Epigenetic Alterations in Hepatocellular Carcinoma: Mechanisms, Biomarkers, and Therapeutic Implications
Abstract
1. Introduction
2. Epigenetics
2.1. Epigenetic Modifiers
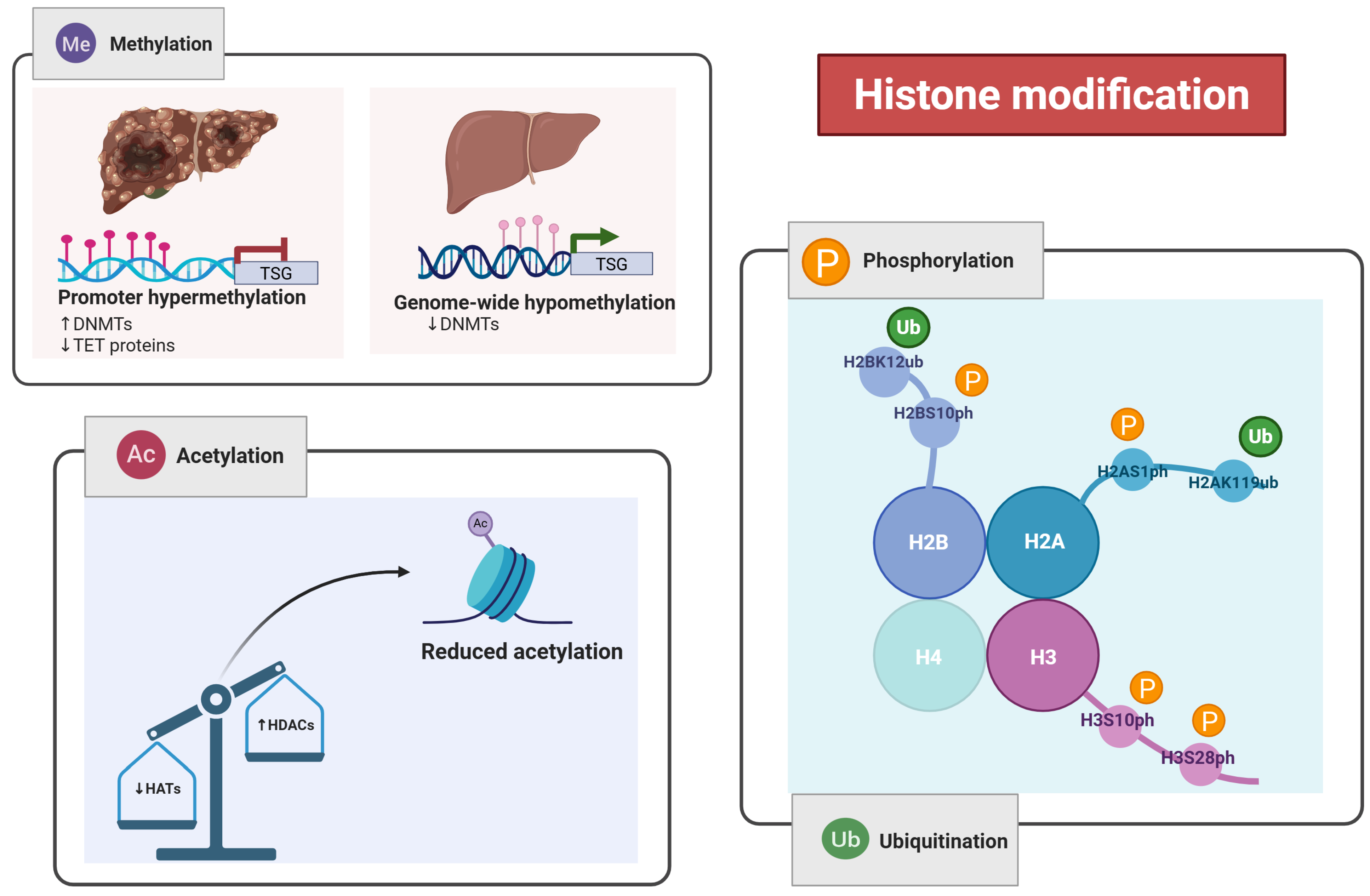
2.1.1. Histone Modification
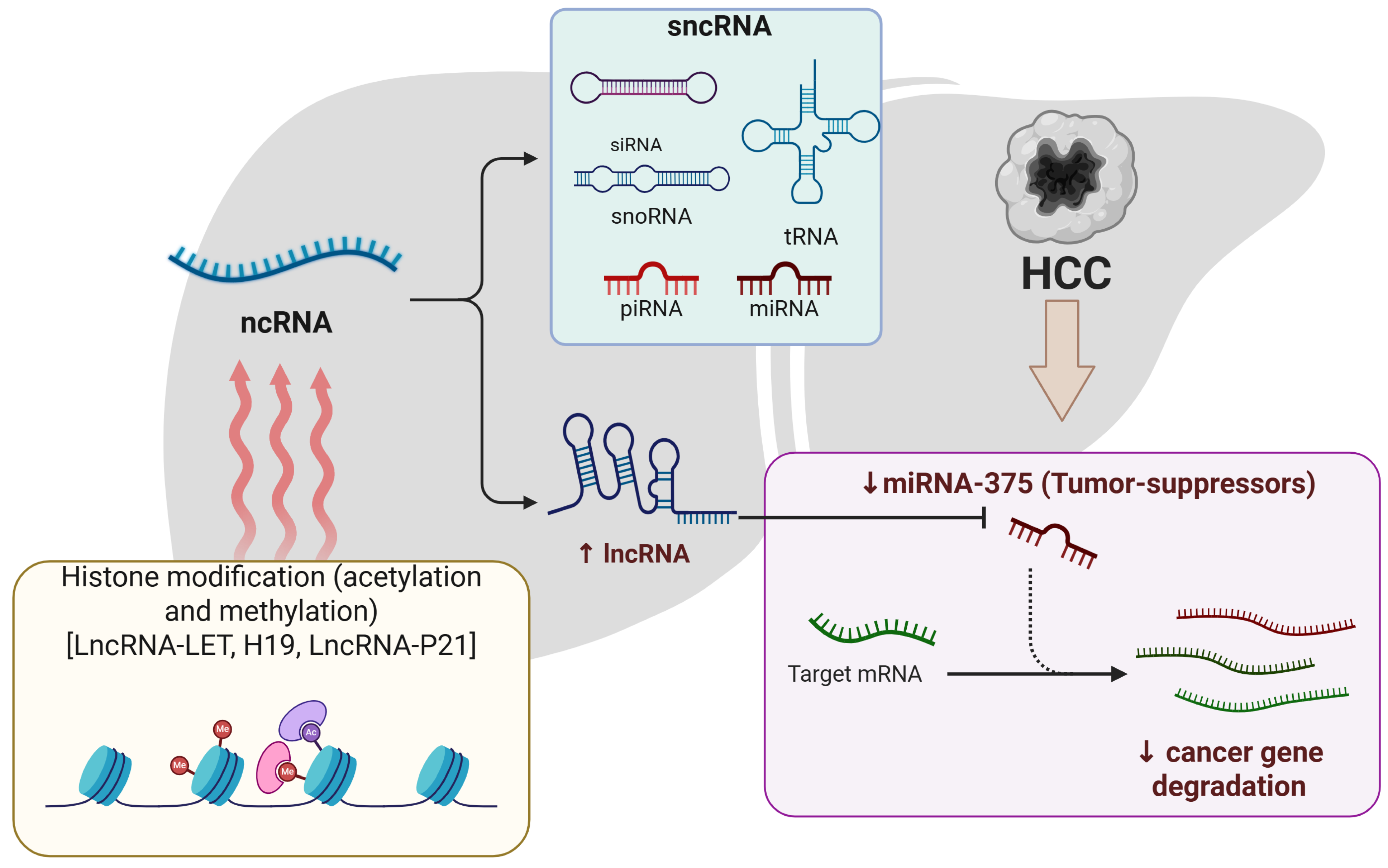
2.1.2. Epigenetic Association of ncRNA in HCC
2.1.3. DNA Methylation
2.1.4. mRNA Methylation
2.2. Bioinformatics Tools and Databases for Epigenetic Target Discovery
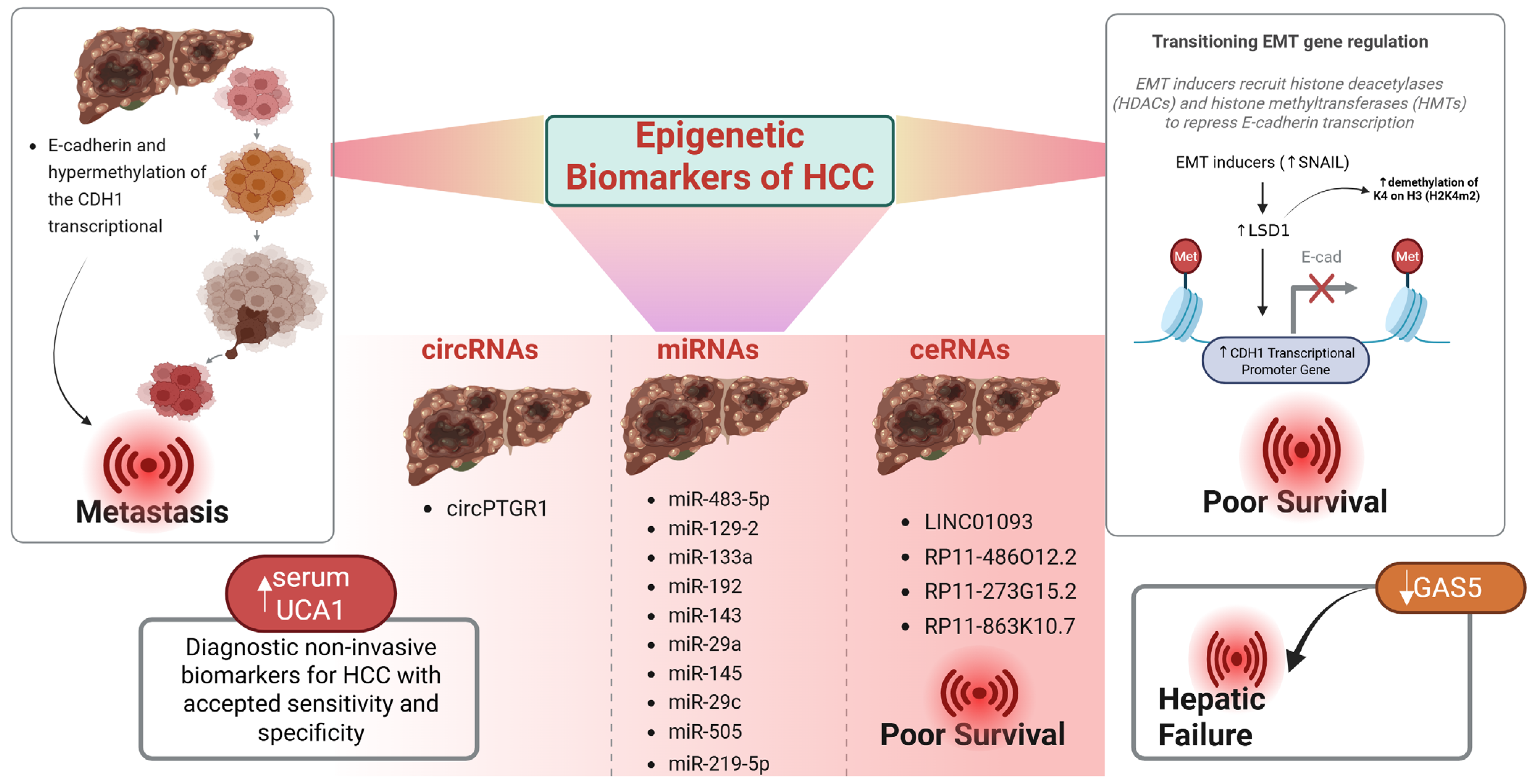
3. Epigenetic Biomarkers
miRNA Biomarkers in Diagnosis and Prognosis
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Guo, Q.; Zhu, X.; Beeraka, N.M.; Zhao, R.; Li, S.; Li, F.; Mahesh, P.A.; Nikolenko, V.N.; Fan, R.; Liu, J. Projected epidemiological trends and burden of liver cancer by 2040 based on GBD, CI5plus, and WHO data. Sci. Rep. 2024, 14, 28131. [Google Scholar] [CrossRef]
- Hussain, M.S.; Moglad, E.; Afzal, M.; Gupta, G.; Hassan Almalki, W.; Kazmi, I.; Alzarea, S.I.; Kukreti, N.; Gupta, S.; Kumar, D.; et al. Non-coding RNA mediated regulation of PI3K/Akt pathway in hepatocellular carcinoma: Therapeutic perspectives. Pathol. Res. Pract. 2024, 258, 155303. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Wang, X.; Zhang, W.; Chen, Y.; Lu, X.; Jin, C.; Tu, L.; Jiang, T.; Yang, Y.; et al. Precision Strike Strategy for Liver Diseases Trilogy with Xiao-Chai-Hu Decoction: A Meta-Analysis with Machine Learning. Phytomedicine 2025, 142, 156796. [Google Scholar] [CrossRef]
- Kontomanolis, E.N.; Koutras, A.; Syllaios, A.; Schizas, D.; Mastoraki, A.; Garmpis, N.; Diakosavvas, M.; Angelou, K.; Tsatsaris, G.; Pagkalos, A.; et al. Role of oncogenes and tumor-suppressor genes in carcinogenesis: A review. Anticancer Res. 2020, 40, 6009–6015. [Google Scholar] [CrossRef]
- Wang, J.; Tao, X.; Liu, Z.; Yan, Y.; Cheng, P.; Liu, B.; Du, H.; Niu, B. Noncoding RNAs in sepsis-associated acute liver injury: Roles, mechanisms, and therapeutic applications. Pharmacol. Res. 2025, 212, 107596. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, K.; Wang, M.; He, Z.; Yu, B.; Wang, X.; Pan, X.; Luo, Y.; Xu, S.; Lau, J.T.Y.; et al. VEGF-FGF Signaling Activates Quiescent CD63+ Liver Stem Cells to Proliferate and Differentiate. Adv. Sci. 2024, 11, e2308711. [Google Scholar] [CrossRef]
- Wu, S.; Powers, S.; Zhu, W.; Hannun, Y.A. Substantial contribution of extrinsic risk factors to cancer development. Nature 2016, 529, 43–47. [Google Scholar] [CrossRef]
- Islam, M.R.; Rauf, A.; Alash, S.; Fakir, M.N.H.; Thufa, G.K.; Sowa, M.S.; Mukherjee, D.; Kumar, H.; Hussain, M.S.; Aljohani, A.S.M.; et al. A comprehensive review of phytoconstituents in liver cancer prevention and treatment: Targeting insights into molecular signaling pathways. Med. Oncol. 2024, 41, 134. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xu, D.; Cai, Y.; Zhu, L.; Xiao, Q.; Peng, W.; Chen, B. A multi-omic analysis reveals that Gamabufotalin exerts anti-hepatocellular carcinoma effects by regulating amino acid metabolism through targeting STAMBPL1. Phytomedicine 2024, 135, 156094. [Google Scholar] [CrossRef]
- Riddihough, G.; Zahn, L.M. What is epigenetics? Science 2010, 330, 611. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.H.; Mohi-ud-din, R.; Wani, T.U.; Dar, M.O.; Shah, A.J.; Lone, B.; Pooja, C.; Masoodi, M.H. Indole: A privileged heterocyclic moiety in the management of cancer. Curr. Org. Chem. 2021, 25, 724–736. [Google Scholar]
- Chang, H.; Wang, D.; Xia, W.; Pan, X.; Huo, W.; Xu, S.; Li, Y. Epigenetic disruption and glucose homeostasis changes following low-dose maternal bisphenol A exposure. Toxicol. Res. 2016, 5, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Mattiuzzi, C.; Lippi, G. Current cancer epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef]
- Hassan, R.; Mohi-Ud-Din, R.; Dar, M.O.; Shah, A.J.; Mir, P.A.; Shaikh, M.; Pottoo, F.H. Bioactive heterocyclic compounds as potential therapeutics in the treatment of gliomas: A review. Anti-Cancer Agents Med. Chem. 2022, 22, 551–565. [Google Scholar] [CrossRef]
- Mir, R.H.; Mir, P.A.; Uppal, J.; Chawla, A.; Patel, M.; Bardakci, F.; Adnan, M.; Mohi-Ud-Din, R. Evolution of Natural Product Scaffolds as Potential Proteasome Inhibitors in Developing Cancer Therapeutics. Metabolites 2023, 13, 509. [Google Scholar] [CrossRef]
- Chen, F.; Wang, Z.; Yao, H.; Liu, Q.; Gan, Y.; Xu, S.; Bao, H.; Jin, Y.; Hu, Y.P.; Gao, J.; et al. Large-scale manufacturing of human gallbladder epithelial cell products and derived hepatocytes via a chemically defined approach. Trends Biotechnol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Perisetti, A.; Goyal, H.; Yendala, R.; Thandassery, R.B.; Giorgakis, E. Non-cirrhotic hepatocellular carcinoma in chronic viral hepatitis: Current insights and advancements. World J. Gastroenterol. 2021, 27, 3466–3482. [Google Scholar] [CrossRef]
- Budny, A.; Kozłowski, P.; Kamińska, M.; Jankiewicz, M.; Kolak, A.; Budny, B.; Budny, W.; Niemunis-Sawicka, J.; Szczypiór, G.; Kurniawka, B.; et al. Epidemiology and risk factors of hepatocellular carcinoma. Pol. Merkur. Lek. Organ Pol. Tow. Lek. 2017, 43, 133–139. [Google Scholar]
- Pogribny, I.P.; Rusyn, I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014, 342, 223–230. [Google Scholar] [CrossRef] [PubMed]
- El–Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Mohi-Ud-Din, R.; Mir, R.H.; Sawhney, G.; Dar, M.A.; Bhat, Z.A. Possible pathways of hepatotoxicity caused by chemical agents. Curr. Drug Metab. 2019, 20, 867–879. [Google Scholar] [CrossRef]
- Wallace, M.C.; Preen, D.; Jeffrey, G.P.; Adams, L.A. The evolving epidemiology of hepatocellular carcinoma: A global perspective. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chua, M.; Andrisani, O.; So, S. Epigenetics in hepatocellular carcinoma: An update and future therapy perspectives. World J. Gastroenterol. 2014, 20, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Bayo, J.; Fiore, E.J.; Dominguez, L.M.; Real, A.; Malvicini, M.; Rizzo, M.; Atorrasagasti, C.; García, M.G.; Argemi, J.; Martinez, E.D.; et al. A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J. Hepatol. 2019, 71, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yu, W. Advances in Immune Checkpoint Therapy in Hepatocellular Carcinoma. Br. J. Hosp. Med. 2024, 85, 1–21. [Google Scholar] [CrossRef]
- Liu, W.-R.; Shi, Y.-H.; Peng, Y.-f.; Fan, J. Epigenetics of hepatocellular carcinoma: A new horizon. Chin. Med. J. 2012, 125, 2349–2360. [Google Scholar]
- Yang, H.; Zhou, H.; Fu, M.; Xu, H.; Huang, H.; Zhong, M.; Zhang, M.; Hua, W.; Lv, K.; Zhu, G. TMEM64 aggravates the malignant phenotype of glioma by activating the Wnt/β-catenin signaling pathway. Int. J. Biol. Macromol. 2024, 260, 129332. [Google Scholar] [CrossRef]
- Anway, M.D.; Leathers, C.; Skinner, M.K. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 2006, 147, 5515–5523. [Google Scholar] [CrossRef]
- Weaver, I.C.; Champagne, F.A.; Brown, S.E.; Dymov, S.; Sharma, S.; Meaney, M.J.; Szyf, M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: Altering epigenetic marking later in life. J. Neurosci. 2005, 25, 11045–11054. [Google Scholar] [CrossRef]
- Waddington, C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012, 41, 10–13. [Google Scholar] [CrossRef]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Nanney, D.L. Epigenetic Control Systems. Proc. Natl. Acad. Sci. USA 1958, 44, 712–717. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.; Qi, L. Genetic and epigenetic control of gene expression by CRISPR-Cas systems. F1000Research 2017, 6, 747. [Google Scholar] [CrossRef]
- Dupras, C.; Saulnier, K.M.; Joly, Y. Epigenetics, ethics, law and society: A multidisciplinary review of descriptive, instrumental, dialectical and reflexive analyses. Soc. Stud. Sci. 2019, 49, 785–810. [Google Scholar] [CrossRef]
- Fallet, M.; Blanc, M.; Di Criscio, M.; Antczak, P.; Engwall, M.; Guerrero Bosagna, C.; Rüegg, J.; Keiter, S.H. Present and future challenges for the investigation of transgenerational epigenetic inheritance. Environ. Int. 2023, 172, 107776. [Google Scholar] [CrossRef]
- Prasher, D.; Greenway, S.C.; Singh, R.B. The impact of epigenetics on cardiovascular disease. Biochem. Cell Biol. 2020, 98, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. Canalization of development and genetic assimilation of acquired characters. Nature 1959, 183, 1654–1655. [Google Scholar] [CrossRef]
- Grewal, S.I.; Moazed, D. Heterochromatin and epigenetic control of gene expression. Science 2003, 301, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Abdul, Q.A.; Yu, B.P.; Chung, H.Y.; Jung, H.A.; Choi, J.S. Epigenetic modifications of gene expression by lifestyle and environment. Arch. Pharmacal Res. 2017, 40, 1219–1237. [Google Scholar] [CrossRef]
- Mathers, J.C.; Strathdee, G.; Relton, C.L. Induction of epigenetic alterations by dietary and other environmental factors. Adv. Genet. 2010, 71, 3–39. [Google Scholar] [PubMed]
- Burwell, R.G.; Dangerfield, P.H.; Moulton, A.; Grivas, T.B. Adolescent idiopathic scoliosis (AIS), environment, exposome and epigenetics: A molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Worm, J.; Guldberg, P. DNA methylation: An epigenetic pathway to cancer and a promising target for anticancer therapy. J. Oral Pathol. Med. 2002, 31, 443–449. [Google Scholar] [CrossRef]
- Li, Y.; Tollefsbol, T.O. DNA methylation detection: Bisulfite genomic sequencing analysis. Methods Mol. Biol. 2011, 791, 11–21. [Google Scholar]
- Brunmeir, R.; Lagger, S.; Simboeck, E.; Sawicka, A.; Egger, G.; Hagelkruys, A.; Zhang, Y.; Matthias, P.; Miller, W.J.; Seiser, C.; et al. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 2010, 6, e1000927. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 135. [Google Scholar] [CrossRef]
- Filtz, T.M.; Vogel, W.K.; Leid, M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 2014, 35, 76–85. [Google Scholar] [CrossRef]
- Mehnert, J.M.; Kelly, W.K. Histone deacetylase inhibitors: Biology and mechanism of action. Cancer J. 2007, 13, 23–29. [Google Scholar] [CrossRef]
- Legube, G.; Trouche, D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003, 4, 944–947. [Google Scholar] [CrossRef]
- Yang, J.J.; Tao, H.; Deng, Z.Y.; Lu, C.; Li, J. Non-coding RNA-mediated epigenetic regulation of liver fibrosis. Metab. Clin. Exp. 2015, 64, 1386–1394. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Muntean, A.G.; Hess, J.L. Epigenetic dysregulation in cancer. Am. J. Pathol. 2009, 175, 1353–1361. [Google Scholar] [CrossRef]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Gräff, J.; Mansuy, I.M. Epigenetic dysregulation in cognitive disorders. Eur. J. Neurosci. 2009, 30, 1–8. [Google Scholar] [CrossRef]
- Kwon, M.J.; Kim, S.; Han, M.H.; Lee, S.B. Epigenetic Changes in Neurodegenerative Diseases. Mol. Cells 2016, 39, 783–789. [Google Scholar] [CrossRef]
- Mohd Murshid, N.; Aminullah Lubis, F.; Makpol, S. Epigenetic Changes and Its Intervention in Age-Related Neurodegenerative Diseases. Cell. Mol. Neurobiol. 2022, 42, 577–595. [Google Scholar] [CrossRef]
- Duthie, S.J. Epigenetic modifications and human pathologies: Cancer and CVD. Proc. Nutr. Soc. 2011, 70, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.B.; Occean, J.R.; Sen, P. Epigenetic dysregulation in cardiovascular aging and disease. J. Cardiovasc. Aging 2021, 1, 10. [Google Scholar] [CrossRef]
- Zhang, W.; Song, M.; Qu, J.; Liu, G.H. Epigenetic Modifications in Cardiovascular Aging and Diseases. Circ. Res. 2018, 123, 773–786. [Google Scholar] [CrossRef]
- Flashner, B.M.; Russo, M.E.; Boileau, J.E.; Leong, D.W.; Gallicano, G.I. Epigenetic factors and autism spectrum disorders. Neuromol. Med. 2013, 15, 339–350. [Google Scholar] [CrossRef]
- Larizza, L.; Finelli, P. Developmental disorders with intellectual disability driven by chromatin dysregulation: Clinical overlaps and molecular mechanisms. Clin. Genet. 2019, 95, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, E.; Boardman, J.P.; Drake, A.J. Preterm Birth and the Risk of Neurodevelopmental Disorders—Is There a Role for Epigenetic Dysregulation? Curr. Genom. 2018, 19, 507–521. [Google Scholar] [CrossRef]
- Alaskhar Alhamwe, B.; Khalaila, R.; Wolf, J.; von Bülow, V.; Harb, H.; Alhamdan, F.; Hii, C.S.; Prescott, S.L.; Ferrante, A.; Renz, H.; et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. 2018, 14, 39. [Google Scholar] [CrossRef]
- Healy, S.; Khan, P.; He, S.; Davie, J.R. Histone H3 phosphorylation, immediate-early gene expression, and the nucleosomal response: A historical perspective. Biochem. Cell Biol. 2012, 90, 39–54. [Google Scholar] [CrossRef]
- Sawicka, A.; Seiser, C. Histone H3 phosphorylation—A versatile chromatin modification for different occasions. Biochimie 2012, 94, 2193–2201. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Swygert, S.G.; Peterson, C.L. Chromatin dynamics: Interplay between remodeling enzymes and histone modifications. Biochim. Biophys. Acta 2014, 1839, 728–736. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. In Histone Mutations and Cancer; Book Series Advances in Experimental Medicine and Biology; Springer: Singapore, 2021; Volume 1283, pp. 1–16. [Google Scholar] [CrossRef]
- Kurdistani, S.K.; Tavazoie, S.; Grunstein, M. Mapping global histone acetylation patterns to gene expression. Cell 2004, 117, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Clayton, A.L.; Hebbes, T.R.; Thorne, A.W.; Crane-Robinson, C. Histone acetylation and gene induction in human cells. FEBS Lett. 1993, 336, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Pogo, B.G.; Allfrey, V.G.; Mirsky, A.E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl. Acad. Sci. USA 1966, 55, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef]
- Wang, Y.; Miao, X.; Liu, Y.; Li, F.; Liu, Q.; Sun, J.; Cai, L. Dysregulation of histone acetyltransferases and deacetylases in cardiovascular diseases. Oxid. Med. Cell. Longev. 2014, 2014, 641979. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Murray, K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry 1964, 3, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef]
- Magistri, M.; Faghihi, M.A.; St Laurent, G., III; Wahlestedt, C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012, 28, 389–396. [Google Scholar] [CrossRef]
- Nakamura, M.; Chiba, T.; Kanayama, K.; Kanzaki, H.; Saito, T.; Kusakabe, Y.; Kato, N. Epigenetic dysregulation in hepatocellular carcinoma: An up-to-date review. Hepatol. Res. 2019, 49, 3–13. [Google Scholar] [CrossRef]
- North, J.A.; Šimon, M.; Ferdinand, M.B.; Shoffner, M.A.; Picking, J.W.; Howard, C.J.; Mooney, A.M.; van Noort, J.; Poirier, M.G.; Ottesen, J.J. Histone H3 phosphorylation near the nucleosome dyad alters chromatin structure. Nucleic Acids Res. 2014, 42, 4922–4933. [Google Scholar] [CrossRef]
- Cohen, P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 1989, 58, 453–508. [Google Scholar] [CrossRef] [PubMed]
- Bollen, M.; Stalmans, W. The structure, role, and regulation of type 1 protein phosphatases. Crit. Rev. Biochem. Mol. Biol. 1992, 27, 227–281. [Google Scholar] [CrossRef]
- Ivaldi, M.S.; Karam, C.S.; Corces, V.G. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 2007, 21, 2818–2831. [Google Scholar] [CrossRef]
- Mahmoud, S.A.; Poizat, C. Epigenetics and chromatin remodeling in adult cardiomyopathy. J. Pathol. 2013, 231, 147–157. [Google Scholar] [CrossRef]
- Josefowicz, S.Z.; Shimada, M.; Armache, A.; Li, C.H.; Miller, R.M.; Lin, S.; Yang, A.; Dill, B.D.; Molina, H.; Park, H.S.; et al. Chromatin Kinases Act on Transcription Factors and Histone Tails in Regulation of Inducible Transcription. Mol. Cell 2016, 64, 347–361. [Google Scholar] [CrossRef]
- Ivanovska, I.; Orr-Weaver, T.L. Histone modifications and the chromatin scaffold for meiotic chromosome architecture. Cell Cycle 2006, 5, 2064–2071. [Google Scholar] [CrossRef] [PubMed]
- Saul, D.; Kosinsky, R.L. Epigenetics of Aging and Aging-Associated Diseases. Int. J. Mol. Sci. 2021, 22, 401. [Google Scholar] [CrossRef]
- Qin, J.; Wen, B.; Liang, Y.; Yu, W.; Li, H. Histone Modifications and their Role in Colorectal Cancer (Review). Pathol. Oncol. Res. POR 2020, 26, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003, 17, 2733–2740. [Google Scholar] [CrossRef]
- Ai, H.; Peng, S.; Li, J.B. Chemical methods for studying the crosstalk between histone H2B ubiquitylation and H3 methylation. J. Pept. Sci. 2022, 28, e3381. [Google Scholar] [CrossRef]
- Mattiroli, F.; Penengo, L. Histone Ubiquitination: An Integrative Signaling Platform in Genome Stability. Trends Genet. 2021, 37, 566–581. [Google Scholar] [CrossRef]
- Oss-Ronen, L.; Sarusi, T.; Cohen, I. Histone Mono-Ubiquitination in Transcriptional Regulation and Its Mark on Life: Emerging Roles in Tissue Development and Disease. Cells 2022, 11, 2404. [Google Scholar] [CrossRef]
- Machour, F.E.; Ayoub, N. Transcriptional Regulation at DSBs: Mechanisms and Consequences. Trends Genet. 2020, 36, 981–997. [Google Scholar] [CrossRef]
- Beyer, J.N.; Raniszewski, N.R.; Burslem, G.M. Advances and Opportunities in Epigenetic Chemical Biology. Chembiochem 2021, 22, 17–42. [Google Scholar] [CrossRef]
- Fujimoto, M.; Takii, R.; Matsumoto, M.; Okada, M.; Nakayama, K.I.; Nakato, R.; Fujiki, K.; Shirahige, K.; Nakai, A. HSF1 phosphorylation establishes an active chromatin state via the TRRAP-TIP60 complex and promotes tumorigenesis. Nat. Commun. 2022, 13, 4355. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Shilatifard, A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345. [Google Scholar] [CrossRef]
- Reddington, C.J.; Fellner, M.; Burgess, A.E.; Mace, P.D. Molecular Regulation of the Polycomb Repressive-Deubiquitinase. Int. J. Mol. Sci. 2020, 21, 7837. [Google Scholar] [CrossRef]
- Cohen, I.; Bar, C.; Ezhkova, E. Activity of PRC1 and Histone H2AK119 Monoubiquitination: Revising Popular Misconceptions. BioEssays News Rev. Mol. Cell. Dev. Biol. 2020, 42, e1900192. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, M.; Matsushita, N. DNA Damage Response Regulation by Histone Ubiquitination. Int. J. Mol. Sci. 2022, 23, 8187. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef]
- Minnoye, L.; Marinov, G.K.; Krausgruber, T.; Pan, L.; Marand, A.P.; Secchia, S.; Greenleaf, W.J.; Furlong, E.E.M.; Zhao, K.; Schmitz, R.J.; et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primers 2021, 1, 10. [Google Scholar] [CrossRef]
- Chodurska, B.; Kunej, T. Long non-coding RNAs in humans: Classification, genomic organization and function. Non-Coding RNA Res. 2025, 11, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, R.; Suzuki, H. Long noncoding RNA involvement in cancer. BMB Rep. 2012, 45, 604–611. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Xiao, R.D.; Luo, R.G.; Xie, J.B.; Su, Z.D.; Wang, Y. Construction of long non-coding RNA- and microRNA-mediated competing endogenous RNA networks in alcohol-related esophageal cancer. PLoS ONE 2022, 17, e0269742. [Google Scholar] [CrossRef]
- Li, A.; Yu, W.H.; Hsu, C.L.; Huang, H.C.; Juan, H.F. Modular signature of long non-coding RNA association networks as a prognostic biomarker in lung cancer. BMC Med. Genom. 2021, 14, 290. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, M.; Zhang, L.; Lou, J.; Zhou, F.; Fang, M. Non-coding RNA in thyroid cancer—Functions and mechanisms. Cancer Lett. 2021, 496, 117–126. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Wong, C.M.; Tsang, F.H.; Ng, I.O. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027. [Google Scholar] [CrossRef]
- Klingenberg, M.; Matsuda, A.; Diederichs, S.; Patel, T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017, 67, 603–618. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Shen, J.; Zhang, L.; Li, M.; Hu, W.; Cho, C. Therapeutic targeting of noncoding RNAs in hepatocellular carcinoma: Recent progress and future prospects. Oncol. Lett. 2018, 15, 3395–3402. [Google Scholar] [CrossRef]
- Han Li, C.; Chen, Y. Small and Long Non-Coding RNAs: Novel Targets in Perspective Cancer Therapy. Curr. Genom. 2015, 16, 319–326. [Google Scholar] [CrossRef]
- Tutar, L. Introductory Chapter: Noncoding RNAs–A Brief Introduction. In Recent Advances in Noncoding RNAs; IntechOpen: London, UK, 2022. [Google Scholar]
- Huang, X.; Fejes Tóth, K.; Aravin, A.A. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017, 33, 882–894. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, P.C. MicroRNA: A small molecule with a big biological impact. MicroRNA 2012, 1, 1. [Google Scholar] [CrossRef]
- Freeman, J.A.; Espinosa, J.M. The impact of post-transcriptional regulation in the p53 network. Brief. Funct. Genom. 2013, 12, 46–57. [Google Scholar] [CrossRef]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Gupta, J.; Abdulsahib, W.K.; Turki Jalil, A.; Saadi Kareem, D.; Aminov, Z.; Alsaikhan, F.; Ramírez-Coronel, A.A.; Ramaiah, P.; Farhood, B. Prostate cancer and microRNAs: New insights into apoptosis. Pathol. Res. Pract. 2023, 245, 154436. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.L.; Lehmann, U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 7894–7913. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Q.; Wen, J.; Wu, Y.; Man, C. MiR-375: A novel multifunctional regulator. Life Sci. 2021, 275, 119323. [Google Scholar] [CrossRef]
- Yan, J.W.; Lin, J.S.; He, X.X. The emerging role of miR-375 in cancer. Int. J. Cancer 2014, 135, 1011–1018. [Google Scholar] [CrossRef]
- Li, J.; Xuan, Z.; Liu, C. Long non-coding RNAs and complex human diseases. Int. J. Mol. Sci. 2013, 14, 18790–18808. [Google Scholar] [CrossRef]
- Kugel, J.F.; Goodrich, J.A. Non-coding RNAs: Key regulators of mammalian transcription. Trends Biochem. Sci. 2012, 37, 144–151. [Google Scholar] [CrossRef]
- Saw, P.E.; Xu, X.; Chen, J.; Song, E.W. Non-coding RNAs: The new central dogma of cancer biology. Sci. China Life Sci. 2021, 64, 22–50. [Google Scholar] [CrossRef]
- Yang, X.; Xie, X.; Xiao, Y.F.; Xie, R.; Hu, C.J.; Tang, B.; Li, B.S.; Yang, S.M. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015, 360, 119–124. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, P.S.; Zhang, L.; Hussain, M.S.; Khan, G.; Mawkili, W.; Hanbashi, A.; Gupta, G.; Balakrishnan, P.; Arumugam, S. Non-coding RNAs as key regulators in hepatitis B virus-related hepatocellular carcinoma. Front. Immunol. 2025, 16, 1602252. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, L.; Wang, Y.; Li, H.; Ren, X.; Wei, F.; Yu, W.; Wang, X.; Zhang, L.; Yu, J.; et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014, 35, 9531–9538. [Google Scholar] [CrossRef]
- Yuan, C.; Ning, Y.; Pan, Y. Emerging roles of HOTAIR in human cancer. J. Cell. Biochem. 2020, 121, 3235–3247. [Google Scholar] [CrossRef]
- Yang, F.; Huo, X.S.; Yuan, S.X.; Zhang, L.; Zhou, W.P.; Wang, F.; Sun, S.H. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 2013, 49, 1083–1096. [Google Scholar] [CrossRef]
- Wu, S.C.; Kallin, E.M.; Zhang, Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010, 20, 1109–1116. [Google Scholar] [CrossRef]
- Tierno, D.; Grassi, G.; Scaggiante, B. New challenges in hepatocellular carcinoma: A role for PIWI-interacting RNAs? World J. Gastroenterol. 2024, 30, 2843–2848. [Google Scholar] [CrossRef]
- Liao, Z.; Yang, L.; Cheng, X.; Huang, X.; Zhang, Q.; Wen, D.; Song, Z.; Li, Y.; Wen, S.; Li, Y.; et al. pir-hsa-216911 inhibit pyroptosis in hepatocellular carcinoma by suppressing TLR4 initiated GSDMD activation. Cell Death Discov. 2025, 11, 11. [Google Scholar] [CrossRef]
- Hussain, M.S.; Moglad, E.; Afzal, M.; Bansal, P.; Kaur, H.; Deorari, M.; Ali, H.; Shahwan, M.; Hassan Almalki, W.; Kazmi, I.; et al. Circular RNAs in the KRAS pathway: Emerging players in cancer progression. Pathol. Res. Pract. 2024, 256, 155259. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; He, R.; Dang, Y.; Wu, H.; Feng, Z.; Chen, G. The Latest Overview of circRNA in the Progression, Diagnosis, Prognosis, Treatment, and Drug Resistance of Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 608257. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, W.; Wang, L.; Zhang, Z.; Zhang, F.; Zheng, S.; Kong, D. The effects of epigenetic modification on the occurrence and progression of liver diseases and the involved mechanism. Expert Rev. Gastroenterol. Hepatol. 2020, 14, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.; Mann, D.A. Epigenetics in liver disease: From biology to therapeutics. Gut 2016, 65, 1895–1905. [Google Scholar] [CrossRef]
- Wang, S.; Wu, W.; Claret, F.X. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 2017, 12, 187–197. [Google Scholar] [CrossRef]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef]
- Ferreira, H.J.; Esteller, M. Non-coding RNAs, epigenetics, and cancer: Tying it all together. Cancer Metastasis Rev. 2018, 37, 55–73. [Google Scholar] [CrossRef]
- Marchese, F.P.; Huarte, M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Hanif, H.; Ali, M.J.; Susheela, A.T.; Khan, I.W.; Luna-Cuadros, M.A.; Khan, M.M.; Lau, D.T. Update on the applications and limitations of alpha-fetoprotein for hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 216–229. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Y.; Sun, L.; Qiao, G.; Song, Y.; Liu, B. Exosomal MicroRNAs as Liquid Biopsy Biomarkers in Hepatocellular Carcinoma. OncoTargets Ther. 2020, 13, 2021–2030. [Google Scholar] [CrossRef]
- Lumkul, L.; Jantaree, P.; Jaisamak, K.; Wongkummool, W.; Lapisatepun, W.; Orrapin, S.; Udomruk, S.; Lo Piccolo, L.; Chaiyawat, P. Combinatorial Gene Expression Profiling of Serum HULC, HOTAIR, and UCA1 lncRNAs to Differentiate Hepatocellular Carcinoma from Liver Diseases: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2024, 25, 1258. [Google Scholar] [CrossRef]
- Guerriero, P.; Moshiri, F.; Lupini, L.; Sabbioni, S.; Negrini, M.; Callegari, E. Circulating tumor DNAs and non-coding RNAs as potential biomarkers for hepatocellular carcinoma diagnosis, prognosis and response to therapy. Hepatoma Res. 2019, 5, 6. [Google Scholar] [CrossRef]
- Choudhuri, S.; Cui, Y.; Klaassen, C.D. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol. Appl. Pharmacol. 2010, 245, 378–393. [Google Scholar] [CrossRef] [PubMed]
- Mosca, L.; Vitiello, F.; Borzacchiello, L.; Coppola, A.; Tranchese, R.V.; Pagano, M.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. Mutual Correlation between Non-Coding RNA and S-Adenosylmethionine in Human Cancer: Roles and Therapeutic Opportunities. Cancers 2021, 13, 3264. [Google Scholar] [CrossRef]
- Du, D.; He, J.; Ju, C.; Wang, C.; Li, H.; He, F.; Zhou, M. When N7-methyladenosine modification meets cancer: Emerging frontiers and promising therapeutic opportunities. Cancer Lett. 2023, 562, 216165. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sheng, Y.; Yang, J.; Wang, C.; Zhang, R.; Zhu, Y.; Zhang, Z.; Zhang, K.; Yan, S.; Sun, H.; et al. Osteopontin alters DNA methylation through up-regulating DNMT1 and sensitizes CD133+/CD44+ cancer stem cells to 5 azacytidine in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 179. [Google Scholar] [CrossRef] [PubMed]
- Hlady, R.A.; Sathyanarayan, A.; Thompson, J.J.; Zhou, D.; Wu, Q.; Pham, K.; Lee, J.H.; Liu, C.; Robertson, K.D. Integrating the epigenome to identify drivers of hepatocellular carcinoma. Hepatology 2019, 69, 639–652. [Google Scholar] [CrossRef]
- Hama, N.; Totoki, Y.; Miura, F.; Tatsuno, K.; Saito-Adachi, M.; Nakamura, H.; Arai, Y.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Epigenetic landscape influences the liver cancer genome architecture. Nat. Commun. 2018, 9, 1643. [Google Scholar] [CrossRef]
- Zhang, Y.; Chou, J.B.; Li, J.; Li, H.; Du, Q.; Yadav, A.; Zhou, S.; Shalaginov, M.Y.; Fang, Z.; Zhong, H.; et al. Broadband transparent optical phase change materials for high-performance nonvolatile photonics. Nat. Commun. 2019, 10, 4279. [Google Scholar] [CrossRef]
- Shao, F.; Yang, X.; Wang, W.; Wang, J.; Guo, W.; Feng, X.; Shi, S.; Xue, Q.; Gao, S.; Gao, Y.; et al. Associations of PGK1 promoter hypomethylation and PGK1-mediated PDHK1 phosphorylation with cancer stage and prognosis: A TCGA pan-cancer analysis. Cancer Commun. 2019, 39, 1–17. [Google Scholar] [CrossRef]
- Hua, D.; Hu, Y.; Wu, Y.-Y.; Cheng, Z.-H.; Yu, J.; Du, X.; Huang, Z.-H. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp. Mol. Pathol. 2011, 91, 455–460. [Google Scholar] [CrossRef]
- Cui, H.; Kong, Y.; Xu, M.; Zhang, H. Notch3 Functions as a Tumor Suppressor by Controlling Cellular Senescence. Cancer Res. 2013, 73, 3451–3459. [Google Scholar] [CrossRef]
- Herceg, Z.; Paliwal, A. Epigenetic mechanisms in hepatocellular carcinoma: How environmental factors influence the epigenome. Mutat. Res./Rev. Mutat. Res. 2011, 727, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Arechederra, M.; Daian, F.; Yim, A.; Bazai, S.K.; Richelme, S.; Dono, R.; Saurin, A.J.; Habermann, B.H.; Maina, F. Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat. Commun. 2018, 9, 3164. [Google Scholar] [CrossRef]
- Bárcena-Varela, M.; Caruso, S.; Llerena, S.; Álvarez-Sola, G.; Uriarte, I.; Latasa, M.U.; Urtasun, R.; Rebouissou, S.; Alvarez, L.; Jimenez, M.; et al. Dual targeting of histone methyltransferase G9a and DNA-methyltransferase 1 for the treatment of experimental hepatocellular carcinoma. Hepatology 2019, 69, 587–603. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-S.; Ban, H.S.; Hur, K.; Cho, H.-S. The epigenetic regulation of HCC metastasis. Int. J. Mol. Sci. 2018, 19, 3978. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.H.; Zhou, L.; Ng, K.Y.; Wong, T.L.; Lee, T.K.; Sharma, R.; Loong, J.H.; Ching, Y.P.; Yuan, Y.-F.; Xie, D.; et al. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep. 2018, 25, 690–701.e8. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Fu, M.; Ge, C.; Zhou, H.; Huang, H.; Zhong, M.; Zhang, M.; Xu, H.; Zhu, G.; Hua, W.; et al. m6A-Mediated Upregulation of lncRNA CHASERR Promotes the Progression of Glioma by Modulating the miR-6893-3p/TRIM14 Axis. Mol. Neurobiol. 2024, 61, 5418–5440. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, C.; Chen, J.; Chen, D.; Yang, B.; He, B.; Hu, W.; Zhang, Y.; Liu, H.; Dai, L.; et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 2019, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wei, L.; Law, C.T.; Tsang, F.H.C.; Shen, J.; Cheng, C.L.H.; Tsang, L.H.; Ho, D.W.H.; Chiu, D.K.C.; Lee, J.M.F.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef]
- Li, J.; Xie, H.; Ying, Y.; Chen, H.; Yan, H.; He, L.; Xu, M.; Xu, X.; Liang, Z.; Liu, B.; et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer 2020, 19, 152. [Google Scholar] [CrossRef]
- Zhou, T.; Li, S.; Xiang, D.; Liu, J.; Sun, W.; Cui, X.; Ning, B.; Li, X.; Cheng, Z.; Jiang, W.; et al. m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct. Target. Ther. 2020, 5, 296. [Google Scholar] [CrossRef]
- Bernasconi, A.; Canakoglu, A.; Masseroli, M.; Ceri, S. The road towards data integration in human genomics: Players, steps and interactions. Brief. Bioinform. 2021, 22, 30–44. [Google Scholar] [CrossRef]
- Zheng, Y.; Jun, J.; Brennan, K.; Gevaert, O. EpiMix: An integrative tool for epigenomic subtyping using DNA methylation. bioRxiv 2023. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, C.; Zhao, J.; Wang, C.; Xu, Y.; Han, Z.; Jiang, G.; Guo, X.; Li, R.; Bu, X.; et al. Correlation of CpG island methylator phenotype with poor prognosis in hepatocellular carcinoma. Exp. Mol. Pathol. 2010, 88, 112–117. [Google Scholar] [CrossRef]
- Lin, T.; Ponn, A.; Lu, J. Requirement of the histone demethylase LSD1 in Snail-mediated transcriptional repression during epithelial-mesenchymal transition. Cancer Res. 2011, 71, 3422. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, X.; Zhang, D.; Chen, X.; Zhu, X. High expression of Snail and NF-κB; predicts poor survival in Chinese hepatocellular carcinoma patients. Oncotarget 2017, 8, 4543. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.; Wu, X.; Zhang, C.; Li, G. Diagnostic value of lncRNAs as biomarker in hepatocellular carcinoma: An updated meta-analysis. Can. J. Gastroenterol. Hepatol. 2018, 2018, 8410195. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Wu, X.; Li, Y. Identification and validation of potential long non-coding RNA biomarkers in predicting survival of patients with head and neck squamous cell carcinoma. Oncol. Lett. 2019, 17, 5642–5652. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-H.; Lee, J.H.; Kim, G.; Lee, E.; Lee, Y.R.; Jang, S.Y.; Lee, H.W.; Chun, J.M.; Han, Y.S.; Yoon, J.S.; et al. Expression of the long noncoding RNA GAS5 correlates with liver fibrosis in patients with nonalcoholic fatty liver disease. Genes 2020, 11, 545. [Google Scholar] [CrossRef]
- Shang, X.; Li, G.; Liu, H.; Li, T.; Liu, J.; Zhao, Q.; Wang, C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine 2016, 95, e3811. [Google Scholar] [CrossRef]
- Jin, Y.; Ren, Y.; Gao, Y.; Zhang, L.; Ding, Z. Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3311–3319. [Google Scholar]
- Chen, D.; Zhang, C.; Lin, J.; Song, X.; Wang, H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 1275. [Google Scholar] [CrossRef]
- Fu, L.; Yao, T.; Chen, Q.; Mo, X.; Hu, Y.; Guo, J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget 2017, 8, 58405. [Google Scholar] [CrossRef]
- Wang, M.; Gu, B.; Yao, G.; Li, P.; Wang, K. Circular RNA expression profiles and the pro-tumorigenic function of circRNA_10156 in hepatitis B virus-related liver cancer. Int. J. Med Sci. 2020, 17, 1351. [Google Scholar] [CrossRef]
- Dai, X.; Chen, C.; Yang, Q.; Xue, J.; Chen, X.; Sun, B.; Luo, F.; Liu, X.; Xiao, T.; Xu, H.; et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018, 9, 454. [Google Scholar] [CrossRef]
- Wang, G.; Liu, W.; Zou, Y.; Wang, G.; Deng, Y.; Luo, J.; Zhang, Y.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a–MET pathway. EBioMedicine 2019, 40, 432–445. [Google Scholar] [CrossRef]
- Lin, X.-J.; Chong, Y.; Guo, Z.-W.; Xie, C.; Yang, X.-J.; Zhang, Q.; Li, S.-P.; Xiong, Y.; Yuan, Y.; Min, J.; et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: A multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015, 16, 804–815. [Google Scholar] [CrossRef]
- Yang, J.; Sheng, Y.-Y.; Wei, J.-W.; Gao, X.-M.; Zhu, Y.; Jia, H.-L.; Dong, Q.-Z.; Qin, L.-X. MicroRNA-219-5p promotes tumor growth and metastasis of hepatocellular carcinoma by regulating cadherin 1. BioMed Res. Int. 2018, 2018, 4793971. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-y.; Tang, Y.-p.; Liu, J.-j.; Liang, R.; Luo, X.-l. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J. Cancer 2020, 11, 2993. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Banaudha, K.K.; Verma, M. The role of microRNAs in the management of liver cancer. Methods Mol. Biol. 2012, 863, 241–251. [Google Scholar]
- Anwar, S.L.; Albat, C.; Krech, T.; Hasemeier, B.; Schipper, E.; Schweitzer, N.; Vogel, A.; Kreipe, H.; Lehmann, U. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int. J. Cancer 2013, 133, 660–670. [Google Scholar] [CrossRef]
- Zhang, Z.; Ge, S.; Wang, X.; Yuan, Q.; Yan, Q.; Ye, H.; Che, Y.; Lin, Y.; Zhang, J.; Liu, P. Serum miR-483-5p as a potential biomarker to detect hepatocellular carcinoma. Hepatol. Int. 2013, 7, 199–207. [Google Scholar] [CrossRef]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of Genome-Wide Circulating MicroRNA in Hepatocellular Carcinoma: MiR-483-5p as a Potential Biomarker. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Zhang, T.; Hao, M.; Zhang, X.; Zhang, J.; Xie, Q.; Wang, Y.; Guo, M.; Zhuang, H.; et al. Methylation-mediated repression of micro RNA 129-2 enhances oncogenic SOX 4 expression in HCC. Liver Int. 2013, 33, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Tsunedomi, R.; Iizuka, N.; Yoshimura, K.; Iida, M.; Tsutsui, M.; Hashimoto, N.; Kanekiyo, S.; Sakamoto, K.; Tamesa, T.; Oka, M. ABCB6 mRNA and DNA methylation levels serve as useful biomarkers for prediction of early intrahepatic recurrence of hepatitis C virus-related hepatocellular carcinoma. Int. J. Oncol. 2013, 42, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Ozen, C.; Yildiz, G.; Dagcan, A.T.; Cevik, D.; Ors, A.; Keles, U.; Topel, H.; Ozturk, M. Genetics and epigenetics of liver cancer. New Biotechnol. 2013, 30, 381–384. [Google Scholar] [CrossRef] [PubMed]
| Epigenetic Mechanism | Molecular Players | Functional Role in HCC | Biomarkers/Targets | Clinical Implication |
|---|---|---|---|---|
| DNA Methylation | DNMT1, DNMT3A, DNMT3B, TET1/2/3 | Gene silencing, genomic instability | RASSF1, APC, CDKN2A, GATA4, CDKL2 | Diagnostic and therapeutic value |
| Histone Modification | HATs, HDACs, HMTs, LSDs, PRMT6 | Chromatin remodeling, DNA accessibility, gene regulation | H3K27me3, H3S10ph, H2AK119ub, H2BK120ub | Therapeutic value |
| Non-coding RNAs (ncRNAs) | miR-375, miR-219-5p, HOTAIR, GAS5, UCA1, lncRNA-LET | Regulation of transcription/translation, chromatin targeting, and metastasis | H19, circPTGR1, circ_0004018 | Diagnostic and prognostic value |
| mRNA Methylation (m6A) | METTL3, WTAP, YTHDF2 | Regulation of mRNA stability, cell cycle, suppressor gene silencing | SOCS2, ETS1 | Therapeutic value |
| Epigenetic Biomarkers | miRNAs, lncRNAs, circRNAs | EMT, drug resistance, prognosis | miR-483-5p, LINC01093, has_circ_0005075 | Diagnostic and prognostic value |
| Drug Name | Target | Epigenetic Mechanism | Trial Phase | Combination Tested | Outcome Summary |
|---|---|---|---|---|---|
| Guadecitabine | DNMT1 | DNA hypomethylation | Phase II | Sorafenib | Limited efficacy as monotherapy |
| Vorinostat | HDAC | Histone deacetylase inhibition | Phase I/II | Nivolumab | Enhances T-cell infiltration |
| Belinostat | HDAC | Histone acetylation | Phase I | Chemotherapy | Under investigation |
| SGI-110 | DNMT | DNA demethylation | Preclinical | Checkpoint inhibitors | Shows potential synergy |
| Romidepsin | HDAC1/2 | Histone deacetylation | Phase I | N/A | Toxicity limits standalone application |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wali, A.F.; Ansari, A.R.; Mir, P.A.; El-Tanani, M.; Babiker, R.; Hussain, M.S.; Uppal, J.; Zargar, A.I.; Mir, R.H. Epigenetic Alterations in Hepatocellular Carcinoma: Mechanisms, Biomarkers, and Therapeutic Implications. Pharmaceuticals 2025, 18, 1281. https://doi.org/10.3390/ph18091281
Wali AF, Ansari AR, Mir PA, El-Tanani M, Babiker R, Hussain MS, Uppal J, Zargar AI, Mir RH. Epigenetic Alterations in Hepatocellular Carcinoma: Mechanisms, Biomarkers, and Therapeutic Implications. Pharmaceuticals. 2025; 18(9):1281. https://doi.org/10.3390/ph18091281
Chicago/Turabian StyleWali, Adil Farooq, Abid Reza Ansari, Prince Ahad Mir, Mohamed El-Tanani, Rasha Babiker, Md Sadique Hussain, Jasreen Uppal, Asma Ishrat Zargar, and Reyaz Hassan Mir. 2025. "Epigenetic Alterations in Hepatocellular Carcinoma: Mechanisms, Biomarkers, and Therapeutic Implications" Pharmaceuticals 18, no. 9: 1281. https://doi.org/10.3390/ph18091281
APA StyleWali, A. F., Ansari, A. R., Mir, P. A., El-Tanani, M., Babiker, R., Hussain, M. S., Uppal, J., Zargar, A. I., & Mir, R. H. (2025). Epigenetic Alterations in Hepatocellular Carcinoma: Mechanisms, Biomarkers, and Therapeutic Implications. Pharmaceuticals, 18(9), 1281. https://doi.org/10.3390/ph18091281







