregulated in TNBC, restoration induces apoptosis and unsensitizeses tumors to therapy. | [55] | |
| miR-205 | Targets HER2 and other oncogenic pathways; inhibits tumor growth. | Downregulated in TNBC; restoration reduces tumor aggressiveness. | [56] | |
| Circular RNAs (circRNAs) | circTADA2A | Acts as a miRNA sponge; regulates gene expression. | Promotes tumor growth and metastasis in TNBC. | [57] |
| circHER2 | Derived from the HER2 gene, it regulates HER2 signaling. | Associated with HER2-positive BCs; potential biomarker for therapy response. | [58] | |
| circSMARCA5 | Regulates alternative splicing and gene expression. | Downregulated in TNBC, restoration inhibits tumor growth and metastasis. | [59] | |
| Small Nucleolar RNAs (snoRNAs) | SNORD50A/B | Regulates RNA processing and modification. | Deletions associated with poor prognosis in luminal BCs. | [60] |
| SNHG1 | Acts as a scaffold for chromatin-modifying enzymes. | Promotes tumor growth and therapy resistance in TNBC. | [61] | |
| Piwi-Interacting RNAs (piRNAs) | piR-823 | Regulates DNA methylation and gene silencing. | Overexpressed in TNBC; promotes tumor growth and chemoresistance. | [62] |
2.5. A Multi-Omic Approach to Prognostic and Therapeutic Stratification in Breast Cancer
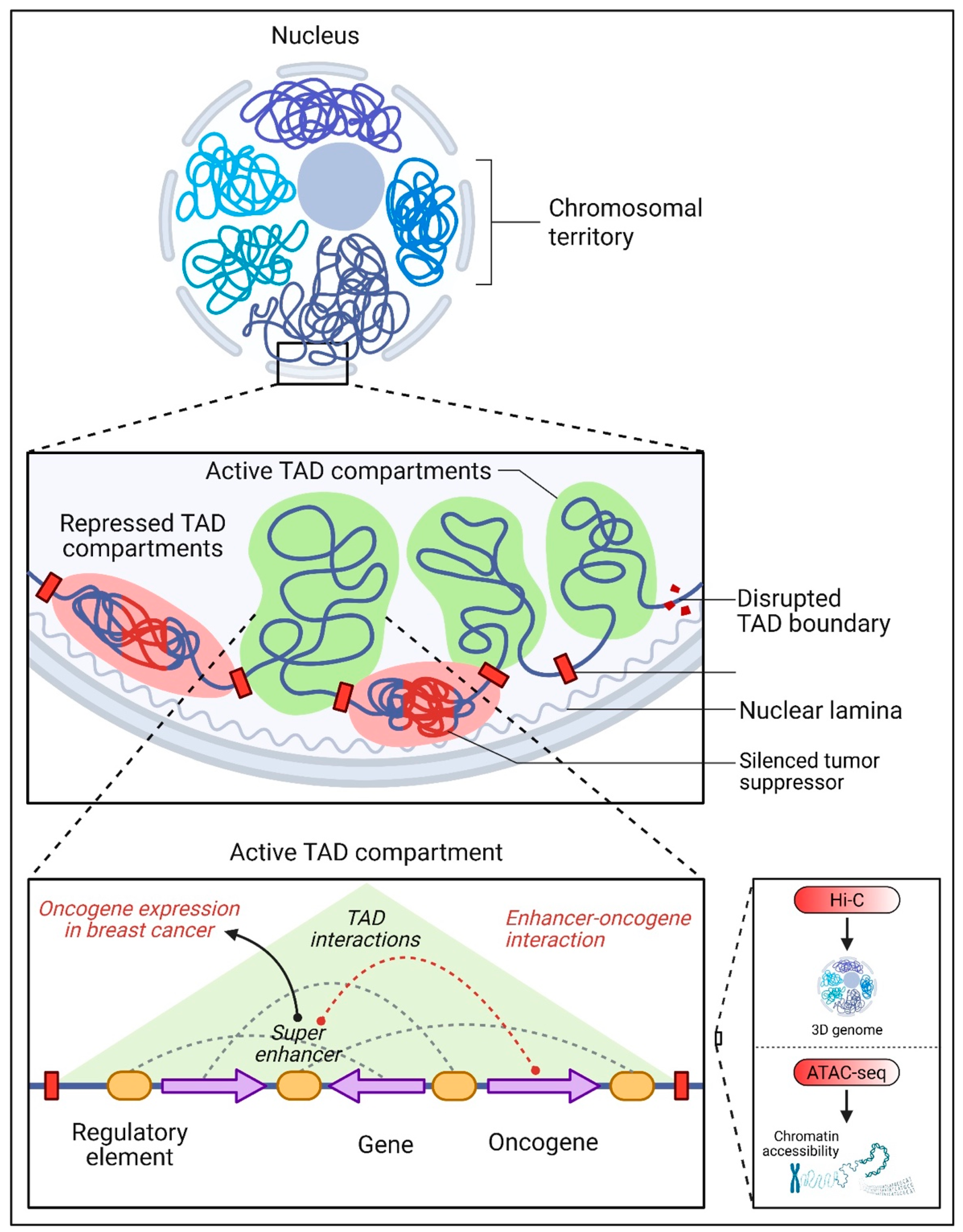
3. The Interplay Between Non-Coding DNA and Chromatin Organization
3.1. Chromatin Accessibility and 3D Genome Architecture
3.2. Transcription Factor Recruitment
4. Epigenetic Regulation by Non-Coding DNA
4.1. DNA Methylation
4.2. Histone Modifications
4.3. Non-Coding RNAs and Chromatin Remodeling
4.4. Therapeutic Targeting of Epigenetic Regulators
4.5. The Dynamic Interplay of m6A Epitranscriptomics and Cancer Progression
5. Relevance of Non-Coding RNA to BC
5.1. Non-Coding RNAs in Luminal Subtypes
5.2. Non-Coding RNAs in HER2-Enriched Subtypes
5.3. Non-Coding RNAs in Triple-Negative BC (TNBC)
5.4. Non-Coding RNAs as Biomarkers and Therapeutic Targets
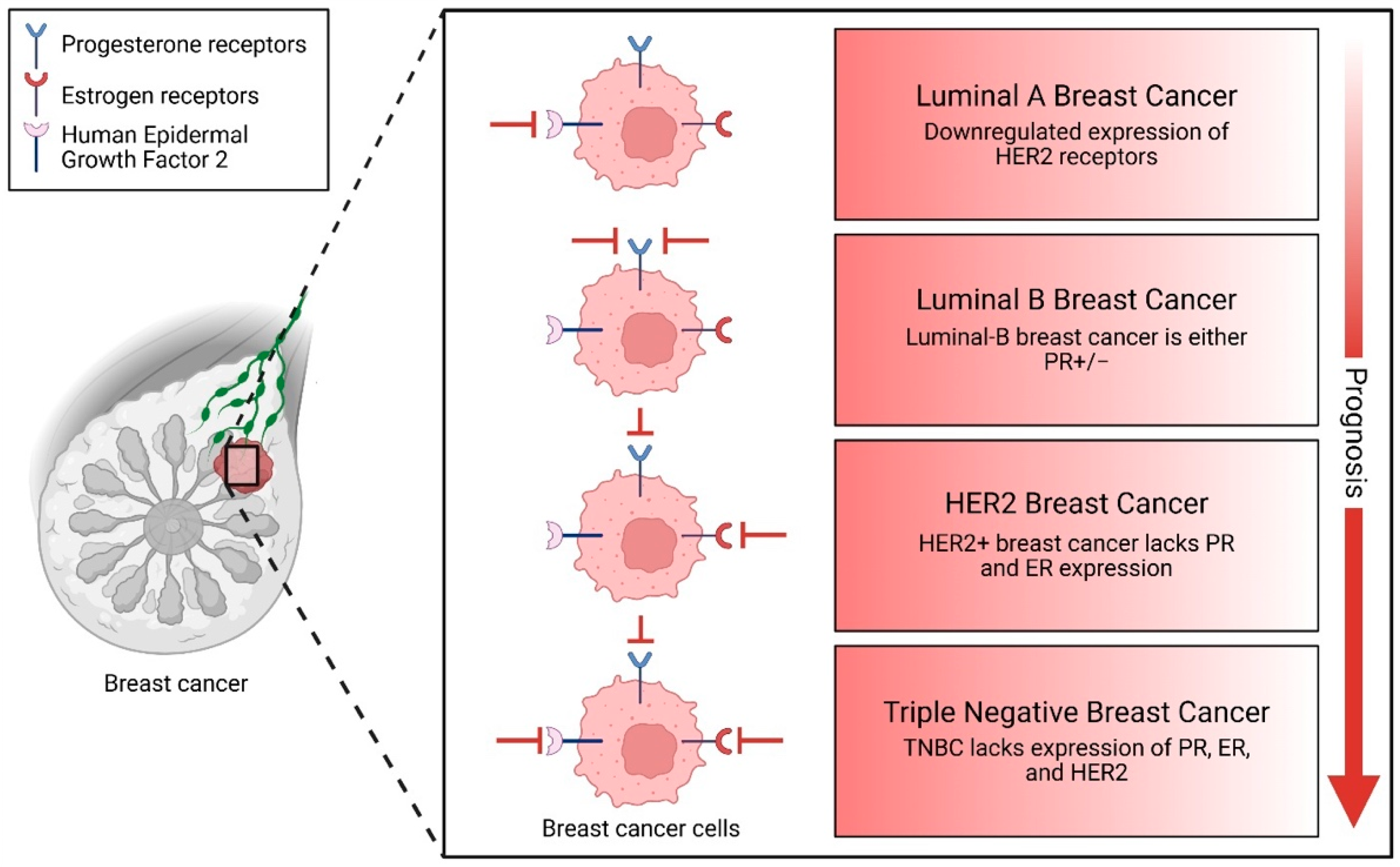
6. Non-Coding Elements in BC Subtypes
6.1. Luminal Subtypes (ER+/PR+)
6.2. HER2-Enriched Subtype
6.2.1. HER2 Amplification and Enhancer Activity
6.2.2. miRNAs in HER2 Regulation and Signaling
6.2.3. Non-Coding Mutations in HER2 Regulatory Regions
6.3. TNBC and Basal-Like (BLBC) Subtype
6.3.1. miRNAs in TNBC/BLBC: The miR-200 Family and EMT
6.3.2. Immune Evasion and Therapeutic Opportunities
7. Therapeutic Challenges and Future Directions in BC Research
7.1. General Challenges and Future Directions
7.2. Endocrine Therapy Resistance: Mechanisms and Implications in Luminal BC
7.3. Therapeutic Opportunities and Future Directions in Luminal BC
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Singh, S.; Saini, H.; Sharma, A.; Gupta, S.; Huddar, V.G.; Tripathi, R. Breast cancer: miRNAs monitoring chemoresistance and systemic therapy. Front. Oncol. 2023, 13, 1155254. [Google Scholar] [CrossRef]
- Singh, T.; Kaushik, M.; Mishra, L.C.; Behl, C.; Singh, V.; Tuli, H.S. Exosomal miRNAs as novel avenues for breast cancer treatment. Front. Genet. 2023, 14, 1134779. [Google Scholar] [CrossRef] [PubMed]
- Sarmento-Ribeiro, A.B.; Scorilas, A.; Gonçalves, A.C.; Efferth, T.; Trougakos, I.P. The emergence of drug resistance to targeted cancer therapies: Clinical evidence. Drug Resist. Updates 2019, 47, 100646. [Google Scholar] [CrossRef]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Non-Coding RNA 2021, 7, 47. [Google Scholar] [CrossRef]
- Elliott, K.; Larsson, E. Non-coding driver mutations in human cancer. Nat. Rev. Cancer 2021, 21, 500–509. [Google Scholar] [CrossRef]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Niederriter, A.R.; Varshney, A.; Parker, S.C.; Martin, D.M. Super Enhancers in Cancers, Complex Disease, and Developmental Disorders. Genes 2015, 6, 1183–1200. [Google Scholar] [CrossRef]
- Matros, E.; Wang, Z.C.; Lodeiro, G.; Miron, A.; Iglehart, J.D.; Richardson, A.L. BRCA1 promoter methylation in sporadic breast tumors: Relationship to gene expression profiles. Breast Cancer Res. Treat. 2005, 91, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Sarhangi, N.; Hajjari, S.; Heydari, S.F.; Ganjizadeh, M.; Rouhollah, F.; Hasanzad, M. Breast cancer in the era of precision medicine. Mol. Biol. Rep. 2022, 49, 10023–10037. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, K.; Liu, R.; Song, Y.; Lv, Y.; Bi, P.; Yang, F.; Li, S.; Zhao, J.; Li, X.; et al. Single-cell transcriptome sequencing of B-cell heterogeneity and tertiary lymphoid structure predicts breast cancer prognosis and neoadjuvant therapy efficacy. Clin. Transl. Med. 2023, 13, e1346. [Google Scholar] [CrossRef]
- Khalid, M.; Paracha, R.Z.; Nisar, M.; Malik, S.; Tariq, S.; Arshad, I.; Siddiqa, A.; Hussain, Z.; Ahmad, J.; Ali, A. Long non-coding RNAs and their targets as potential biomarkers in breast cancer. IET Syst. Biol. 2021, 15, 137–147. [Google Scholar] [CrossRef]
- Cava, C.; Armaos, A.; Lang, B.; Tartaglia, G.G.; Castiglioni, I. Identification of long non-coding RNAs and RNA binding proteins in breast cancer subtypes. Sci. Rep. 2022, 12, 693. [Google Scholar] [CrossRef]
- Escuin, D.; Bell, O.; García-Valdecasas, B.; Clos, M.; Larrañaga, I.; López-Vilaró, L.; Mora, J.; Andrés, M.; Arqueros, C.; Barnadas, A. Small Non-Coding RNAs and Their Role in Locoregional Metastasis and Outcomes in Early-Stage Breast Cancer Patients. Int. J. Mol. Sci. 2024, 25, 3982. [Google Scholar] [CrossRef]
- Arruabarrena-Aristorena, A.; Maag, J.L.; Kittane, S.; Cai, Y.; Karthaus, W.R.; Ladewig, E.; Park, J.; Kannan, S.; Ferrando, L.; Cocco, E.; et al. FOXA1 Mutations Reveal Distinct Chromatin Profiles and Influence Therapeutic Response in Breast Cancer. Cancer Cell 2020, 38, 534–550.e9. [Google Scholar] [CrossRef] [PubMed]
- Jeselsohn, R.; Buchwalter, G.; De Angelis, C.; Brown, M.; Schiff, R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015, 12, 573–583. [Google Scholar] [CrossRef]
- Tomasello, G.; Gambini, D.; Petrelli, F.; Azzollini, J.; Arcanà, C.; Ghidini, M.; Peissel, B.; Manoukian, S.; Garrone, O. Characterization of the HER2 status in BRCA-mutated breast cancer: A single institutional series and systematic review with pooled analysis. ESMO Open 2022, 7, 100531. [Google Scholar] [CrossRef]
- Gaibar, M.; Beltrán, L.; Romero-Lorca, A.; Fernández-Santander, A.; Novillo, A. Somatic Mutations in HER2 and Implications for Current Treatment Paradigms in HER2-Positive Breast Cancer. J. Oncol. 2020, 2020, 6375956. [Google Scholar] [CrossRef]
- Lancho, O.; Herranz, D. The MYC Enhancerome: Long-Range Transcriptional Regulation of MYC in Cancer. Trends Cancer 2018, 4, 810–822. [Google Scholar] [CrossRef]
- Pruszko, M.; Milano, E.; Forcato, M.; Donzelli, S.; Ganci, F.; Di Agostino, S.; De Panfilis, S.; Fazi, F.; O Bates, D.; Bicciato, S.; et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017, 18, 1331–1351. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Zong, X.; Malakar, P.; Ray, T.; Singh, D.K.; Freier, S.M.; Jensen, T.; Prasanth, S.G.; Karni, R.; Ray, P.S.; et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016, 7, 40418–40436. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Q.; Zhang, Z.-C.; Wu, Y.-Y.; Pi, Y.-N.; Lou, S.-H.; Liu, T.-B.; Lou, G.; Yang, C. Bromodomain and extraterminal (BET) proteins: Biological functions, diseases and targeted therapy. Signal Transduct. Target. Ther. 2023, 8, 420. [Google Scholar] [CrossRef]
- Sideris, N.; Dama, P.; Bayraktar, S.; Stiff, T.; Castellano, L. LncRNAs in breast cancer: A link to future approaches. Cancer Gene Ther. 2022, 29, 1866–1877. [Google Scholar] [CrossRef]
- Zhu, Y.; Gujar, A.D.; Wong, C.-H.; Tjong, H.; Ngan, C.Y.; Gong, L.; Chen, Y.-A.; Kim, H.; Liu, J.; Li, M.; et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell 2021, 39, 694–707.e7. [Google Scholar] [CrossRef]
- Deforzh, E.; Kharel, P.; Karelin, A.; Ivanov, P.; Krichevsky, A.M. HOXDeRNA activates a cancerous transcription program and super-enhancers genome-wide. bioRxiv 2023. [Google Scholar] [CrossRef]
- Lewis, M.W.; King, C.M.; Wisniewska, K.; Regner, M.J.; Coffey, A.; Kelly, M.R.; Mendez-Giraldez, R.; Davis, E.S.; Phanstiel, D.H.; Franco, H.L. CRISPR Screening of Transcribed Super-Enhancers Identifies Drivers of Triple-Negative Breast Cancer Progression. Cancer Res. 2024, 84, 3684–3700. [Google Scholar] [CrossRef]
- Mungamuri, S.K.; Murk, W.; Grumolato, L.; Bernstein, E.; Aaronson, S.A. Chromatin Modifications Sequentially Enhance ErbB2 Expression in ErbB2-Positive Breast Cancers. Cell Rep. 2013, 5, 302–313. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Long, W.; Liu, Q. Targeting Super-Enhancers as a Therapeutic Strategy for Cancer Treatment. Front. Pharmacol. 2019, 10, 361. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.; Lai, Y.; Liao, Y.I.; Liu, R.; Qiu, W. Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1. Int. J. Biol. Macromol. 2015, 80, 290–297. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, S.; Yang, J.; Chen, X.; Huang, W. Identification of miR-146a is Associated with the Aggressiveness and Suppresses Proliferation via Targeting CDKN2A in Breast Cancer. J. Cancer 2018, 9, 2200–2208. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Q.; Hu, Z.; Feng, Y.; Fan, L.; Tang, Z.; Yuan, J.; Shan, W.; Li, C.; Hu, X.; et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 2016, 23, 522–530. [Google Scholar] [CrossRef]
- Qin, S.; Ning, M.; Liu, Q.; Ding, X.; Wang, Y.; Liu, Q. Knockdown of long non-coding RNA CDKN2B-AS1 suppresses the progression of breast cancer by miR-122-5p/STK39 axis. Bioengineered 2021, 12, 5125–5137. [Google Scholar]
- Parfenyev, S.; Singh, A.; Fedorova, O.; Daks, A.; Kulshreshtha, R.; Barlev, N.A. Interplay between p53 and non-coding RNAs in the regulation of EMT in breast cancer. Cell Death Dis. 2021, 12, 58. [Google Scholar] [CrossRef]
- Zhai, H.; Zhang, X.; Sun, X.; Zhang, D.; Ma, S. Long Non-coding RNA LINC01420 Contributes to Pancreatic Cancer Progression Through Targeting KRAS Proto-oncogene. Dig. Dis. Sci. 2019, 65, 1042–1052. [Google Scholar] [CrossRef]
- Cheng, T.; Wu, Y.; Liu, Z.; Yu, Y.; Sun, S.; Guo, M.; Sun, B.; Huang, C. CDKN2A-mediated molecular subtypes characterize the hallmarks of tumor microenvironment and guide precision medicine in triple-negative breast cancer. Front. Immunol. 2022, 13, 970950. [Google Scholar] [CrossRef]
- Shi, L.; Magee, P.; Fassan, M.; Sahoo, S.; Leong, H.S.; Lee, D.; Sellers, R.; Brullé-Soumaré, L.; Cairo, S.; Monteverde, T.; et al. A KRAS-responsive long non-coding RNA controls microRNA processing. Nat. Commun. 2021, 12, 1–19. [Google Scholar] [CrossRef]
- Benoit-Pilven, C.; Besson, A.; Putoux, A.; Benetollo, C.; Saccaro, C.; Guguin, J.; Sala, G.; Cologne, A.; Delous, M.; Lesca, G.; et al. Clinical interpretation of variants identified in RNU4ATAC, a non-coding spliceosomal gene. PLoS ONE 2020, 15, e0235655. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, S.; Song, J.; Han, S.R.; Kim, J.H.; Lee, S.-W. Efficient circular RNA engineering by end-to-end self-targeting and splicing reaction using Tetrahymena group I intron ribozyme. Mol. Ther.-Nucleic Acids 2023, 33, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Chorev, M.; Carmel, L. The Function of Introns. Front. Genet. 2012, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Lee, K.S.; Choi, J.K. Comprehensive characterisation of intronic mis-splicing mutations in human cancers. Oncogene 2021, 40, 1347–1361. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Su, J.; Deng, L.; Wang, Y.-D. Roles and Mechanisms of Long Non-Coding RNAs in Breast Cancer. Int. J. Mol. Sci. 2022, 24, 89. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Lal, P.M.M.; Siddiqui, M.M.H.; Soulat, A.M.; Mohan, A.M.; Tanush, D.M.; Tirath, K.M.; Raja, S.M.; Khan, M.M.K.; Raja, A.M.; Chaulagain, A.M.; et al. MicroRNAs as promising biomarkers and potential therapeutic agents in breast cancer management: A comprehensive review. Ann. Med. Surg. 2024, 86, 3543–3550. [Google Scholar] [CrossRef]
- Arun, G.; Spector, D.L. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019, 16, 860–863. [Google Scholar] [CrossRef]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.-Y.; Siu, M.-T.; Ho, C.-W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Baljon, K.J.; Ramaiah, P.; Saleh, E.A.M.; Al-Dolaimy, F.; Al-Dami, F.H.; Gandla, K.; Alkhafaji, A.T.; Abbas, A.H.R.; Alsaalamy, A.H.; Bisht, Y.S. LncRNA PVT1: As a therapeutic target for breast cancer. Pathol. Res. Pract. 2023, 248, 154675. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Nemtsova, M.V. Mutual Regulation of ncRNAs and Chromatin Remodeling Complexes in Normal and Pathological Conditions. Int. J. Mol. Sci. 2024, 24, 7848. [Google Scholar] [CrossRef]
- Vo, T.H.; Abdelaal, E.E.-S.; Jordan, E.; O’DOnovan, O.; McNeela, E.A.; Mehta, J.P.; Rani, S. miRNAs as biomarkers of therapeutic response to HER2-targeted treatment in breast cancer: A systematic review. Biochem. Biophys. Rep. 2023, 37, 101588. [Google Scholar] [CrossRef]
- Klicka, K.; Grzywa, T.M.; Mielniczuk, A.; Klinke, A.; Włodarski, P.K. The role of miR-200 family in the regulation of hallmarks of cancer. Front. Oncol. 2022, 12, 965231. [Google Scholar] [CrossRef]
- Piergentili, R.; Marinelli, E.; Cucinella, G.; Lopez, A.; Napoletano, G.; Gullo, G.; Zaami, S. miR-125 in Breast Cancer Etiopathogenesis: An Emerging Role as a Biomarker in Differential Diagnosis, Regenerative Medicine, and the Challenges of Personalized Medicine. Non-Coding RNA 2024, 10, 16. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Liu, R.; Kasinski, A.L.; Shen, H.; Slack, F.J.; Tang, D.G. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front. Cell Dev. Biol. 2021, 9, 640587. [Google Scholar] [CrossRef]
- Plantamura, I.; Cataldo, A.; Cosentino, G.; Iorio, M.V. miR-205 in Breast Cancer: State of the Art. Int. J. Mol. Sci. 2020, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.; Zhang, S.; Deng, Y.; Wang, M.; Deng, X.; Yang, S.; Wu, Y.; Dai, Z. Regulatory mechanisms, functions, and clinical significance of CircRNAs in triple-negative breast cancer. J. Hematol. Oncol. 2021, 14, 41. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X. A Comprehensive Review of HER2 in Cancer Biology and Therapeutics. Genes 2024, 15, 903. [Google Scholar] [CrossRef]
- Xue, C.; Wei, J.; Li, M.; Chen, S.; Zheng, L.; Zhan, Y.; Duan, Y.; Deng, H.; Xiong, W.; Li, G.; et al. The Emerging Roles and Clinical Potential of circSMARCA5 in Cancer. Cells 2022, 11, 3074. [Google Scholar] [CrossRef]
- Siprashvili, Z.; Webster, D.; Johnston, D.; Shenoy, R.M.; Ungewickell, A.J.; Bhaduri, A.; Flockhart, R.J.; Zarnegar, B.J.; Che, Y.; Meschi, F.; et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat. Genet. 2016, 48, 53–58. [Google Scholar] [CrossRef]
- Deng, L.; Wang, J.; Song, J.; Wu, Q.; Gong, Z.; Song, J.; Hou, L. Long noncoding RNA SNHG1 promotes breast cancer progression by regulating the miR-641/RRS1 axis. Sci. Rep. 2024, 14, 3265. [Google Scholar] [CrossRef]
- Öner, Ç.; Köser, F.; Çolak, E. The association of piR-651 and piR-823 on metastatic and invasive characteristics of triple negative breast cancer cells. Nucleosides Nucleotides Nucleic Acids 2024, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, L.; Zhao, J.; Du, X.; Yu, Q.; Wu, J.; Wang, B.; Ou, R. Construction and Verification of a Hypoxia-Related 4-lncRNA Model for Prediction of Breast Cancer. Int. J. Gen. Med. 2021, 14, 4611–4623. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lv, Q.; Huang, H.; Sun, C.; Pang, D.; Wu, J. Identification of a four-long non-coding RNA signature in predicting breast cancer survival. Oncol. Lett. 2019, 19, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Chen, P.; Chen, J.; Deng, Y.; Huang, C. Landscape Analysis of Matrix Metalloproteinases Unveils Key Prognostic Markers for Patients with Breast Cancer. Front. Genet. 2022, 12, 809600. [Google Scholar] [CrossRef]
- Deng, M.; Chen, X.; Qiu, J.; Liu, G.; Huang, C. A Neural Network-Based Scoring System for Predicting Prognosis and Therapy in Breast Cancer. Curr. Protoc. 2024, 4, e1122. [Google Scholar] [CrossRef]
- Huang, C.; Deng, M.; Leng, D.; Sun, B.; Zheng, P.; Zhang, X.D. MIRS: An AI scoring system for predicting the prognosis and therapy of breast cancer. iScience 2023, 26, 108322. [Google Scholar] [CrossRef]
- Dill, C.D.; Dammer, E.B.; Griffen, T.L.; Seyfried, N.T.; Lillard, J.W. A network approach reveals driver genes associated with survival of patients with triple-negative breast cancer. iScience 2021, 24, 102451. [Google Scholar] [CrossRef]
- Huang, W.; Li, H.; Yu, Q.; Xiao, W.; Wang, D.O. LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022, 41, 100. [Google Scholar] [CrossRef]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef]
- Zufferey, M.; Tavernari, D.; Ciriello, G. Methods for the Analysis of Topologically Associating Domains (TADs). Methods Mol. Biol. 2022, 2301, 39–59. [Google Scholar] [PubMed]
- Maisuradze, L.; King, M.C.; Surovtsev, I.V.; Mochrie, S.G.J.; Shattuck, M.D.; O’hErn, C.S.; Dunbrack, R.L. Identifying topologically associating domains using differential kernels. PLOS Comput. Biol. 2024, 20, e1012221. [Google Scholar] [CrossRef]
- Dixon, J.R.; Jung, I.; Selvaraj, S.; Shen, Y.; Antosiewicz-Bourget, J.E.; Lee, A.Y.; Ye, Z.; Kim, A.; Rajagopal, N.; Xie, W.; et al. Chromatin architecture reorganization during stem cell differentiation. Nature 2015, 518, 331–336. [Google Scholar] [CrossRef]
- Wang, X.; Yan, J.; Ye, Z.; Zhang, Z.; Wang, S.; Hao, S.; Shen, B.; Wei, G. Reorganization of 3D chromatin architecture in doxorubicin-resistant breast cancer cells. Front. Cell Dev. Biol. 2022, 10, 974750. [Google Scholar] [CrossRef]
- Llinàs-Arias, P.; Ensenyat-Mendez, M.; Íñiguez-Muñoz, S.; Orozco, J.I.J.; Valdez, B.; Salomon, M.P.; Matsuba, C.; Solivellas-Pieras, M.; Bedoya-López, A.F.; Sesé, B.; et al. Chromatin insulation orchestrates matrix metalloproteinase gene cluster expression reprogramming in aggressive breast cancer tumors. Mol. Cancer 2023, 22, 190. [Google Scholar] [CrossRef]
- Ramírez-Cuéllar, J.; Ferrari, R.; Sanz, R.T.; Valverde-Santiago, M.; García-García, J.; Nacht, A.S.; Castillo, D.; Le Dily, F.; Neguembor, M.V.; Malatesta, M.; et al. LATS1 controls CTCF chromatin occupancy and hormonal response of 3D-grown breast cancer cells. EMBO J. 2024, 43, 1770–1798. [Google Scholar] [CrossRef] [PubMed]
- Taberlay, P.C.; Achinger-Kawecka, J.; Lun, A.T.; Buske, F.A.; Sabir, K.; Gould, C.M.; Zotenko, E.; Bert, S.A.; Giles, K.A.; Bauer, D.C.; et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016, 26, 719–731. [Google Scholar] [CrossRef]
- Verma, P.; Zhou, Y.; Cao, Z.; Deraska, P.V.; Deb, M.; Arai, E.; Li, W.; Shao, Y.; Puentes, L.; Li, Y.; et al. ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nat. Cell Biol. 2021, 23, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Iglesia, M.D.; Jayasinghe, R.G.; Chen, S.; Terekhanova, N.V.; Herndon, J.M.; Storrs, E.; Karpova, A.; Zhou, D.C.; Al Deen, N.N.; Shinkle, A.T.; et al. Differential chromatin accessibility and transcriptional dynamics define breast cancer subtypes and their lineages. Nat. Cancer 2024, 5, 1713–1736. [Google Scholar] [CrossRef]
- Sundaram, L.; Kumar, A.; Zatzman, M.; Salcedo, A.; Ravindra, N.; Shams, S.; Louie, B.H.; Bagdatli, S.T.; Myers, M.A.; Sarmashghi, S.; et al. Single-cell chromatin accessibility reveals malignant regulatory programs in primary human cancers. Science 2024, 385, eadk9217. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21.29.1–21.29.9. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef]
- Khurana, E.; Fu, Y.; Chakravarty, D.; Demichelis, F.; Rubin, M.A.; Gerstein, M. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016, 17, 93–108. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. In Epigenetics: Development and Disease; Kundu, T.K., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 289–317. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Y.; Olopade, O.I. MYC and Breast Cancer. Genes Cancer 2010, 1, 629–640. [Google Scholar] [CrossRef]
- Pavithran, H.; Kumavath, R. Emerging role of pioneer transcription factors in targeted ERα positive breast cancer. Explor. Target. Anti-Tumor Ther. 2021, 2, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Anestis, A.; Zoi, I.; Papavassiliou, A.G.; Karamouzis, M.V. Androgen Receptor in Breast Cancer—Clinical and Preclinical Research Insights. Molecules 2020, 25, 358. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, I.; Gasparri, M.L.; De Marco, M.P.; Costanzi, F.; Besharat, A.R.; Papadia, A.; Kuehn, T.; Gentilini, O.D.; Bellati, F.; Caserta, D. The Clinical and Pathological Profile of BRCA1 Gene Methylated Breast Cancer Women: A Meta-Analysis. Cancers 2021, 13, 1391. [Google Scholar] [CrossRef]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. Adv. Exp. Med. Biol. 2016, 945, 151–172. [Google Scholar]
- Shi, Y.; Huang, Q.; Kong, X.; Zhao, R.; Chen, X.; Zhai, Y.; Xiong, L. Current Knowledge of Long Non-Coding RNA HOTAIR in Breast Cancer Progression and Its Application. Life 2021, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Yang, C.; Gao, A.; Sun, M.; Lv, D. MiR-101: An Important Regulator of Gene Expression and Tumor Ecosystem. Cancers 2022, 14, 5861. [Google Scholar] [CrossRef]
- Yan, B.; Guo, Q.; Fu, F.-J.; Wang, Z.; Yin, Z.; Wei, Y.B.; Yang, J.R. The role of miR-29b in cancer: Regulation, function, and signaling. OncoTargets Ther. 2015, 8, 539–548. [Google Scholar] [CrossRef]
- Chowdhury, M.R.; Bahadur, R.P.; Basak, J. Genome-wide prediction of cauliflower miRNAs and lncRNAs and their roles in post-transcriptional gene regulation. Planta 2021, 254, 72. [Google Scholar] [CrossRef] [PubMed]
- Pisignano, G.; Ladomery, M. Post-Transcriptional Regulation Through Long Non-Coding RNAs (lncRNAs). Non-Coding RNA 2021, 7, 29. [Google Scholar] [CrossRef]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef]
- Zolota, V.; Tzelepi, V.; Piperigkou, Z.; Kourea, H.; Papakonstantinou, E.; Argentou, M.-I.; Karamanos, N.K. Epigenetic Alterations in Triple-Negative Breast Cancer—The Critical Role of Extracellular Matrix. Cancers 2021, 13, 713. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Ma, T.; Yu, B. Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target. Ther. 2023, 8, 69. [Google Scholar] [CrossRef]
- Artz, O.; White, J.R.; Rousseau, B.; Argiles, G.; Foote, M.B.; Johannet, P.; Patel, M.; Abdelfattah, S.; Patel, S.; Wilde, C.A. Role of recurrent somatic mutations that alter conserved m6A motifs in human cancer. Nucleic Acids Res. Cancer 2024, 7, zcaf014. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Yang, L.; Han, L.; Hu, Z.; Gu, Z.; Deng, X. Prevalence of synonymous mutations in m6A modification sites in human cancers. Genes Dis. 2025, 12, 101373. [Google Scholar] [CrossRef]
- Lan, Y.; Xia, Z.; Shao, Q.; Lin, P.; Lu, J.; Xiao, X.; Zheng, M.; Chen, D.; Dou, Y.; Xie, Q. Synonymous mutations promote tumorigenesis by disrupting m6A-dependent mRNA metabolism. Cell 2025, 188, 1828–1841.e15. [Google Scholar] [CrossRef]
- Hodara, E.; Mades, A.; Swartz, L.; Iqbal, M.; Xu, T.; Bsteh, D.; Goldkorn, A. m6A epitranscriptome analysis reveals differentially methylated transcripts that drive early chemoresistance in bladder cancer. Nucleic Acids Res. Cancer 2023, 5, zcad054. [Google Scholar] [CrossRef]
- Lobo, J.; Barros-Silva, D.; Henrique, R.; Jerónimo, C. The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors. Genes 2018, 9, 552. [Google Scholar] [CrossRef]
- Fong, A.S.; Pan, Y.; Yu, J.; Wong, C.C. Interplay between the m6A Epitranscriptome and Tumor Metabolism: Mechanisms and Therapeutic Implications. Biomedicines 2022, 10, 2589. [Google Scholar] [CrossRef]
- Wang, S.; Jin, J.; Chen, J.; Lou, W. MUC14-Related ncRNA-mRNA Network in Breast Cancer. Genes 2021, 12, 1677. [Google Scholar] [CrossRef]
- Xia, Q.; Shen, J.; Wang, Q.; Chen, R.; Zheng, X.; Yan, Q.; Du, L.; Li, H.; Duan, S.; Kumar, V. Cuproptosis-associated ncRNAs predict breast cancer subtypes. PLoS ONE 2024, 19, e0299138. [Google Scholar] [CrossRef]
- Zhao, S.; Ma, D.; Xiao, Y.; Li, X.-M.; Ma, J.-L.; Zhang, H.; Xu, X.-L.; Lv, H.; Jiang, W.-H.; Yang, W.-T.; et al. Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncology 2020, 25, e1481–e1491. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, R.; Mori, H.; Ha, D.; Wu, X.; Wang, J.; Wang, X.; Saeki, K.; Chang, G.; Shim, H.J.; Chan, Y.; et al. Molecular features of luminal breast cancer defined through spatial and single-cell transcriptomics. Clin. Transl. Med. 2024, 14, e1548. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tsouko, E.; Jonsson, P.; Bergh, J.; Hartman, J.; Aydogdu, E.; Williams, C. miR-206 inhibits cell migration through direct targeting of the actin-binding protein Coronin 1C in triple-negative breast cancer. Mol. Oncol. 2014, 8, 1690–1702. [Google Scholar] [CrossRef]
- Wei, J.; Lu, Y.; Wang, R.; Xu, X.; Liu, Q.; He, S.; Pan, H.; Liu, X.; Yuan, B.; Ding, Y.; et al. MicroRNA-375: Potential cancer suppressor and therapeutic drug. Biosci. Rep. 2021, 41, BSR20211494. [Google Scholar] [CrossRef]
- Schettini, F.; Pascual, T.; Conte, B.; Chic, N.; Brasó-Maristany, F.; Galván, P.; Martínez, O.; Adamo, B.; Vidal, M.; Muñoz, M.; et al. HER2-enriched subtype and pathological complete response in HER2-positive breast cancer: A systematic review and meta-analysis. Cancer Treat. Rev. 2020, 84, 101965. [Google Scholar] [CrossRef]
- Choi, W.; Liu, R.; Mak, C.; Maadi, H.; Godbout, R. Overcoming retinoic acid resistance in HER2-enriched breast cancers: Role of MYC. FEBS J. 2024, 291, 3521–3538. [Google Scholar] [CrossRef]
- Gan, F.-J.; Li, Y.; Xu, M.-X.; Zhou, T.; Wu, S.; Hu, K.; Li, Y.; Sun, S.-H.; Luo, Q. LncRNA BCAR4 expression predicts the clinical response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. Cancer Biomark. 2021, 32, 339–351. [Google Scholar] [CrossRef]
- Peng, L.; Jiang, J.; Tang, B.; Nice, E.C.; Zhang, Y.-Y.; Xie, N. Managing therapeutic resistance in breast cancer: From the lncRNAs perspective. Theranostics 2020, 10, 10360–10377. [Google Scholar] [CrossRef]
- Verma, A.; Singh, A.; Singh, M.P.; Nengroo, M.A.; Saini, K.K.; Satrusal, S.R.; Khan, M.A.; Chaturvedi, P.; Sinha, A.; Meena, S.; et al. EZH2-H3K27me3 mediated KRT14 upregulation promotes TNBC peritoneal metastasis. Nat. Commun. 2022, 13, 7344. [Google Scholar] [CrossRef]
- Michaels, E.; Chen, N.; Nanda, R. The Role of Immunotherapy in Triple-Negative Breast Cancer (TNBC). Clin. Breast Cancer 2024, 24, 263–270. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, G.; Liu, X.-L.; Zhang, G.; Zhao, S.-Q.; Zhang, S.-L.; Luo, L.-H.; Yin, D.-C.; Zhang, C.-Y. Progress of non-coding RNAs in triple-negative breast cancer. Life Sci. 2021, 272, 119238. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Di Bonito, M.; Cerrone, M.; Collina, F.; De Laurentiis, M.; Botti, G. Long Non-Coding RNA HOTAIR in Breast Cancer Therapy. Cancers 2020, 12, 1197. [Google Scholar] [CrossRef]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011, 13, R45. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Arbitrio, M.; Caracciolo, D.; Cordua, A.; Cuomo, O.; Grillone, K.; Riillo, C.; Caridà, G.; Scionti, F.; Labanca, C.; et al. miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: A systematic review. Mol. Ther. Nucleic Acids 2022, 27, 1191–1224. [Google Scholar] [CrossRef] [PubMed]
- Dobre, E.-G.; Dinescu, S.; Costache, M. Connecting the Missing Dots: ncRNAs as Critical Regulators of Therapeutic Susceptibility in Breast Cancer. Cancers 2020, 12, 2698. [Google Scholar] [CrossRef]
- Amer, H.T.; Eissa, R.A.; El Tayebi, H.M. A cutting-edge immunomodulatory interlinkage between HOTAIR and MALAT1 in tumor-associated macrophages in breast cancer: A personalized immunotherapeutic approach. Front. Mol. Biosci. 2022, 9, 1032517. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 2021, 11, 340–354. [Google Scholar] [CrossRef]
- Han, X.; Ren, C.; Lu, C.; Qiao, P.; Yang, T.; Yu, Z. Deubiquitination of MYC by OTUB1 contributes to HK2 mediated glycolysis and breast tumorigenesis. Cell Death Differ. 2022, 29, 1864–1873. [Google Scholar] [CrossRef]
- Zhao, N.; Cao, J.; Xu, L.; Tang, Q.; Dobrolecki, L.E.; Lv, X.; Talukdar, M.; Lu, Y.; Wang, X.; Hu, D.Z.; et al. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J. Clin. Investig. 2018, 128, 1283–1299. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Y.; Xu, G.; Tian, Z.; Yang, Q.; Gong, Y.; Wu, G. The initial expression alterations occurring to transcription factors during the formation of breast cancer: Evidence from bioinformatics. Cancer Med. 2022, 11, 1371–1395. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Ma, D.; Xu, X.-E.; Jin, X.; Yu, K.-D.; Jiang, Y.-Z.; Shao, Z.-M. Genomic Landscape and Endocrine-Resistant Subgroup in Estrogen Receptor-Positive, Progesterone Receptor-Negative, and HER2-Negative Breast Cancer. Theranostics 2018, 8, 6386–6399. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-X.; Wang, L.-J.; Hou, J.; Liu, H.-Y.; Wang, R.; Wang, C.; Xie, W.-H.; Buijs, J.T. Identification of Long Noncoding RNAs as Predictors of Survival in Triple-Negative Breast Cancer Based on Network Analysis. BioMed Res. Int. 2020, 2020, 8970340. [Google Scholar] [CrossRef]
- Xiping, Z.; Bo, C.; Shifeng, Y.; Feijiang, Y.; Hongjian, Y.; Qihui, C.; Binbin, T. Roles of MALAT1 in development and migration of triple negative and Her-2 positive breast cancer. Oncotarget 2018, 9, 2255–2267. [Google Scholar] [CrossRef]
- Arisan, E.D.; Rencuzogullari, O.; Cieza-Borrella, C.; Arenas, F.M.; Dwek, M.; Lange, S.; Uysal-Onganer, P. MiR-21 Is Required for the Epithelial–Mesenchymal Transition in MDA-MB-231 Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 1557. [Google Scholar] [CrossRef] [PubMed]
- Desouky, E.; Khaliefa, A.; Hozayen, W.; Shaaban, S.; Hasona, N. Signature of miR-21 and MEG-2 and their correlation with TGF-β signaling in breast cancer. Hum. Exp. Toxicol. 2023, 42, 9603271231159799. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-C.; Cheng, T.-C.; Tu, S.-H.; Chang, J.; Guo, P.; Chen, L.-C.; Ho, Y.-S. Tumor targeting and therapeutic assessments of RNA nanoparticles carrying α9-nAChR aptamer and anti-miR-21 in triple-negative breast cancers. Mol. Ther.-Nucleic Acids 2023, 33, 351–366. [Google Scholar] [CrossRef]
- Ruiz de Garibay, G.; Fernandez-Garcia, I.; Mazoyer, S.; Leme de Calais, F.; Ameri, P.; Vijayakumar, S.; Martinez-Ruiz, H.; Damiola, F.; Barjhoux, L.; Thomassen, M.; et al. Altered regulation of BRCA1 exon 11 splicing is associated with breast cancer risk in carriers of BRCA1 pathogenic variants. Hum. Mutat. 2021, 42, 1488–1502. [Google Scholar] [CrossRef]
- Ryu, J.; Lee, H.; Cho, E.H.; Yoon, K.; Kim, M.; Joo, J.; Lee, E.; Kang, H.; Lee, S.; Lee, D.O.; et al. Exon splicing analysis of intronic variants in multigene cancer panel testing for hereditary breast/ovarian cancer. Cancer Sci. 2020, 111, 3912–3925. [Google Scholar] [CrossRef]
- Lai, J.; Chen, B.; Li, Y.; Lin, X.; Li, M.; Liu, J.; Liao, N. Integrated analysis of cell cycle-related genes in HR+/HER2− breast cancer. Breast Cancer 2022, 29, 121–130. [Google Scholar] [CrossRef]
- Nishimura, F.G.; Sampaio, B.B.; Komoto, T.T.; da Silva, W.J.; da Costa, M.M.G.; Haddad, G.I.; Peronni, K.C.; Evangelista, A.F.; Hossain, M.; Dimmock, J.R.; et al. Exploring CDKN1A Upregulation Mechanisms: Insights into Cell Cycle Arrest Induced by NC2603 Curcumin Analog in MCF-7 Breast Cancer Cells. Int. J. Mol. Sci. 2024, 25, 4989. [Google Scholar] [CrossRef]
- Nasif, D.; Laurito, S.; Real, S.; Branham, M.T. Exploring the epigenetic profile of ID4 in breast cancer: Bioinformatic insights into methylation patterns and chromatin accessibility dynamics. Breast Cancer Res. Treat. 2024, 207, 91–101. [Google Scholar] [CrossRef]
- Ni, J.; Xi, X.; Xiao, S.; Xiao, X. Silencing of circHIPK3 Sensitizes Paclitaxel-Resistant Breast Cancer Cells to Chemotherapy by Regulating HK2 Through Targeting miR-1286. Cancer Manag. Res. 2021, 13, 5573–5585. [Google Scholar] [CrossRef]
- Gao, X.; Qin, T.; Mao, J.; Zhang, J.; Fan, S.; Lu, Y.; Sun, Z.; Zhang, Q.; Song, B.; Li, L. PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 2019, 38, 256. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Silveira, D.A.; Lorenzoni, P.R.; Mombach, J.C.M.; Hashimoto, R.F. LncRNA PTENP1/miR-21/PTEN Axis Modulates EMT and Drug Resistance in Cancer: Dynamic Boolean Modeling for Cell Fates in DNA Damage Response. Int. J. Mol. Sci. 2024, 25, 8264. [Google Scholar] [CrossRef] [PubMed]
- Yndestad, S.; Austreid, E.; Skaftnesmo, K.O.; Lønning, P.E.; Eikesdal, H.P. Divergent Activity of the Pseudogene PTENP1 in ER-Positive and Negative Breast Cancer. Mol. Cancer Res. 2018, 16, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, Y.; Zheng, W.; Michailidou, K.; Ghoussaini, M.; Bolla, M.K.; Wang, Q.; Dennis, J.; Lush, M.; Milne, R.L.; et al. Fine-scale mapping of 8q24 locus identifies multiple independent risk variants for breast cancer. Int. J. Cancer 2016, 139, 1303–1317. [Google Scholar] [CrossRef]
- Silvestri, V.; Rizzolo, P.; Scarnò, M.; Chillemi, G.; Navazio, A.S.; Valentini, V.; Zelli, V.; Zanna, I.; Saieva, C.; Masala, G.; et al. Novel and known genetic variants for male breast cancer risk at 8q24.21, 9p21.3, 11q13.3 and 14q24.1: Results from a multicenter study in Italy. Eur. J. Cancer 2015, 51, 2289–2295. [Google Scholar] [CrossRef]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121. [Google Scholar] [CrossRef]
- Orafidiya, F.; Deng, L.; Bevan, C.L.; Fletcher, C.E. Crosstalk between Long Non Coding RNAs, microRNAs and DNA Damage Repair in Prostate Cancer: New Therapeutic Opportunities? Cancers 2022, 14, 755. [Google Scholar] [CrossRef] [PubMed]
- Alkhathami, A.G.; Hadi, A.; Alfaifi, M.; Alshahrani, M.Y.; Verma, A.K.; Beg, M.M.A. Serum-Based lncRNA ANRIL, TUG1, UCA1, and HIT Expressions in Breast Cancer Patients. Dis. Markers 2022, 2022, 9997212. [Google Scholar] [CrossRef]
- Khorshidi, H.R.; Noroozi, R.; Sarrafzadeh, S.; Sayad, A.; Taheri, M.; Ghafouri-Fard, S. ANRIL genetic variants in Iranian breast cancer patients. Cell J. 2017, 19 (Suppl. S1), 72–78. [Google Scholar] [CrossRef]
- Mehta-Mujoo, P.M.; Cunliffe, H.E.; Hung, N.A.; Slatter, T.L. Long Non-Coding RNA ANRIL in the Nucleus Associates with Periostin Expression in Breast Cancer. Front. Oncol. 2019, 9, 885. [Google Scholar] [CrossRef]
- Islam, S.S.; Al-Tweigeri, T.; Al-Harbi, L.; Ujjahan, S.; Al-Mozaini, M.; Tulbah, A.; Aboussekhra, A. Long noncoding RNA DLEU2 and ROR1 pathway induces epithelial-to-mesenchymal transition and cancer stem cells in breast cancer. Cell Death Discov. 2024, 10, 61. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, B.; Cai, Y.; Guo, C.; Liu, K.; Yuan, C. DLEU2: A Meaningful Long Noncoding RNA in Oncogenesis. Curr. Pharm. Des. 2021, 27, 2337–2343. [Google Scholar] [CrossRef]
- Awada, Z.; Bouaoun, L.; Nasr, R.; Tfayli, A.; Cuenin, C.; Akika, R.; Boustany, R.-M.; Makoukji, J.; Tamim, H.; Zgheib, N.K.; et al. LINE-1 methylation mediates the inverse association between body mass index and breast cancer risk: A pilot study in the Lebanese population. Environ. Res. 2021, 197, 111094. [Google Scholar] [CrossRef] [PubMed]
- Miret, N.; Zappia, C.D.; Altamirano, G.; Pontillo, C.; Zárate, L.; Gómez, A.; Lasagna, M.; Cocca, C.; Kass, L.; Monczor, F.; et al. AhR ligands reactivate LINE-1 retrotransposon in triple-negative breast cancer cells MDA-MB-231 and non-tumorigenic mammary epithelial cells NMuMG. Biochem. Pharmacol. 2020, 175, 113904. [Google Scholar] [CrossRef]
- Shi, X.; Si, X.; Zhang, E.; Zang, R.; Yang, N.; Cheng, H.; Zhang, Z.; Pan, B.; Sun, Y. Paclitaxel-induced stress granules increase LINE-1 mRNA stability to promote drug resistance in breast cancer cells. J. Biomed. Res. 2021, 35, 411–424. [Google Scholar] [CrossRef]
- Park, J.-W.; Rhee, J.-K. Integrative Analysis of ATAC-Seq and RNA-Seq Through Machine Learning Identifies 10 Signature Genes for Breast Cancer Intrinsic Subtypes. Biology 2024, 13, 799. [Google Scholar] [CrossRef]
- Hodgkinson, K.; Forrest, L.A.; Vuong, N.; Garson, K.; Djordjevic, B.; Vanderhyden, B.C. GREB1 is an estrogen receptor-regulated tumour promoter that is frequently expressed in ovarian cancer. Oncogene 2018, 37, 5873–5886. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Z.; Cenciarini, M.E.; Proietti, C.J.; Amasino, M.; Hong, T.; Yang, M.; Liao, Y.; Chiang, H.-C.; Kaklamani, V.G.; et al. Tamoxifen Resistance in Breast Cancer Is Regulated by the EZH2-ERα-GREB1 Transcriptional Axis. Cancer Res. 2018, 78, 671–684. [Google Scholar] [CrossRef]
- Ma, Q.; Yang, L.; Tolentino, K.; Wang, G.; Zhao, Y.; Litzenburger, U.M.; Shi, Q.; Zhu, L.; Yang, C.; Jiao, H.; et al. Inducible lncRNA transgenic mice reveal continual role of HOTAIR in promoting breast cancer metastasis. eLife 2022, 11, e79126. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Li, Y.; Zhou, Z.; Ma, M.; Fu, K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed. Pharmacother. 2017, 95, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Dustin, D.; Gu, G.; Fuqua, S.A. Mechanisms of Endocrine Resistance in Breast Cancer. Curr. Oncol. Rep. 2019, 21, 92. [Google Scholar]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Shimoi, T.; Yoshida, M.; Kitamura, Y.; Yoshino, T.; Kawachi, A.; Shimomura, A.; Noguchi, E.; Yunokawa, M.; Yonemori, K.; Shimizu, C.; et al. TERT promoter hotspot mutations in breast cancer. Breast Cancer 2017, 25, 292–296. [Google Scholar] [CrossRef]
- Grisanzio, C.; Freedman, M.L. Chromosome 8q24-Associated Cancers and MYC. Genes Cancer 2010, 1, 555–559. [Google Scholar] [CrossRef]
- Guo, J.; Wei, Z.; Jia, T.; Wang, L.; Nama, N.; Liang, J.; Liao, X.; Liu, X.; Gao, Y.; Liu, X.; et al. Dissecting transcription of the 8q24-MYC locus in prostate cancer recognizes the equilibration between androgen receptor direct and indirect dual-functions. J. Transl. Med. 2023, 21, 716. [Google Scholar] [CrossRef]
- Tuupanen, S.; Yan, J.; Turunen, M.; Gylfe, A.E.; Kaasinen, E.; Li, L.; Eng, C.; Culver, D.A.; Kalady, M.F.; Pennison, M.J.; et al. Characterization of the colorectal cancer–associated enhancer MYC-335 at 8q24: The role of rs67491583. Cancer Genet. 2012, 205, 25–33. [Google Scholar] [CrossRef]
- Chauhan, S.S.; Casillas, A.L.; Vizzerra, A.D.; Liou, H.; Clements, A.N.; Flores, C.E.; Prevost, C.T.; Kashatus, D.F.; Snider, A.J.; Snider, J.M.; et al. PIM1 drives lipid droplet accumulation to promote proliferation and survival in prostate cancer. Oncogene 2024, 43, 406–419. [Google Scholar]
- Chen, X.; Zhou, J.; Wang, Y.; Wang, X.; Chen, K.; Chen, Q.; Huang, D.; Jiang, R. PIM1/NF-κB/CCL2 blockade enhances anti-PD-1 therapy response by modulating macrophage infiltration and polarization in tumor microenvironment of NSCLC. Oncogene 2024, 43, 2517–2530. [Google Scholar] [CrossRef]
- Ko, R.; Seo, J.; Park, H.; Lee, N.; Lee, S.Y. Pim1 promotes IFN-β production by interacting with IRF3. Exp. Mol. Med. 2022, 54, 2092–2103. [Google Scholar] [CrossRef]
- Zhao, W.; Qiu, R.; Li, P.; Yang, J. PIM1: A promising target in patients with triple-negative breast cancer. Med. Oncol. 2017, 34, 142. [Google Scholar] [CrossRef]
- Al-Showimi, M.; Al-Yousef, N.; Alharbi, W.; Alkhezayem, S.; Almalik, O.; Alhusaini, H.; Alghamdi, A.; Al-Moghrabi, N. MicroRNA-126 expression in the peripheral white blood cells of patients with breast and ovarian cancer is a potential biomarker for the early prediction of cancer risk in the carriers of methylated BRCA1. Oncol. Lett. 2022, 24, 276. [Google Scholar] [CrossRef] [PubMed]
- Gambacurta, A.; Tullio, V.; Savini, I.; Mauriello, A.; Catani, M.V.; Gasperi, V. Identification of the EBF1/ETS2/KLF2-miR-126-Gene Feed-Forward Loop in Breast Carcinogenesis and Stemness. Int. J. Mol. Sci. 2025, 26, 328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, P.; Sun, T.; Li, D.; Xu, X.; Rui, Y.; Li, C.; Chong, M.; Ibrahim, T.; Mercatali, L.; et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013, 15, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Rangel, L.B.; Huang, T.H. Estrogen response in luminal breast cancer. Oncotarget 2013, 4, 1548–1549. [Google Scholar] [CrossRef]
- Jeffreys, S.A.; Powter, B.; Balakrishnar, B.; Mok, K.; Soon, P.; Franken, A.; Neubauer, H.; de Souza, P.; Becker, T.M. Endocrine Resistance in Breast Cancer: The Role of Estrogen Receptor Stability. Cells 2020, 9, 2077. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yu, C.-W. Epigenetic modulations in triple-negative breast cancer: Therapeutic implications for tumor microenvironment. Pharmacol. Res. 2024, 204, 107205. [Google Scholar] [CrossRef]
- Herzog, S.K.; Fuqua, S.A.W. ESR1 mutations and therapeutic resistance in metastatic breast cancer: Progress and remaining challenges. Br. J. Cancer 2021, 126, 174–186. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2020, 348, 56–61. [Google Scholar] [CrossRef]
- Schettini, F.; Prat, A. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast 2021, 59, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, W. Non-coding RNAs in breast cancer: Biological functions and therapeutic implications. Mol. Cancer 2021, 20, 24. [Google Scholar]
- Liu, Q.; Kulak, M.V.; Borcherding, N.; Maina, P.K.; Zhang, W.; Weigel, R.J.; Qi, H.H. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene 2017, 37, 687–694. [Google Scholar] [CrossRef]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Ding, Y.; Li, C.; Zhao, H.; Wang, J.; Meng, S. Posttranscriptional upregulation of HER3 by HER2 mRNA induces trastuzumab resistance in breast cancer. Mol. Cancer 2018, 17, 113. [Google Scholar] [CrossRef]
- Kumar, M.; Sahu, R.K.; Goyal, A.; Sharma, S.; Kaur, N.; Mehrotra, R.; Singh, U.R.; Hedau, S. BRCA1 Promoter Methylation and Expression—Associations with ER+, PR+ and HER2+ Subtypes of Breast Carcinoma. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 3293–3299. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Ortiz, A.; Sanchez-Muñoz, A.; Parrado, M.R.C.; Álvarez, M.; Ribelles, N.; Dominguez, A.R.; Alba, E. Deciphering HER2 Breast Cancer Disease: Biological and Clinical Implications. Front. Oncol. 2019, 9, 1124. [Google Scholar] [CrossRef]
- Blas, P.E.; Rodriguez, E.S.R.; Williams, H.L.; Levin, M.K.; Bell, J.S.K.; Pierobon, M.; Barrett, A.S.; Petricoin, E.F.; O’SHaughnessy, J.A. Targeting HER2/HER3 co-mutations in metastatic breast cancer: Case reports of exceptional responders to trastuzumab and pertuzumab therapy. Cancer Rep. 2024, 7, e1954. [Google Scholar] [CrossRef]
- Fang, H.; Xie, J.; Zhang, M.; Zhao, Z.; Wan, Y.; Yao, Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am. J. Transl. Res. 2017, 9, 953–961. [Google Scholar]
- Bonetti, P.; Climent, M.; Panebianco, F.; Tordonato, C.; Santoro, A.; Marzi, M.J.; Pelicci, P.G.; Ventura, A.; Nicassio, F. Dual role for miR-34a in the control of early progenitor proliferation and commitment in the mammary gland and in breast cancer. Oncogene 2019, 38, 360–374. [Google Scholar] [CrossRef]
- Cencioni, C.; Spallotta, F.; Savoia, M.; Kuenne, C.; Guenther, S.; Re, A.; Wingert, S.; Rehage, M.; Sürün, D.; Siragusa, M.; et al. Zeb1-Hdac2-eNOS circuitry identifies early cardiovascular precursors in naive mouse embryonic stem cells. Nat. Commun. 2018, 9, 1281. [Google Scholar] [CrossRef]
- Feng, Z.-M.; Qiu, J.; Chen, X.-W.; Liao, R.-X.; Liao, X.-Y.; Zhang, L.-P.; Chen, X.; Li, Y.; Chen, Z.-T.; Sun, J.-G. Essential role of miR-200c in regulating self-renewal of breast cancer stem cells and their counterparts of mammary epithelium. BMC Cancer 2015, 15, 645. [Google Scholar] [CrossRef] [PubMed]
- Baylie, T.; Kasaw, M.; Getinet, M.; Getie, G.; Jemal, M.; Nigatu, A.; Ahmed, H.; Bogale, M. The role of miRNAs as biomarkers in breast cancer. Front. Oncol. 2024, 14, 1374821. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-T.; Zhang, L.-L.; Li, F.-B.; Zhang, J.; Zhang, Z.-B.; Mi, D.-Z.; Sun, J.; Zhang, H.-Y.; Wang, C.-Y.; Chen, Y.-H.; et al. LN-439A, a novel BAP1 inhibitor, suppresses the growth of basal-like breast cancer by degrading KLF5. Acta Pharmacol. Sin. 2025, 46, 715–727. [Google Scholar] [CrossRef]
- Zhang, J.; Miao, N.; Lao, L.; Deng, W.; Wang, J.; Zhu, X.; Huang, Y.; Lin, H.; Zeng, W.; Zhang, W.; et al. Activation of Bivalent Gene POU4F1 Promotes and Maintains Basal-like Breast Cancer. Adv. Sci. 2024, 11, e2307660. [Google Scholar] [CrossRef]
- El-Fattah, A.A.A.; Sadik, N.A.H.; Shaker, O.G.; Mohamed Kamal, A.; Shahin, N.N. Serum Long Non-Coding RNAs PVT1, HOTAIR, and NEAT1 as Potential Biomarkers in Egyptian Women with Breast Cancer. Biomolecules 2021, 11, 301. [Google Scholar] [CrossRef]
- Patel, N.A.; Lui, A.; Trujillo, A.N.; Motawe, Z.Y.; Bader, D.; Schuster, J.; Burgess, A.; Alves, N.G.; Jo, M.; Breslin, J.W. Female and male obese Zucker rats display differential inflammatory mediator and long non-coding RNA profiles. Life Sci. 2023, 335, 122285. [Google Scholar] [CrossRef]
- Kim, N.H.; Song, S.H.; Choi, Y.H.; Hwang, K.H.; Yun, J.S.; Song, H.; Cha, S.Y.; Cho, S.B.; Lee, I.; Kim, H.S.; et al. Competing Endogenous RNA of Snail and Zeb1 UTR in Therapeutic Resistance of Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 9589. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Z.; Hu, L. The regulatory effects of metformin on the [SNAIL/miR-34]:[ZEB/miR-200] system in the epithelial-mesenchymal transition (EMT) for colorectal cancer (CRC). Eur. J. Pharmacol. 2018, 834, 45–53. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2019, 14, 622–628. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, C.; Xiong, H.; Shen, Y.; Lu, Y.; Zhou, J.; Wang, L. Dysregulation of long non-coding RNA in breast cancer: An overview of mechanism and clinical implication. Oncotarget 2016, 8, 5508–5522. [Google Scholar] [CrossRef] [PubMed]
- Kerr, A.J.; Dodwell, D.; McGale, P.; Holt, F.; Duane, F.; Mannu, G.; Darby, S.C.; Taylor, C.W. Adjuvant and neoadjuvant breast cancer treatments: A systematic review of their effects on mortality. Cancer Treat. Rev. 2022, 105, 102375. [Google Scholar] [CrossRef]
- Tripathy, D.; Im, S.-A.; Colleoni, M.; Franke, F.; Bardia, A.; Harbeck, N.; Hurvitz, S.A.; Chow, L.; Sohn, J.; Lee, K.S.; et al. Ribociclib plus endocrine therapy for premenopausal women with hormone-receptor-positive, advanced breast cancer (MONALEESA-7): A randomised phase 3 trial. Lancet Oncol. 2018, 19, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jin, H.; Zhou, Y. ESR1 mutations and their roles in endocrine therapy resistance in hormone receptor-positive breast cancer. Explor. Target. Anti-Tumor Ther. 2024, 5, 100284. [Google Scholar]
- Baselga, J.; Hortobagyi, G.N. Targeting the PI3K pathway in breast cancer: Rationale and advances. In American Society of Clinical Oncology Educational Book; ASCO: Alexandria, VA, USA, 2013; Volume 33, pp. e37–e47. [Google Scholar]
- Nikita, N.; Sun, Z.; Sharma, S.; Shaver, A.; Seewaldt, V.; Lu-Yao, G. Epigenetic Landscapes of Aging in Breast Cancer Survivors: Unraveling the Impact of Therapeutic Interventions—A Scoping Review. Cancers 2025, 17, 866. [Google Scholar] [CrossRef]
- Li, Y.; Wang, R.; Tan, X. Breast cancer stem cells: Emerging mechanisms of resistance and therapeutic strategies. Cancers 2022, 14, 5206. [Google Scholar]
- Saatci, O.; Huynh-Dam, K.-T.; Sahin, O. Endocrine resistance in breast cancer: From molecular mechanisms to therapeutic strategies. J. Mol. Med. 2021, 99, 1691–1710. [Google Scholar] [CrossRef]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.-W.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016, 35, 2746–2755. [Google Scholar] [CrossRef]
- Tufail, M. The MALAT1-breast cancer interplay: Insights and implications. Expert Rev. Mol. Diagn. 2023, 23, 665–678. [Google Scholar] [CrossRef]
- Ocaña, A.; Nieto-Jiménez, C.; Pandiella, A. BET inhibitors as novel therapeutic agents in breast cancer. Oncotarget 2017, 8, 71285–71291. [Google Scholar] [CrossRef]
- González-Woge, M.; Contreras-Espinosa, L.; García-Gordillo, J.A.; Aguilar-Villanueva, S.; Bargallo-Rocha, E.; Cabrera-Galeana, P.; Vasquez-Mata, T.; Cervantes-López, X.; Vargas-Lías, D.S.; Montiel-Manríquez, R.; et al. The Expression Profiles of lncRNAs Are Associated with Neoadjuvant Chemotherapy Resistance in Locally Advanced, Luminal B-Type Breast Cancer. Int. J. Mol. Sci. 2024, 25, 8077. [Google Scholar] [CrossRef]
- Horie, K.; Takagi, K.; Takeiwa, T.; Mitobe, Y.; Kawabata, H.; Suzuki, T.; Ikeda, K.; Inoue, S. Estrogen-Inducible LncRNA BNAT1 Functions as a Modulator for Estrogen Receptor Signaling in Endocrine-Resistant Breast Cancer Cells. Cells 2022, 11, 3610. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Olvera, S.I.; Aguilar-Arnal, L.; Cisneros-Villanueva, M.; Hidalgo-Miranda, A.; Marchat, L.A.; Salinas-Vera, Y.M.; Ramos-Payán, R.; Pérez-Plasencia, C.; Carlos-Reyes, Á.; Puente-Rivera, J.; et al. Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment. Cells 2022, 11, 3458. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.; Avniel-Polak, S.; Abu-Kamel, S.; Antman, I.; Saadoun, T.; Brim, C.; Jumaa, M.; Maron, Y.; Maimon, O.; Bel-Ange, A.; et al. Enhancer landscape of lung neuroendocrine tumors reveals regulatory and developmental signatures with potential theranostic implications. Proc. Natl. Acad. Sci. USA 2024, 121, e2405001121. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Willi, M.; Shin, H.Y.; Liu, C.; Hennighausen, L. Progressing super-enhancer landscape during mammary differentiation controls tissue-specific gene regulation. Nucleic Acids Res. 2018, 46, 10796–10809. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, Z.; Zhang, J.; Hao, Z.; He, Y.; Wu, Z.; Song, Y.; Yuan, K.; Zheng, S.; Zhao, Q.; et al. lncRNA MALAT1 participates in metformin inhibiting the proliferation of breast cancer cell. J. Cell. Mol. Med. 2021, 25, 7135–7145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, X.; Wang, X.; Xie, Y.; Wang, Z.; Xu, Y.; You, X.; Liang, Z.; Cao, H. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res. Treat. 2015, 152, 199–208. [Google Scholar] [CrossRef] [PubMed]

| Non-Coding Region | Mutation Type | Associated Gene/Pathway | Functional Impact | BC Subtype | Clinical Relevance | References |
|---|---|---|---|---|---|---|
| Enhancer | SNP | MYC | Increased MYC expression | Triple-negative | Potential drug target | [124,125] |
| Promoter | Deletion | TP53 | Reduced gene expression | Luminal | Diagnostic biomarker | [23,126,127] |
| lncRNA | Overexpression | MALAT1 | Promotes metastasis | HER2+ | Prognostic indicator | [23,128,129] |
| miRNA | Mutation | miR-21 | Disrupted mRNA regulation | All subtypes | Therapeutic target (anti-miR) | [130,131,132] |
| Intronic Region | Insertion | BRCA1 | Aberrant splicing | Hereditary cases | Risk prediction | [133,134] |
| 3′ UTR | SNP | CDKN1A | Alters miRNA binding | Triple-negative | Regulatory variant affecting cell cycle | [135,136] |
| 5′ UTR | Methylation | FOXA1 | Transcriptional repression | Luminal A | Epigenetic marker of prognosis | [137] |
| Circular RNA | Overexpression | circHIPK3 | Sponges tumor suppressor miRNAs | TNBC | Modulates chemoresistance | [138] |
| Pseudogene | Amplification | PTENP1 | Competes for miRNA binding | HER2- | Regulates tumor suppressor PTEN | [139,140,141] |
| Intergenic | SNP | 8q24 locus | Modulates enhancer–promoter looping | ER+ | Associated with familial risk | [142,143] |
| Ultraconserved Region | SNP | TUC338 | Enhances oncogenic signaling | TNBC | Putative diagnostic biomarker | [144,145] |
| Antisense RNA | Overexpression | ANRIL | Silences CDKN2A/B via PRC2 recruitment | Luminal B | Epigenetic driver of proliferation | [146,147,148] |
| Transcribed Spacer | Insertion | DLEU2 | Affects miRNA cluster stability | HER2+ | miRNA network regulator | [149,150] |
| Repeat Element | Expansion | LINE-1 | Induces genomic instability | Basal-like | Potential target for genome stabilization | [151,152,153] |
| Bidirectional Promoter | Deletion | ESR1/GREB1 | Alters co-regulated gene expression | ER+ | Estrogen response marker | [154,155,156] |
| Non-Coding Mutation | Location | Nucleotide Change | Pathogenicity/Significance | Reference |
|---|---|---|---|---|
| TERT promoter | Chromosome 5p15.33 | C228T or C250T | The mutations create a new binding site for ETS/TCF transcription factors, leading to the overexpression of telomerase, a key hallmark of cancer, which promotes unlimited cell proliferation. While rare in breast cancer, these mutations are considered pathogenic. | [161] |
| MYC distal enhancer | Chromosome 8q24 | Point mutations and SNPs (e.g., rs13281615) | Mutations in this region, which is ~2 Mb from the MYC gene, can increase the risk of breast cancer. They are believed to alter the binding of transcription factors, leading to the deregulation and overexpression of the MYC oncogene. | [162,163,164] |
| PIM1 super-enhancer | Chromosome 6p21.2 | Various mutations | Mutations within the super-enhancer of the PIM1 oncogene, particularly in luminal breast cancer, are linked to increased PIM1 expression. This is often associated with a more aggressive phenotype and poorer prognosis. | [165,166,167,168] |
| miR-126 promoter | Chromosome 3p21 | DNA methylation | While not a direct mutation, hypermethylation of the miR-126 promoter silences its expression. Since miR-126 acts as a tumor suppressor, its loss promotes metastasis and is associated with poor prognosis in breast cancer patients. | [169,170,171] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabit, H.; Sobhy, S.; Abdel-Ghany, S.; Wadan, A.-H.S.; Ayodele, O.; Albrahim, Y.; Banerjee, H.N.; Elhashash, A.; Arneth, B. The Role of Non-Coding Regions in Breast Cancer: From Gene Regulation to Therapeutic Implications. Pharmaceuticals 2025, 18, 1370. https://doi.org/10.3390/ph18091370
Sabit H, Sobhy S, Abdel-Ghany S, Wadan A-HS, Ayodele O, Albrahim Y, Banerjee HN, Elhashash A, Arneth B. The Role of Non-Coding Regions in Breast Cancer: From Gene Regulation to Therapeutic Implications. Pharmaceuticals. 2025; 18(9):1370. https://doi.org/10.3390/ph18091370
Chicago/Turabian StyleSabit, Hussein, Sara Sobhy, Shaimaa Abdel-Ghany, Al-Hassan Soliman Wadan, Olubukola Ayodele, Yasser Albrahim, Hirendra N. Banerjee, Ahmed Elhashash, and Borros Arneth. 2025. "The Role of Non-Coding Regions in Breast Cancer: From Gene Regulation to Therapeutic Implications" Pharmaceuticals 18, no. 9: 1370. https://doi.org/10.3390/ph18091370
APA StyleSabit, H., Sobhy, S., Abdel-Ghany, S., Wadan, A.-H. S., Ayodele, O., Albrahim, Y., Banerjee, H. N., Elhashash, A., & Arneth, B. (2025). The Role of Non-Coding Regions in Breast Cancer: From Gene Regulation to Therapeutic Implications. Pharmaceuticals, 18(9), 1370. https://doi.org/10.3390/ph18091370









