Towards Dual-Tracer SPECT for Prostate Cancer Imaging Using [99mTc]Tc-PSMA-I&S and [111In]In-RM2
Abstract
1. Introduction
2. Results
2.1. Syntheses and Radiolabeling
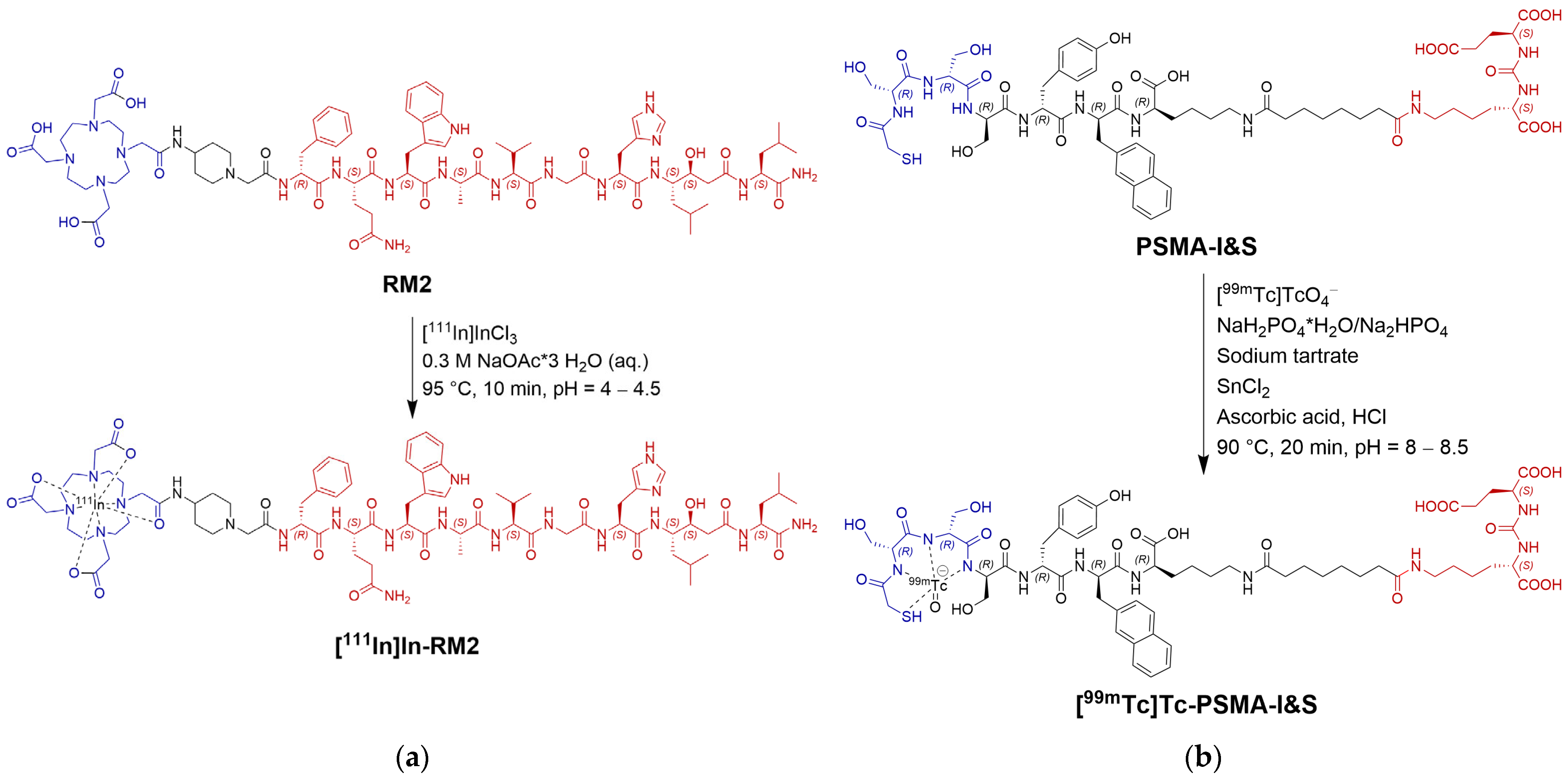
2.1.1. RM2 Synthesis and Radiolabeling
2.1.2. PSMA-I&S Synthesis and Radiolabeling
2.2. CHO-K1 Cell Transduction with Human GRPR or Human PSMA
2.3. In Vitro Evaluation
2.4. Dual-Isotope SPECT Phantom Measurements
2.5. In/Ex Vivo Evaluation
2.5.1. µSPECT/CT Imaging



2.5.2. Biodistribution Studies
2.5.3. Western Blot Analysis of Cell and Tissue Lysates
3. Discussion
4. Materials and Methods
4.1. Syntheses
4.2. Cell Culture and In Vitro Evaluation
4.2.1. Viral Transduction of CHO-K1 Cells for GRPR or PSMA Overexpression
4.2.2. Internalization Experiments
4.2.3. Saturation Assays
4.3. γ-Counter Set-Up for Dual-Isotope Measurements
4.4. Dual-Isotope SPECT (Phantom) Measurements
4.5. In/Ex Vivo Evaluation
4.5.1. µSPECT/CT Imaging
4.5.2. Biodistribution Studies
4.5.3. Protein Extraction and Western Blot Analyses of Cell and Tissue Lysates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Tolkach, Y.; Kristiansen, G. The Heterogeneity of Prostate Cancer: A Practical Approach. Pathobiology 2018, 85, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Wibmer, A.G.; Burger, I.A.; Sala, E.; Hricak, H.; Weber, W.A.; Vargas, H.A. Molecular Imaging of Prostate Cancer. Radiographics 2016, 36, 142–159. [Google Scholar] [CrossRef]
- Hofman, M.S.; Hicks, R.J.; Maurer, T.; Eiber, M. Prostate-specific Membrane Antigen PET: Clinical Utility in Prostate Cancer, Normal Patterns, Pearls, and Pitfalls. RadioGraphics 2018, 38, 200–217. [Google Scholar] [CrossRef]
- Farolfi, A.; Calderoni, L.; Mattana, F.; Mei, R.; Telo, S.; Fanti, S.; Castellucci, P. Current and Emerging Clinical Applications of PSMA PET Diagnostic Imaging for Prostate Cancer. J. Nucl. Med. 2021, 62, 596–604. [Google Scholar] [CrossRef]
- Paschalis, A.; Sheehan, B.; Riisnaes, R.; Rodrigues, D.N.; Gurel, B.; Bertan, C.; Ferreira, A.; Lambros, M.B.K.; Seed, G.; Yuan, W.; et al. Prostate-specific Membrane Antigen Heterogeneity and DNA Repair Defects in Prostate Cancer. Eur. Urol. 2019, 76, 469–478. [Google Scholar] [CrossRef]
- Shetty, D.; Patel, D.; Le, K.; Bui, C.; Mansberg, R. Pitfalls in Gallium-68 PSMA PET/CT Interpretation—A Pictorial Review. Tomography 2018, 4, 182–193. [Google Scholar] [CrossRef]
- Ceci, F.; Castellucci, P.; Cerci, J.J.; Fanti, S. New aspects of molecular imaging in prostate cancer. Methods 2017, 130, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Stott Reynolds, T.J.; Smith, C.J.; Lewis, M.R. Peptide-Based Radiopharmaceuticals for Molecular Imaging of Prostate Cancer. Adv. Exp. Med. Biol. 2018, 1096, 135–158. [Google Scholar] [CrossRef]
- Hoshi, S.; Yaginuma, K.; Meguro, S.; Onagi, A.; Matsuoka, K.; Hata, J.; Sato, Y.; Akaihata, H.; Kataoka, M.; Ogawa, S.; et al. PSMA Targeted Molecular Imaging and Radioligand Therapy for Prostate Cancer: Optimal Patient and Treatment Issues. Curr. Oncol. 2023, 30, 7286–7302. [Google Scholar] [CrossRef]
- Surasi, D.S.; Eiber, M.; Maurer, T.; Preston, M.A.; Helfand, B.T.; Josephson, D.; Tewari, A.K.; Somford, D.M.; Rais-Bahrami, S.; Koontz, B.F.; et al. Diagnostic Performance and Safety of Positron Emission Tomography with 18F-rhPSMA-7.3 in Patients with Newly Diagnosed Unfavourable Intermediate- to Very-high-risk Prostate Cancer: Results from a Phase 3, Prospective, Multicentre Study (LIGHTHOUSE). Eur. Urol. 2023, 84, 361–370. [Google Scholar] [CrossRef]
- Mitran, B.; Varasteh, Z.; Abouzayed, A.; Rinne, S.S.; Puuvuori, E.; De Rosa, M.; Larhed, M.; Tolmachev, V.; Orlova, A.; Rosenström, U. Bispecific GRPR-Antagonistic Anti-PSMA/GRPR Heterodimer for PET and SPECT Diagnostic Imaging of Prostate Cancer. Cancers 2019, 11, 1371. [Google Scholar] [CrossRef]
- Fernández, R.; Soza-Ried, C.; Iagaru, A.; Stephens, A.; Müller, A.; Schieferstein, H.; Sandoval, C.; Amaral, H.; Kramer, V. Imaging GRPr Expression in Metastatic Castration-Resistant Prostate Cancer with [68Ga]Ga-RM2—A Head-to-Head Pilot Comparison with [68Ga]Ga-PSMA-11. Cancers 2024, 16, 173. [Google Scholar] [CrossRef] [PubMed]
- Iagaru, A. Will GRPR Compete with PSMA as a Target in Prostate Cancer? J. Nucl. Med. 2017, 58, 1883–1884. [Google Scholar] [CrossRef] [PubMed]
- Minamimoto, R.; Hancock, S.; Schneider, B.; Chin, F.T.; Jamali, M.; Loening, A.; Vasanawala, S.; Gambhir, S.S.; Iagaru, A. Pilot Comparison of 68Ga-RM2 PET and 68Ga-PSMA-11 PET in Patients with Biochemically Recurrent Prostate Cancer. J. Nucl. Med. 2016, 57, 557–562. [Google Scholar] [CrossRef]
- Eder, M.; Schäfer, M.; Bauder-Wüst, U.; Haberkorn, U.; Eisenhut, M.; Kopka, K. Preclinical evaluation of a bispecific low-molecular heterodimer targeting both PSMA and GRPR for improved PET imaging and therapy of prostate cancer. Prostate 2014, 74, 659–668. [Google Scholar] [CrossRef]
- Bandari, R.P.; Carmack, T.L.; Malhotra, A.; Watkinson, L.; Fergason Cantrell, E.A.; Lewis, M.R.; Smith, C.J. Development of Heterobivalent Theranostic Probes Having High Affinity/Selectivity for the GRPR/PSMA. J. Med. Chem. 2021, 64, 2151–2166. [Google Scholar] [CrossRef]
- Lundmark, F.; Abouzayed, A.; Rinne, S.S.; Timofeev, V.; Sipkina, N.; Naan, M.; Kirichenko, A.; Vasyutina, M.; Ryzhkova, D.; Tolmachev, V.; et al. Preclinical Characterisation of PSMA/GRPR-Targeting Heterodimer [68Ga]Ga-BQ7812 for PET Diagnostic Imaging of Prostate Cancer: A Step towards Clinical Translation. Cancers 2023, 15, 442. [Google Scholar] [CrossRef] [PubMed]
- Barone-Rochette, G.; Leclere, M.; Calizzano, A.; Vautrin, E.; Céline, G.-C.; Broisat, A.; Ghezzi, C.; Baguet, J.P.; Machecourt, J.; Vanzetto, G.; et al. Stress thallium-201/rest technetium-99m sequential dual-isotope high-speed myocardial perfusion imaging validation versus invasive coronary angiography. J. Nucl. Cardiol. 2015, 22, 513–522. [Google Scholar] [CrossRef]
- Sharir, T.; Slomka, P. Dual-isotope myocardial perfusion SPECT imaging: Past, present, and future. J. Nucl. Cardiol. 2018, 25, 2024–2028. [Google Scholar] [CrossRef]
- Sommerauer, M.; Graf, C.; Schäfer, N.; Huber, G.; Schneider, P.; Wüthrich, R.; Schmid, C.; Steinert, H. Sensitivity and Specificity of Dual-Isotope 99mTc-Tetrofosmin and 123I Sodium Iodide Single Photon Emission Computed Tomography (SPECT) in Hyperparathyroidism. PLoS ONE 2015, 10, e0129194. [Google Scholar] [CrossRef]
- Quintana, J.C.; Blend, M.J. The Dual-Isotope ProstaScint Imaging Procedure: Clinical Experience and Staging Results in 145 Patients. Clin. Nucl. Med. 2000, 25, 33. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.C.; Lee, I.H.; Yeh, T.L.; Chen, K.C.; Huang, H.C.; Chen, P.S.; Yang, Y.K.; Yao, W.J.; Lu, R.-B.; Chiu, N.T. Distribution volume ratio of serotonin and dopamine transporters in euthymic patients with a history of major depression—A dual-isotope SPECT study. Psychiatry Res. Neuroimaging 2010, 184, 157–161. [Google Scholar] [CrossRef]
- Guo, Z.; Gao, M.; Zhang, D.; Li, Y.; Song, M.; Zhuang, R.; Su, X.; Chen, G.; Liu, T.; Liu, P.; et al. Simultaneous SPECT imaging of multi-targets to assist in identifying hepatic lesions. Sci. Rep. 2016, 6, 28812. [Google Scholar] [CrossRef]
- van der Kroon, I.; Joosten, L.; Nock, B.A.; Maina, T.; Boerman, O.C.; Brom, M.; Gotthardt, M. Improved Quantification of the Beta Cell Mass after Pancreas Visualization with 99mTc-demobesin-4 and Beta Cell Imaging with 111In-exendin-3 in Rodents. Mol. Pharm. 2016, 13, 3478–3483. [Google Scholar] [CrossRef] [PubMed]
- Hijnen, N.M.; de Vries, A.; Nicolay, K.; Grüll, H. Dual-isotope 111In/177Lu SPECT imaging as a tool in molecular imaging tracer design. Contrast Media Mol. Imaging 2012, 7, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, D.J.; Zhang, J.; Kazules, T.; Vandehei, T.; Bolle, E.; Chowdhury, S.; Parnham, K.; Patt, B.E. In Vivo Dual-Isotope SPECT Imaging with Improved Energy Resolution. In Proceedings of the 2006 IEEE Nuclear Science Symposium Conference Record, San Diego, CA, USA, 29 October–1 November 2006; pp. 3821–3826. [Google Scholar]
- Available online: https://www-nds.iaea.org/relnsd/vcharthtml/VChartHTML.html (accessed on 4 January 2024).
- Herrero Álvarez, N.; Bauer, D.; Hernández-Gil, J.; Lewis, J.S. Recent Advances in Radiometals for Combined Imaging and Therapy in Cancer. ChemMedChem 2021, 16, 2909–2941. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.d.A.; Fuscaldi, L.L.; Townsend, D.M.; Rubello, D.; Barros, A.L.B.d. Radiolabeled bombesin derivatives for preclinical oncological imaging. Biomed. Pharmacother. 2017, 87, 58–72. [Google Scholar] [CrossRef]
- Sobel, R.E.; Sadar, M.D. Cell lines used in prostate cancer research: A compendium of old and new lines—Part 1. J. Urol. 2005, 173, 342–359. [Google Scholar] [CrossRef]
- Namekawa, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Application of Prostate Cancer Models for Preclinical Study: Advantages and Limitations of Cell Lines, Patient-Derived Xenografts, and Three-Dimensional Culture of Patient-Derived Cells. Cells 2019, 8, 74. [Google Scholar] [CrossRef]
- Wu, X.; Gong, S.; Roy-Burman, P.; Lee, P.; Culig, Z. Current mouse and cell models in prostate cancer research. Endocr. Relat Cancer 2013, 20, R155–R170. [Google Scholar] [CrossRef] [PubMed]
- Liolios, C.; Schafer, M.; Haberkorn, U.; Eder, M.; Kopka, K. Novel Bispecific PSMA/GRPr Targeting Radioligands with Optimized Pharmacokinetics for Improved PET Imaging of Prostate Cancer. Bioconjug. Chem. 2016, 27, 737–751. [Google Scholar] [CrossRef]
- Abouzayed, A.; Yim, C.-B.; Mitran, B.; Rinne, S.S.; Tolmachev, V.; Larhed, M.; Rosenström, U.; Orlova, A. Synthesis and Preclinical Evaluation of Radio-Iodinated GRPR/PSMA Bispecific Heterodimers for the Theranostics Application in Prostate Cancer. Pharmaceutics 2019, 11, 358. [Google Scholar] [CrossRef]
- Gödecke, N.; Hauser, H.; Wirth, D. Stable Expression by Lentiviral Transduction of Cells. In Recombinant Protein Expression in Mammalian Cells: Methods and Protocols; Hacker, D.L., Ed.; Springer New York: New York, NY, USA, 2018; pp. 43–55. [Google Scholar]
- Benya, R.V.; Kusui, T.; Katsuno, T.; Tsuda, T.; Mantey, S.A.; Battey, J.F.; Jensen, R.T. Glycosylation of the Gastrin-Releasing Peptide Receptor and Its Effect on Expression, G Protein Coupling, and Receptor Modulatory Processes. Mol. Pharmacol. 2000, 58, 1490–1501. [Google Scholar] [CrossRef] [PubMed]
- Mansi, R.; Wang, X.; Forrer, F.; Waser, B.; Cescato, R.; Graham, K.; Borkowski, S.; Reubi, J.C.; Maecke, H.R. Development of a potent DOTA-conjugated bombesin antagonist for targeting GRPr-positive tumours. Eur. J. Nucl. Med. Mol. Imaging 2011, 38, 97–107. [Google Scholar] [CrossRef]
- Gourni, E.; Canovas, C.; Goncalves, V.; Denat, F.; Meyer, P.T.; Maecke, H.R. (R)-NODAGA-PSMA: A Versatile Precursor for Radiometal Labeling and Nuclear Imaging of PSMA-Positive Tumors. PLoS ONE 2015, 10, e0145755. [Google Scholar] [CrossRef] [PubMed]
- Gourni, E.; Del Pozzo, L.; Bartholomä, M.; Kiefer, Y.; Meyer, P.T.; Maecke, H.R.; Holland, J.P. Radiochemistry and Preclinical PET Imaging of 68Ga-Desferrioxamine Radiotracers Targeting Prostate-Specific Membrane Antigen. Mol. Imaging 2017, 16, 1536012117737010. [Google Scholar] [CrossRef]
- Xu, N.; Ma, C.; Ou, J.; Sun, W.W.; Zhou, L.; Hu, H.; Liu, X.M. Comparative proteomic analysis of three Chinese hamster ovary (CHO) host cells. Biochem. Eng. J. 2017, 124, 122–129. [Google Scholar] [CrossRef]
- Lim, D.J.; Liu, X.-L.; Sutkowski, D.M.; Braun, E.J.; Lee, C.; Kozlowski, J.M. Growth of an androgen-sensitive human prostate cancer cell line, LNCaP, in nude mice. Prostate 1993, 22, 109–118. [Google Scholar] [CrossRef]
- Robu, S.; Schottelius, M.; Eiber, M.; Maurer, T.; Gschwend, J.; Schwaiger, M.; Wester, H.-J. Preclinical Evaluation and First Patient Application of 99mTc-PSMA-I&S for SPECT Imaging and Radioguided Surgery in Prostate Cancer. J. Nucl. Med. 2017, 58, 235–242. [Google Scholar] [CrossRef]
- Balber, T.; Singer, J.; Berroterán-Infante, N.; Dumanic, M.; Fetty, L.; Fazekas-Singer, J.; Vraka, C.; Nics, L.; Bergmann, M.; Pallitsch, K.; et al. Preclinical In Vitro and In Vivo Evaluation of [18F]FE@SUPPY for Cancer PET Imaging: Limitations of a Xenograft Model for Colorectal Cancer. Contrast Media Mol. Imaging 2018, 2018, 1269830. [Google Scholar] [CrossRef]
- Liu, C.; Hasegawa, K.; Russell, S.J.; Sadelain, M.; Peng, K.-W. Prostate-specific membrane antigen retargeted measles virotherapy for the treatment of prostate cancer. Prostate 2009, 69, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Høyvik, A.J.K.; Kvassheim, M.; Ma, L.-W.; Wiig, E.; Hillestad, T.; Revheim, M.-E.; Liukaityte, R.; Bruland, Ø.; Juzeniene, A. Therapeutic evaluation of [212Pb]Pb-AB001 and [177Lu]Lu-PSMA-617 in a mouse model of disseminated prostate cancer. Eur. J. Nucl. Med. Mol. Imaging, 2025; online ahead of print. [Google Scholar] [CrossRef]
- Chan, W.; White, P. Fmoc Solid Phase Peptide Synthesis: A Practical Approach; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Konrad, M.; Rinscheid, A.; Wienand, G.; Nittbaur, B.; Wester, H.-J.; Janzen, T.; Lapa, C.; Pfob, C.H.; Schottelius, M. [99mTc]Tc-PentixaTec: Development, extensive pre-clinical evaluation, and first human experience. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 3937–3948. [Google Scholar] [CrossRef] [PubMed]
- Giammei, C.; Balber, T.; Benčurová, K.; Cardinale, J.; Berroterán-Infante, N.; Brandt, M.; Jouini, N.; Hacker, M.; Mitterhauser, M.; Mindt, T.L. Sorbitol as a Polar Pharmacological Modifier to Enhance the Hydrophilicity of 99mTc-Tricarbonyl-Based Radiopharmaceuticals. Molecules 2020, 25, 2680. [Google Scholar] [CrossRef] [PubMed]
- Hassler, M.R.; Klisaroska, A.; Kollmann, K.; Steiner, I.; Bilban, M.; Schiefer, A.-I.; Sexl, V.; Egger, G. Antineoplastic activity of the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine in anaplastic large cell lymphoma. Biochimie 2012, 94, 2297–2307. [Google Scholar] [CrossRef]
- Hendrikx, G.; De Saint-Hubert, M.; Dijkgraaf, I.; Bauwens, M.; Douma, K.; Wierts, R.; Pooters, I.; Van den Akker, N.M.; Hackeng, T.M.; Post, M.J.; et al. Molecular imaging of angiogenesis after myocardial infarction by (111)In-DTPA-cNGR and (99m)Tc-sestamibi dual-isotope myocardial SPECT. EJNMMI Res. 2015, 5, 2. [Google Scholar] [CrossRef]







| Compound | Total Cell Uptake % AD/106 Cells [a] | Kd [nM] [b] | Bmax [nM] [b] | N° of Binding Sites per Cell |
|---|---|---|---|---|
| [111In]In-RM2 | 40.9 ± 2.4 | 2.33 ± 0.40 | 0.84 ± 0.03 | 5.06 × 105 |
| [99mTc]Tc-PSMA-I&S | 31.5 ± 1.9 | 3.84 ± 0.44 | 0.64 ± 0.02 | 3.85 × 105 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giammei, C.; Balber, T.; Felber, V.; Dillinger, T.; Cardinale, J.; Brandt, M.R.; Stingeder, A.; Mitterhauser, M.; Egger, G.; Mindt, T.L. Towards Dual-Tracer SPECT for Prostate Cancer Imaging Using [99mTc]Tc-PSMA-I&S and [111In]In-RM2. Pharmaceuticals 2025, 18, 1002. https://doi.org/10.3390/ph18071002
Giammei C, Balber T, Felber V, Dillinger T, Cardinale J, Brandt MR, Stingeder A, Mitterhauser M, Egger G, Mindt TL. Towards Dual-Tracer SPECT for Prostate Cancer Imaging Using [99mTc]Tc-PSMA-I&S and [111In]In-RM2. Pharmaceuticals. 2025; 18(7):1002. https://doi.org/10.3390/ph18071002
Chicago/Turabian StyleGiammei, Carolina, Theresa Balber, Veronika Felber, Thomas Dillinger, Jens Cardinale, Marie R. Brandt, Anna Stingeder, Markus Mitterhauser, Gerda Egger, and Thomas L. Mindt. 2025. "Towards Dual-Tracer SPECT for Prostate Cancer Imaging Using [99mTc]Tc-PSMA-I&S and [111In]In-RM2" Pharmaceuticals 18, no. 7: 1002. https://doi.org/10.3390/ph18071002
APA StyleGiammei, C., Balber, T., Felber, V., Dillinger, T., Cardinale, J., Brandt, M. R., Stingeder, A., Mitterhauser, M., Egger, G., & Mindt, T. L. (2025). Towards Dual-Tracer SPECT for Prostate Cancer Imaging Using [99mTc]Tc-PSMA-I&S and [111In]In-RM2. Pharmaceuticals, 18(7), 1002. https://doi.org/10.3390/ph18071002










