A Classification-Based Blood–Brain Barrier Model: A Comparative Approach
Abstract
1. Introduction
2. Results
2.1. Feature Selection
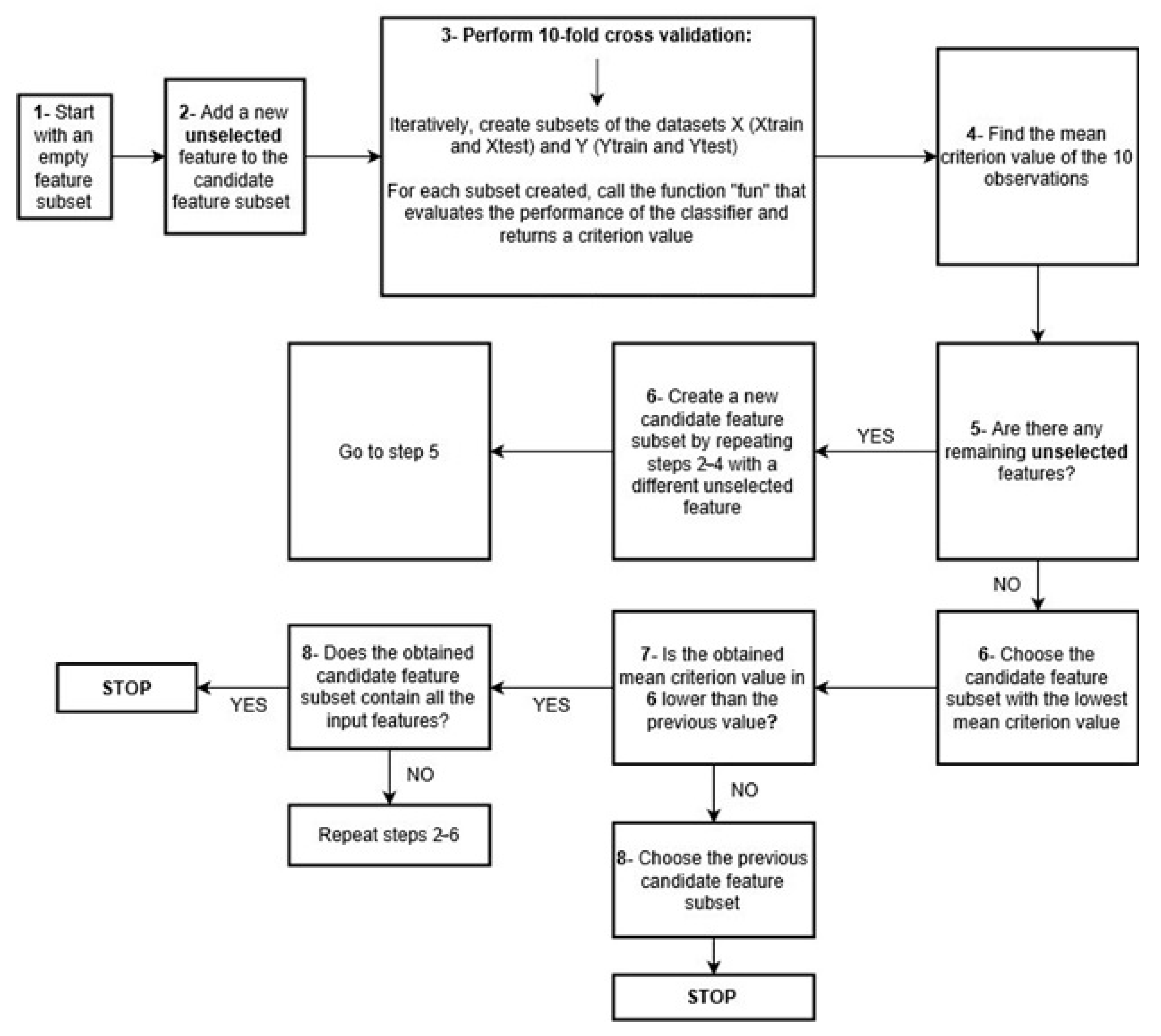
2.2. Part 1 Sequential Feature Selection
2.3. Part 2 Genetic Algorithm
Prediction Performance Evaluation
3. Discussion
4. Materials and Methods
4.1. Dataset Preparation
4.2. Molecular Descriptors
- The molecular weight (MW);
- The polar surface area (PSA);
- The octanol/water partition (logP);
- The number of hydrogen bond acceptors (HAs);
- The number of hydrogen bond donors (HDs);
- pka (strongest acid);
- pka (strongest base);
- The number of rotatable bonds (NRB).
4.3. Feature Selection
4.3.1. Part 1 Sequential Feature Selection
4.3.2. Part 2 Genetic Algorithm
- Selection: the fittest chromosomes of the initial population are preserved for the next generation;
- Cross-over: new chromosomes are created in the new generation by mixing gene subsets of one chromosome with those of another;
- Mutation: A certain gene from a given chromosome is randomly inverted (0 to 1 or vice versa). This allows the algorithm to evaluate new options instead of getting stuck on local minima.
4.4. Classification
- For training, 80% of the dataset was used, and a set of feature vectors with known output was employed to build the classifier.
- The remaining 20% was used as the testing set: A classifier was tested by predicting the outputs of a test set and comparing the predicted results to the actual ones. This step is important to evaluate the performance of any classifier used. Once both sets were ready, the following types of classifiers were applied for performance comparison on the different classifiers: SVM [17] (linear SVM and using polynomial and radial basis function (RBF) kernels), LDA [18] and quadratic discriminant analysis (QDA) [19], and kNN. These classifiers were chosen based on their prevalent use in the literature regarding BBB permeability prediction and their diversity in algorithmic approach: SVM for handling non-linear separation, LDA and QDA for modeling linear and quadratic class boundaries, and kNN as a non-parametric baseline.
4.5. Performance Evaluation
- The sensitivity (SE), which reflects the capacity of the classifier to detect BBB+ drugs in the entire dataset;
- The positive predictive value (PP), which expresses its ability not to deem non-crossing drugs as BBB+;
- The negative predictive value (NP), which reflects its ability not to deem crossing drugs as BBB−;
- The specificity (SP), which expresses the ability of the model to detect BBB- drugs in the dataset;
- The overall accuracy (ACC), which expresses the total true predictions over the total number of prediction.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zlokovic, B.V. The Blood-Brain Barrier in Health and Chronic Neurodegenerative Disorders. Neuron 2008, 57, 178–201. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Shi, Y.; Azevedo, H.S. In vitro blood–brain barrier models for drug research: State-of-the-art and new perspectives on reconstituting these models on artificial basement membrane platforms. Drug Discov. Today 2016, 21, 1367–1386. [Google Scholar] [CrossRef] [PubMed]
- Vastag, M.; Keseru, G.M. Current in vitro and in silico models of blood-brain barrier penetration: A practical view. Curr. Opin. Drug Discov. Dev. 2009, 12, 115. [Google Scholar]
- Saraiva, C.; Praça, C.; Ferreira, R.; Santos, T.; Ferreira, L.; Bernardino, L. Nanoparticle-mediated brain drug delivery: Overcoming blood–brain barrier to treat neurodegenerative diseases. J. Control. Release 2016, 235, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Muehlbacher, M.; Spitzer, G.M.; Liedl, K.R.; Kornhuber, J. Qualitative prediction of blood–brain barrier permeability on a large and refined dataset. J. Comput. Aided Mol. Des. 2011, 25, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Kunwittaya, S.; Nantasenamat, C.; Treeratanapiboon, L.; Srisarin, A.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Influence of logBB cut-off on the prediction of blood-brain barrier permeability. Biomed. Appl. Technol. J. 2013, 1, 16–34. [Google Scholar]
- Castillo-Garit, J.A.; Casanola-Martin, G.M.; Le-Thi-Thu, H.; Barigye, S.J. A simple method to predict blood-brain barrier permeability of drug-like compounds using classification trees. Med. Chem. 2017, 13, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, H.; Wu, Z.; Wang, T.; Li, W.; Tang, Y.; Liu, G. In Silico Prediction of Blood–Brain Barrier Permeability of Compounds by Machine Learning and Resampling Methods. ChemMedChem 2018, 13, 2189–2201. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Divakaran, R.; Konda, L.S.K.; Kristam, R. A classification model for blood brain barrier penetration. J. Mol. Graph. Model. 2019, 96, 107516. [Google Scholar] [CrossRef] [PubMed]
- Adenot, M.; Lahana, R. Blood-Brain Barrier Permeation Models: Discriminating between Potential CNS and Non-CNS Drugs Including PGlycoprotein Substrates. J. Chem. Inf. Comput. Sci. 2004, 44, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xiao, J.; Zhou, N.; Zheng, M.; Luo, X.; Jiang, H.; Chen, K. A Genetic Algorithm Based Support Vector Machine Model for Blood-Brain Barrier Penetration Prediction. Biomed Res. Int. 2015, 2015, 292683. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zheng, F.; Zhan, C. Improved Prediction of Blood–Brain Barrier Permeability Through Machine Learning with Combined Use of Molecular Property-Based Descriptors and Fingerprints. AAPS J. 2018, 20, 54. [Google Scholar] [CrossRef] [PubMed]
- Brito-Sánchez, Y.; Marrero-Ponce, Y.; Barigye, S.J.; Yaber-Goenaga, I.; Morell Perez, C.; Le-Thi-Thu, H.; Cherkasov, A. Towards Better BBB Passage Prediction Using an Extensive and Curated Data Set. Mol. Inform. 2015, 34, 308–330. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Xia, L.Y.; Chen, H.H.; Huang, H.H.; Liang, Y. Improved Classification of Blood-Brain-Barrier Drugs Using Deep Learning. Sci. Rep. 2019, 9, 8802–8811. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Abraham, M.H.; Ibrahim, A.; Fish, P.V.; Cole, S.; Lewis, M.L.; de Groot, M.J.; Reynolds, D.P. Predicting Penetration Across the Blood-Brain Barrier from Simple Descriptors and Fragmentation Schemes. J. Chem. Inf. Model. 2007, 47, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Robu, R.; Holban, S. A genetic algorithm for classification. In Proceedings of the 2011 International Conference on Computers and Computing (ICCC’11), Wuhan, China, 13–14 August 2011. [Google Scholar]
- Ben-Hur, A.; Ong, C.S.; Sonnenburg, S.; Schölkopf, B.; Rätsch, G. Support vector machines and kernels for computational biology. PLoS Comput. Biol. 2008, 4, e1000173. [Google Scholar] [CrossRef] [PubMed]
- Linear Discriminant Analysis. Available online: http://www.saedsayad.com/lda.htm (accessed on 27 April 2025).
- Linear & Quadratic Discriminant Analysis·UC Business Analytics R Programming Guide. Available online: https://uc-r.github.io/discriminant_analysis (accessed on 27 April 2025).
- Artificial Neural Network. Available online: https://www.saedsayad.com/artificial_neural_network.htm (accessed on 27 April 2025).
- Yippy. A Beginner’s Guide to Neural Networks and Deep Learning. Available online: https://skymind.ai/wiki/neural-network (accessed on 27 April 2025).
- Park, S.H.; Goo, J.M.; Jo, C. Receiver Operating Characteristic. (ROC) Curve: Practical Review for Radiologists. Korean J. Radiol. 2004, 5, 11–18. [Google Scholar] [CrossRef] [PubMed]



| SVM (Linear) | SVM (RBF) | SVM (Polynomial) | LDA | QDA | kNN | ANN |
|---|---|---|---|---|---|---|
| 93.28 | 93.35 | 93.03 | 92.72 | 92.78 | 93.10 | 94.6% |
| Classifier Used | Features Chosen |
|---|---|
| SVM (linear) | PSA, logP, HD, pKa (strongest acidic), NRB |
| SVM (RBF) | HD, HA, pKa (strongest acidic) |
| SVM (polynomial) | HD, HA, NRB |
| LDA | All but the HA |
| QDA | MW, PSA, HD, pKa (strongest acidic), pKa (strongest basic) |
| k-NN | All but pKa (strongest basic) and NRB |
| ANN | MW, PSA, HD, pKa (strongest acidic), NRB |
| SVM (Linear) | SVM (RBF) | SVM (Polynomial) | LDA | QDA | kNN | ANN | |
|---|---|---|---|---|---|---|---|
| Without feature selection | 93.28% | 93.35% | 93.03% | 92.72% | 92.78% | 93.10% | 94.6% |
| Backward SFS | 94.67% | 92.79% | 88.4013% | 93.73% | 94.98% | 94.36% | 95.51% |
| GA: KNN based Fitness function | 94.67% | 94.04% | 84.01% | 94.36% | 96.23% | 92.79% | 95.89% |
| GA:SVM based | 93.73% | 94.98% | 96.23% | 94.98% | 95.62% | 93.42% | 96.04% |
| QDA + GA (kNN Based Fitness Function) | SVM + GA (SVM Based Fitness Function) | |
|---|---|---|
| True+ | 256 | 255 |
| True− | 51 | 52 |
| False+ | 11 | 4 |
| False− | 1 | 8 |
| SE | 95.88% | 98.45% |
| PP | 99.61% | 96.95% |
| SP | 98.07% | 86.67% |
| NP | 82.25% | 92.85% |
| ACC | 96.23% | 96.23% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saber, R.; Rihana, S. A Classification-Based Blood–Brain Barrier Model: A Comparative Approach. Pharmaceuticals 2025, 18, 773. https://doi.org/10.3390/ph18060773
Saber R, Rihana S. A Classification-Based Blood–Brain Barrier Model: A Comparative Approach. Pharmaceuticals. 2025; 18(6):773. https://doi.org/10.3390/ph18060773
Chicago/Turabian StyleSaber, Ralph, and Sandy Rihana. 2025. "A Classification-Based Blood–Brain Barrier Model: A Comparative Approach" Pharmaceuticals 18, no. 6: 773. https://doi.org/10.3390/ph18060773
APA StyleSaber, R., & Rihana, S. (2025). A Classification-Based Blood–Brain Barrier Model: A Comparative Approach. Pharmaceuticals, 18(6), 773. https://doi.org/10.3390/ph18060773




