Evidence for the Role of the Mitochondrial ABC Transporter MDL1 in the Uptake of Clozapine and Related Molecules into the Yeast Saccharomyces cerevisiae
Abstract
1. Introduction
- We adapt a comprehensive yeast knock-out library for assessing variation in sensitivity to cytotoxic concentrations of clozapine.
- We exploit modern cheminformatics methods to recognise safranin (and also bilirubin) as a fluorescent analogue of clozapine.
- We determine a mitochondrial location for the chief transporter (MDL1) in terms of physiological effects.
- Using flow cytometry, we determine the massively increased uptake of safranin in cells overexpressing MDL1.
- This mitochondrial transporter is, in fact, and unexpectedly, an ABC transporter acting not as an effluxer but as an influxer (or possibly an antiporter).
- This finding is consistent with other anomalous structural properties of the human homologue (ABCB10) [38] that were seen as being inconsistent with it acting solely as an effluxer.
- The finding of a concentrative uptake via MDL1 provides a simple explanation for the main source of cytotoxicity.
- Overall, the findings underscore the utility of the strategy adopted when seeking transporters for a drug whose transporterome remains unknown.
2. Results
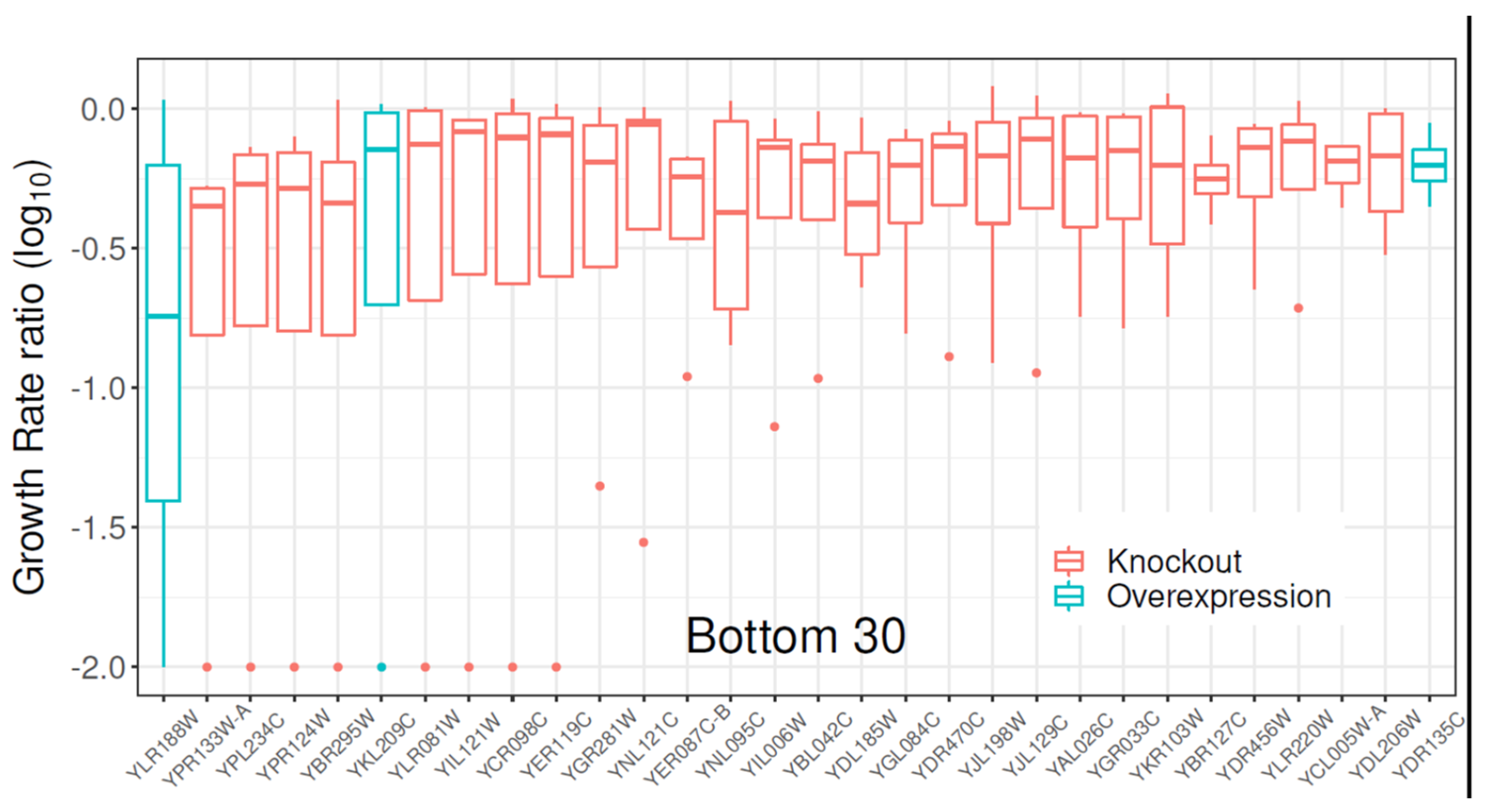
2.1. Gene Library Screening with Clozapine
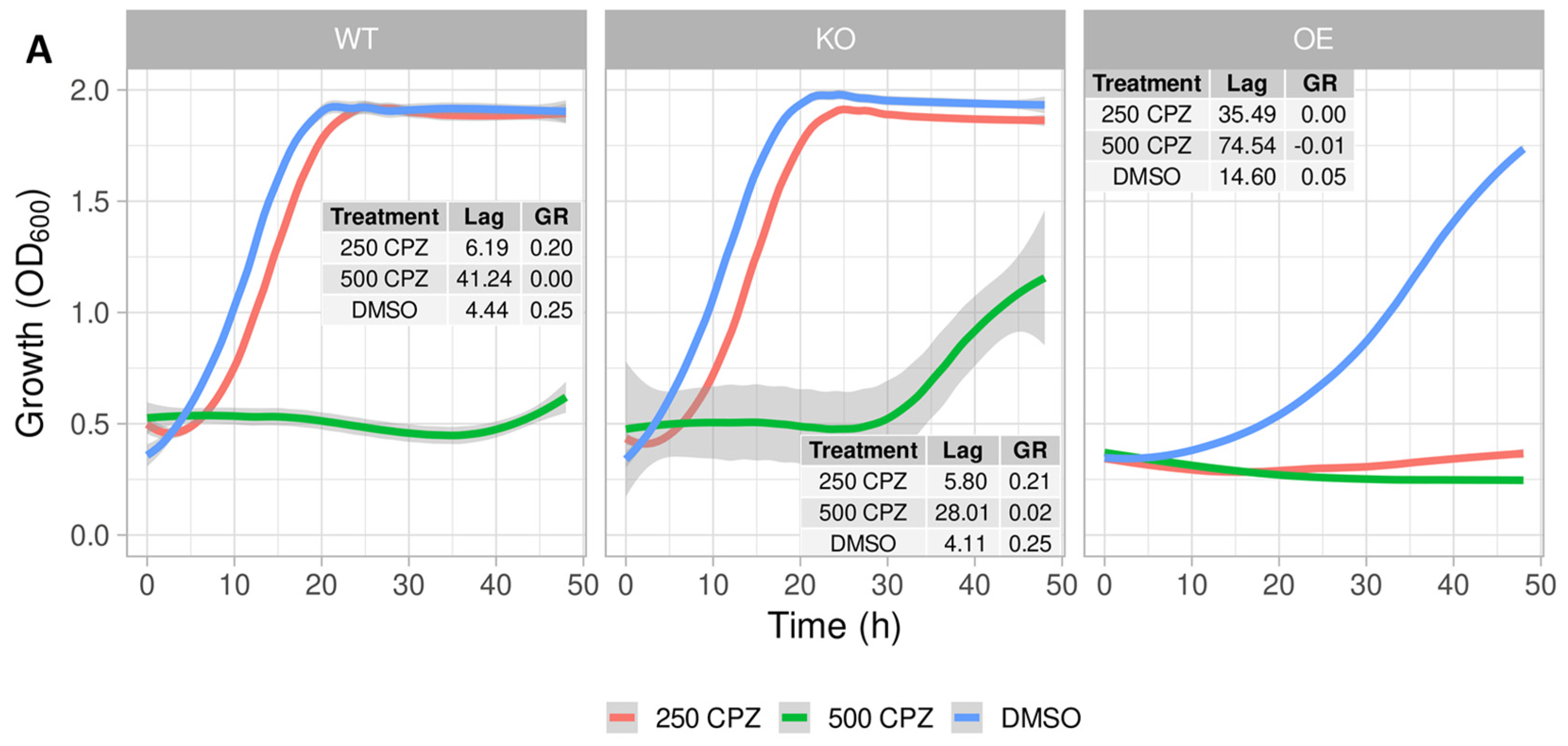
2.2. Growth Profile of S. cerevisiae Expressing the Mitochondrial ABC Transporter YLR188W
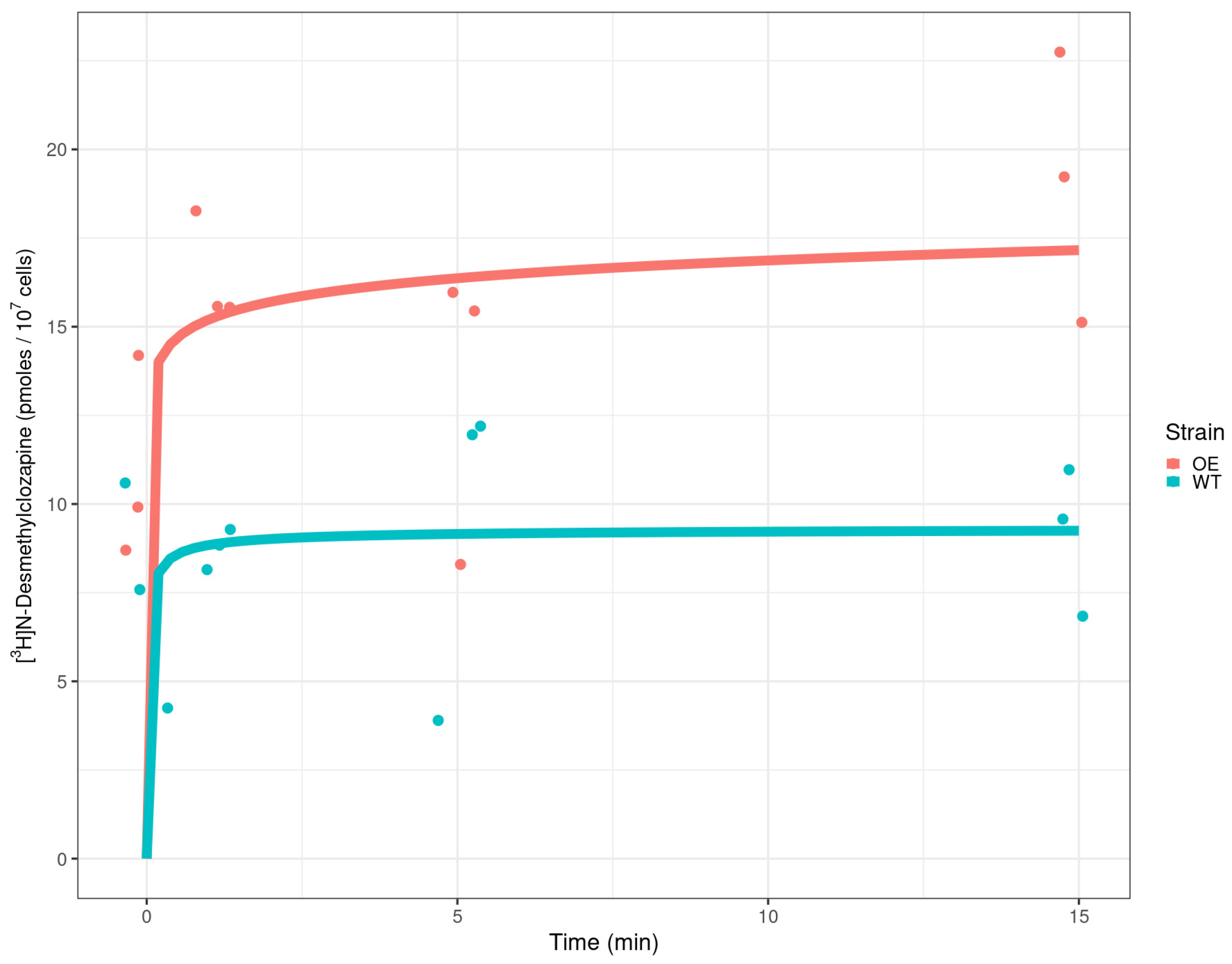
2.3. Detection of Radiolabelled Clozapine in Yeast Cells
2.4. Cheminformatics That Found Safranin O Similarities with Clozapine
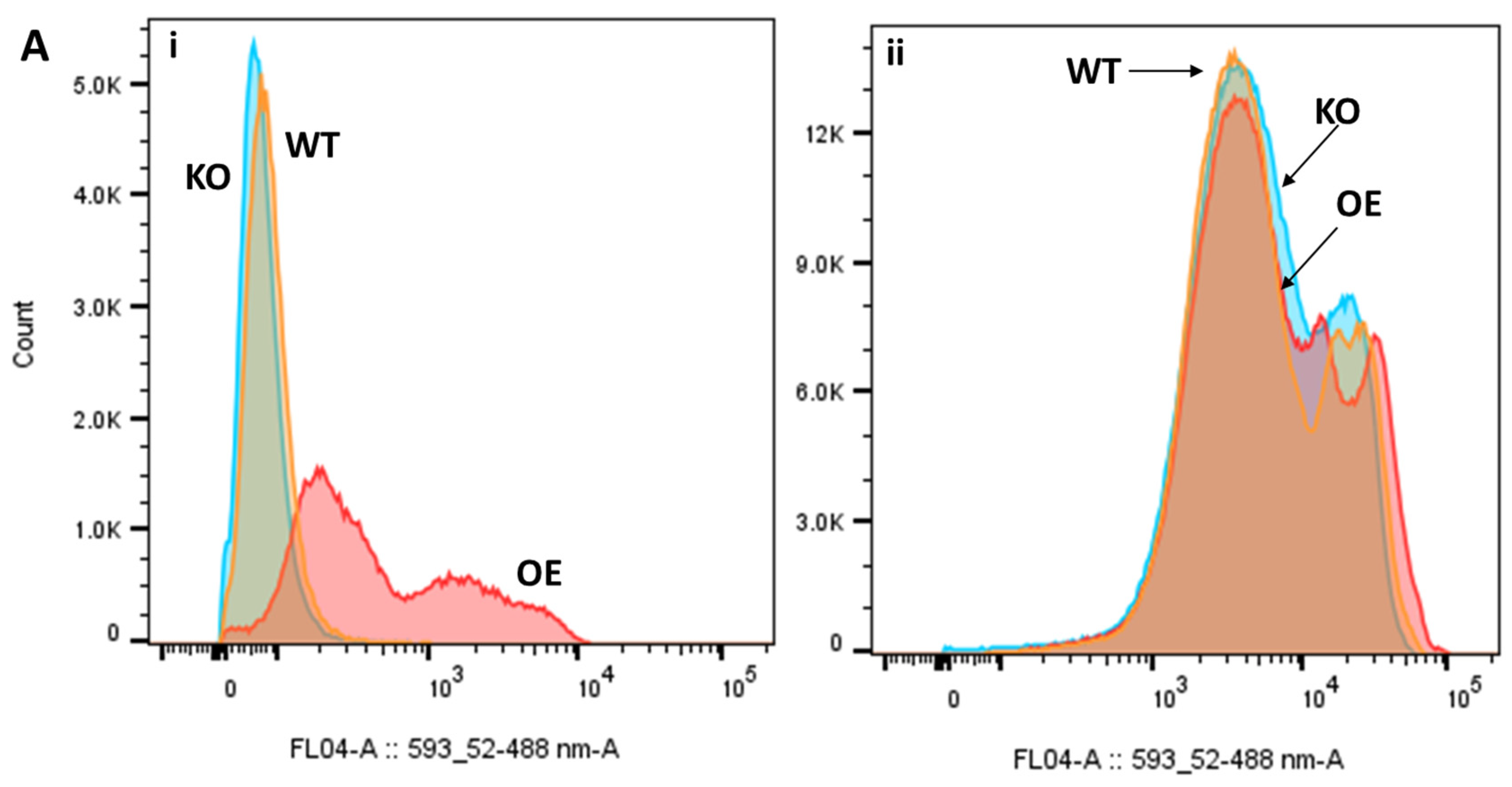
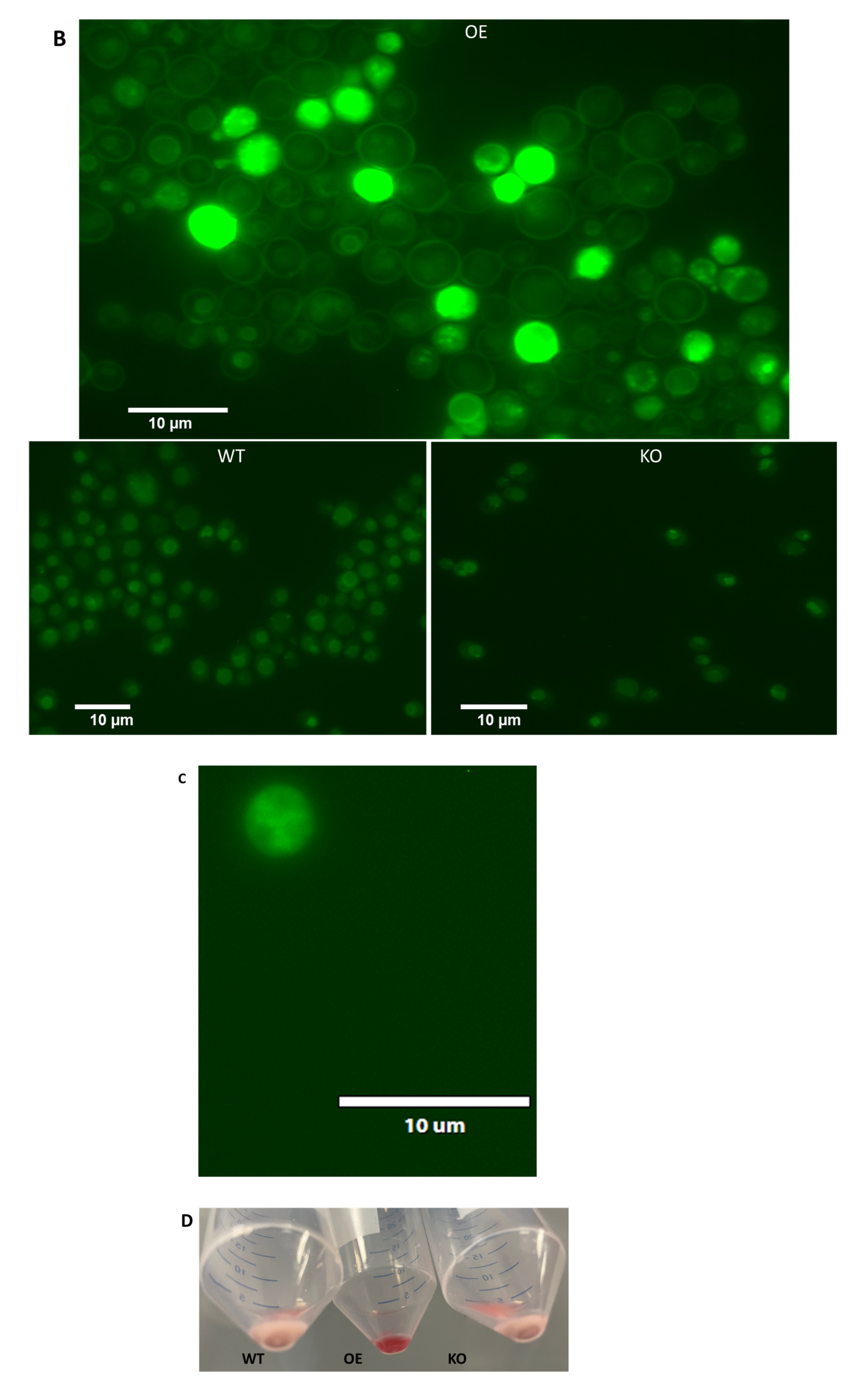
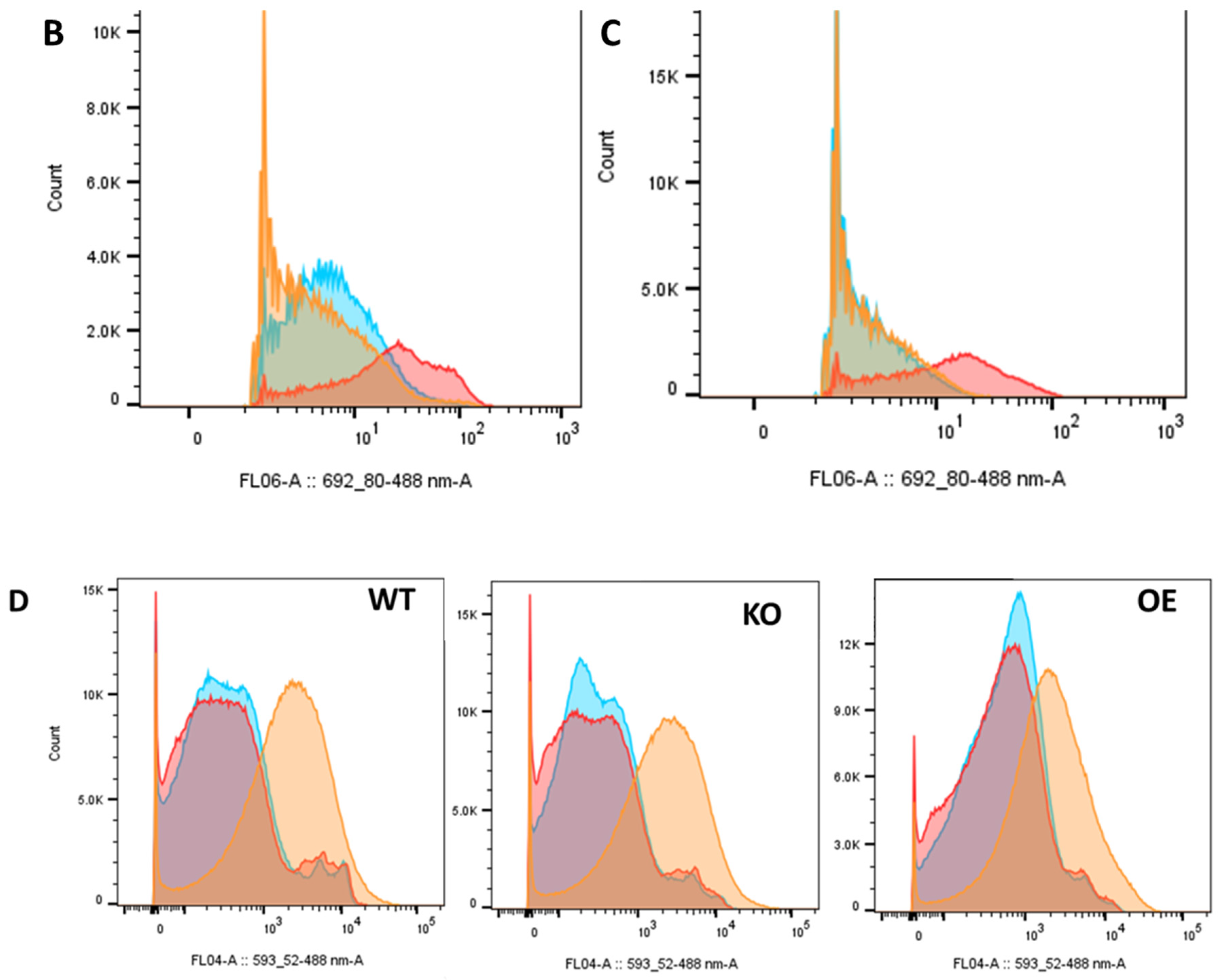
2.5. Differential Uptake of Safranin O as a Molecular Surrogate for Clozapine
2.6. Differential Uptake of Other Molecules
2.6.1. Chlorpromazine
2.6.2. Biliverdin
2.6.3. Prazosin
2.6.4. C-H2DCFDA
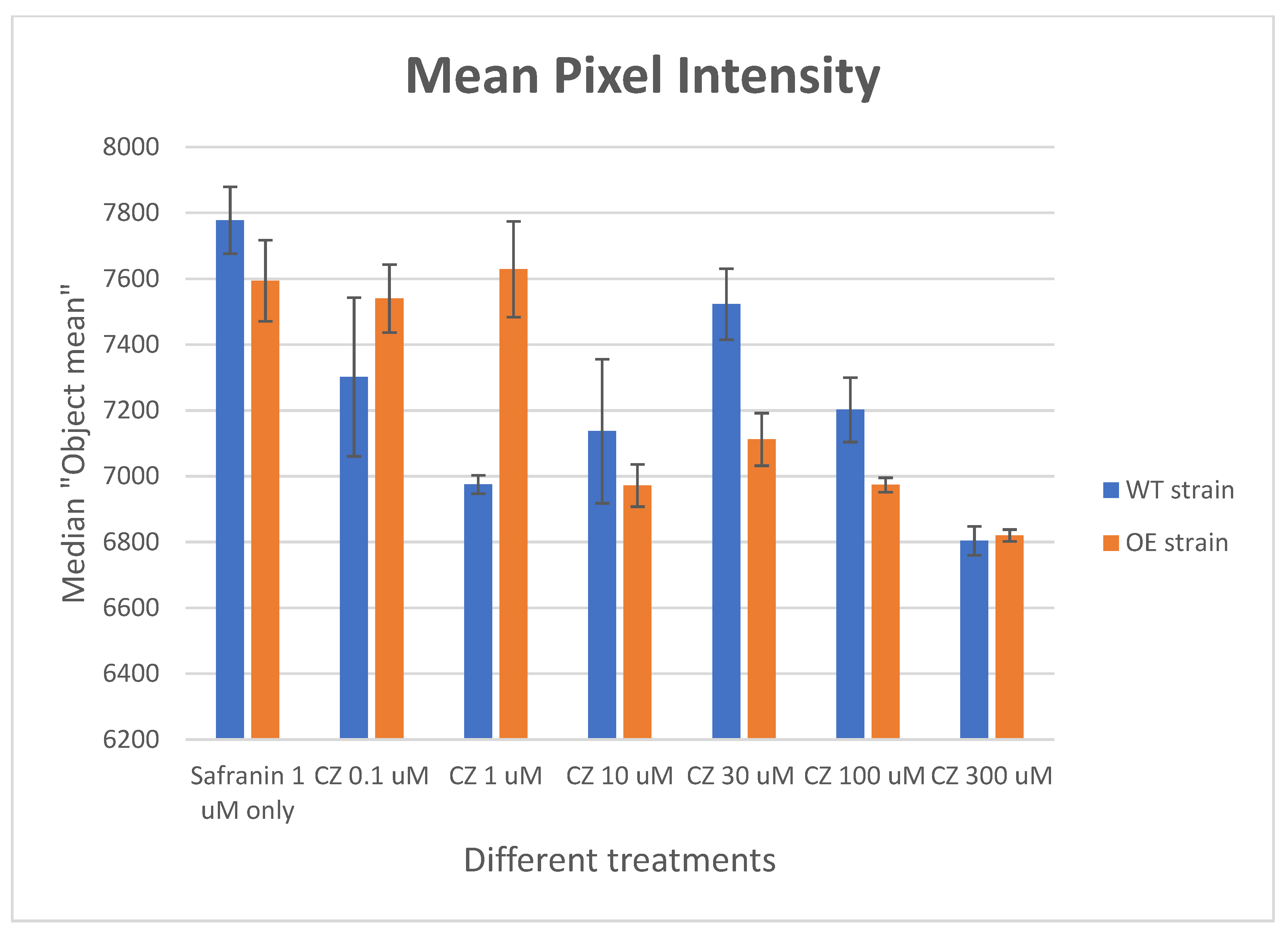
2.7. Effect of Clozapine on Safranin O Uptake
3. Discussion
4. Materials and Methods
4.1. Chemicals and Medium Components
4.2. Culture and Treatment of S. cerevisiae Strains
4.3. Detection of Radiolabelled Clozapine in Yeast Cells
4.4. Cheminformatics That Assessed Safranin O Similarities with Clozapine
4.5. Fluorescence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Correll, C.U.; Agid, O.; Crespo-Facorro, B.; de Bartolomeis, A.; Fagiolini, A.; Seppälä, N.; Howes, O.D. A Guideline and Checklist for Initiating and Managing Clozapine Treatment in Patients with Treatment-Resistant Schizophrenia. CNS Drugs 2022, 36, 659–679. [Google Scholar] [CrossRef] [PubMed]
- Hatano, M.; Kamei, H.; Takeuchi, I.; Gomi, K.; Sakakibara, T.; Hotta, S.; Esumi, S.; Tsubouchi, K.; Shimizu, Y.; Yamada, S. Long-term outcomes of delayed clozapine initiation in treatment-resistant schizophrenia: A multicenter retrospective cohort study. BMC Psychiatry 2023, 23, 673. [Google Scholar] [CrossRef] [PubMed]
- Siskind, D.; McCartney, L.; Goldschlager, R.; Kisely, S. Clozapine v. first- and second-generation antipsychotics in treatment-refractory schizophrenia: Systematic review and meta-analysis. Br. J. Psychiatry 2016, 209, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Qubad, M.; Bittner, R.A. Second to none: Rationale, timing, and clinical management of clozapine use in schizophrenia. Ther. Adv. Psychopharmacol. 2023, 13, 20451253231158152. [Google Scholar] [CrossRef] [PubMed]
- Freibüchler, A.; Seifert, R. Analysis of clinical studies on clozapine from 2012–2022. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Tanzer, T.; Pham, B.; Warren, N.; Barras, M.; Kisely, S.; Siskind, D. Overcoming clozapine’s adverse events: A narrative review of systematic reviews and meta-analyses. Expert. Opin. Drug Saf. 2024, 23, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Crilly, J. The history of clozapine and its emergence in the US market: A review and analysis. Hist. Psychiatry 2007, 18, 39–60. [Google Scholar] [CrossRef] [PubMed]
- Idänpään-Heikkilä, J.; Alhava, E.; Olkinuora, M.; Palva, I.P. Agranulocytosis during treatment with chlozapine. Eur. J. Clin. Pharmacol. 1977, 11, 193–198. [Google Scholar] [CrossRef] [PubMed]
- De Berardis, D.; Rapini, G.; Olivieri, L.; Di Nicola, D.; Tomasetti, C.; Valchera, A.; Fornaro, M.; Di Fabio, F.; Perna, G.; Di Nicola, M.; et al. Safety of antipsychotics for the treatment of schizophrenia: A focus on the adverse effects of clozapine. Ther. Adv. Drug Saf. 2018, 9, 237–256. [Google Scholar] [CrossRef]
- Skokou, M.; Karavia, E.A.; Drakou, Z.; Konstantinopoulou, V.; Kavakioti, C.A.; Gourzis, P.; Kypreos, K.E.; Andreopoulou, O. Adverse Drug Reactions in Relation to Clozapine Plasma Levels: A Systematic Review. Pharmaceuticals 2022, 15, 817. [Google Scholar] [CrossRef]
- Mijovic, A.; MacCabe, J.H. Clozapine-induced agranulocytosis. Ann. Hematol. 2020, 99, 2477–2482. [Google Scholar] [CrossRef] [PubMed]
- Alalawi, A.; Albalawi, E.; Aljohani, A.; Almutairi, A.; Alrehili, A.; Albalawi, A.; Aldhafiri, A. Decoding Clozapine-Induced Agranulocytosis: Unraveling Interactions and Mitigation Strategies. Pharmacy 2024, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Haddad, P.M.; Sharma, S.G. Adverse effects of atypical antipsychotics: Differential risk and clinical implications. CNS Drugs 2007, 21, 911–936. [Google Scholar] [CrossRef] [PubMed]
- Alvir, J.M.; Lieberman, J.A.; Safferman, A.Z.; Schwimmer, J.L.; Schaaf, J.A. Clozapine-induced agranulocytosis. Incidence and risk factors in the United States. N. Engl. J. Med. 1993, 329, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Leucht, S.; Corves, C.; Arbter, D.; Engel, R.R.; Li, C.; Davis, J.M. Second-generation versus first-generation antipsychotic drugs for schizophrenia: A meta-analysis. Lancet 2009, 373, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Li, K.J.; Gurrera, R.J.; Delisi, L.E. Potentially fatal outcomes associated with clozapine. Schizophr. Res. 2018, 199, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Wiciński, M.; Węclewicz, M.M. Clozapine-induced agranulocytosis/granulocytopenia: Mechanisms and monitoring. Curr. Opin. Hematol. 2018, 25, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Fenton, C.; Kang, C. Clozapine is the approved option in treatment-resistant schizophrenia and requires careful management. Drugs Ther. Perspect. 2023, 39, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Young, C.; Ifteni, P.; Kishimoto, T.; Xiang, Y.T.; Schulte, P.F.J.; Correll, C.U.; Taylor, D. Worldwide Differences in Regulations of Clozapine Use. CNS Drugs 2016, 30, 149–161. [Google Scholar] [CrossRef]
- Kar, N.; Barreto, S.; Chandavarkar, R. Clozapine Monitoring in Clinical Practice: Beyond the Mandatory Requirement. Clin. Psychopharmacol. Neurosci. 2016, 14, 323–329. [Google Scholar] [CrossRef]
- Kar, A.; Nutting, T.; Ikram, M.; Sullivan, C. The clozapine conundrum: Navigating neutropenia and the pursuit of effective care in treatment-resistant schizophrenia. Int. J. Psychiatry Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bergemann, N.; Abu-Tair, F.; Aderjan, R.; Kopitz, J. High clozapine concentrations in leukocytes in a patient who developed leukocytopenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 2007, 31, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Oliver, S.G. How drugs get into cells: Tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front. Pharmacol. 2014, 5, 231. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.G. The Structure and Mechanism of Drug Transporters. Methods Mol. Biol. 2021, 2342, 193–234. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Samanta, S.; Swainston, N. Deep learning and generative methods in cheminformatics and chemical biology: Navigating small molecule space intelligently. Biochem. J. 2020, 477, 4559–4580. [Google Scholar] [CrossRef] [PubMed]
- Markowicz-Piasecka, M.; Darlak, P.; Markiewicz, A.; Sikora, J.; Kumar Adla, S.; Bagina, S.; Huttunen, K.M. Current approaches to facilitate improved drug delivery to the central nervous system. Eur. J. Pharm. Biopharm. 2022, 181, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Doherty, E.; Ortega-Prieto, P.; Arizanova, J.; Fets, L. Membrane transporters in cell physiology, cancer metabolism and drug response. Dis. Model. Mech. 2023, 16, dmm050404. [Google Scholar] [CrossRef] [PubMed]
- César-Razquin, A.; Snijder, B.; Frappier-Brinton, T.; Isserlin, R.; Gyimesi, G.; Bai, X.; Reithmeier, R.A.; Hepworth, D.; Hediger, M.A.; Edwards, A.M.; et al. A call for systematic research on solute carriers. Cell 2015, 162, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Superti-Furga, G.; Lackner, D.; Wiedmer, T.; Ingles-Prieto, A.; Barbosa, B.; Girardi, E.; Goldman, U.; Gürtl, B.; Klavins, K.; Klimek, C.; et al. The RESOLUTE consortium: Unlocking SLC transporters for drug discovery. Nat. Rev. Drug Discov. 2020, 19, 429–430. [Google Scholar] [CrossRef]
- Kell, D.B. The transporter-mediated cellular uptake and efflux of pharmaceutical drugs and biotechnology products: How and why phospholipid bilayer transport is negligible in real biomembranes. Molecules 2021, 26, 5629. [Google Scholar] [CrossRef]
- Dickens, D.; Rädisch, S.; Chiduza, G.N.; Giannoudis, A.; Cross, M.J.; Malik, H.; Schaeffeler, E.; Sison-Young, R.L.; Wilkinson, E.L.; Goldring, C.E.; et al. Cellular uptake of the atypical antipsychotic clozapine is a carrier-mediated process. Mol. Pharm. 2018, 15, 3557–3572. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G. Yeast as a navigational aid in genome analysis. Microbiology 1997, 143, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Herrgård, M.J.; Swainston, N.; Dobson, P.; Dunn, W.B.; Arga, K.Y.; Arvas, M.; Blüthgen, N.; Borger, S.; Costenoble, R.; Heinemann, M.; et al. A consensus yeast metabolic network obtained from a community approach to systems biology. Nat. Biotechnol. 2008, 26, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Lanthaler, K.; Bilsland, E.; Dobson, P.; Moss, H.J.; Pir, P.; Kell, D.B.; Oliver, S.G. Genome-wide assessment of the carriers involved in the cellular uptake of drugs: A model system in yeast. BMC Biol. 2011, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.E.; Radic, B.; Mayor-Ruiz, C.; Blomen, V.A.; Trefzer, C.; Kandasamy, R.K.; Huber, K.V.M.; Gridling, M.; Chen, D.; Klampfl, T.; et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 2014, 10, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Girardi, E.; César-Razquin, A.; Lindinger, S.; Papakostas, K.; Lindinger, S.; Konecka, J.; Hemmerich, J.; Kickinger, S.; Kartnig, F.; Gürtl, B.; et al. A widespread role for SLC transmembrane transporters in resistance to cytotoxic drugs. Nat. Chem. Biol. 2020, 16, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Shintre, C.A.; Pike, A.C.; Li, Q.; Kim, J.I.; Barr, A.J.; Goubin, S.; Shrestha, L.; Yang, J.; Berridge, G.; Ross, J.; et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. USA 2013, 110, 9710–9715. [Google Scholar] [CrossRef] [PubMed]
- Young, L.; Leonhard, K.; Tatsuta, T.; Trowsdale, J.; Langer, T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science 2001, 291, 2135–2138. [Google Scholar] [CrossRef]
- Herget, M.; Tampé, R. Intracellular peptide transporters in human—Compartmentalization of the “peptidome”. Pflugers Arch. 2007, 453, 591–600. [Google Scholar] [CrossRef]
- Chloupková, M.; LeBard, L.S.; Koeller, D.M. MDL1 is a high copy suppressor of ATM1: Evidence for a role in resistance to oxidative stress. J. Mol. Biol. 2003, 331, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Radi, M.S.; Munro, L.J.; Sora, J.E.S.; Kim, S.H.; Feist, A.M.; Kell, D.B. Understanding functional redundancy and promiscuity of multidrug transporters in E. coli under lipophilic cation stress. Membranes 2022, 12, 1264. [Google Scholar] [CrossRef] [PubMed]
- Salcedo-Sora, J.E.; Jindal, S.; O’Hagan, S.; Kell, D.B. A palette of fluorophores that are differentially accumulated by wild-type and mutant strains of Escherichia coli: Surrogate ligands for bacterial membrane transporters. Microbiology 2021, 167, 001016. [Google Scholar] [CrossRef]
- Jindal, S.; Yang, L.; Day, P.J.; Kell, D.B. Involvement of multiple influx and efflux transporters in the accumulation of cationic fluorescent dyes by Escherichia coli. BMC Microbiol. 2019, 19, 195. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B. A protet-based, protonic charge transfer model of energy coupling in oxidative and photosynthetic phosphorylation. Adv. Micr Physiol. 2021, 78, 1–177. [Google Scholar] [CrossRef]
- Gasteiger, J. (Ed.) Handbook of Chemoinformatics: From Data to Knowledge; Wiley/VCH: Weinheim, Germany, 2003. [Google Scholar]
- O’Hagan, S.; Swainston, N.; Handl, J.; Kell, D.B. A ‘rule of 0.5’ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics 2015, 11, 323–339. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. The KNIME workflow environment and its applications in Genetic Programming and machine learning. Genetic Progr Evol. Mach. 2015, 16, 387–391. [Google Scholar] [CrossRef][Green Version]
- O’Hagan, S.; Kell, D.B. Analysis of drug-endogenous human metabolite similarities in terms of their maximum common substructures. J. Cheminform 2017, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. Consensus rank orderings of molecular fingerprints illustrate the ‘most genuine’ similarities between marketed drugs and small endogenous human metabolites, but highlight exogenous natural products as the most important ‘natural’ drug transporter substrates. Admet Dmpk 2017, 5, 85–125. [Google Scholar] [CrossRef]
- Mazanetz, M.P.; Marmon, R.J.; Reisser, C.B.T.; Morao, I. Drug discovery applications for KNIME: An open source data mining platform. Curr. Top. Med. Chem. 2012, 12, 1965–1979. [Google Scholar] [CrossRef]
- Roughley, S.D. Five Years of the KNIME Vernalis Cheminformatics Community Contribution. Curr. Med. Chem. 2020, 27, 6495–6522. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; O’Hagan, S.; Swainston, N.; Roberts, T.J.; Kell, D.B. VAE-Sim: A novel molecular similarity measure based on a variational autoencoder. Molecules 2020, 25, 3446. [Google Scholar] [CrossRef]
- Cao, S.; Yang, Y.; He, L.; Hang, Y.; Yan, X.; Shi, H.; Wu, J.; Ouyang, Z. Cryo-EM structures of mitochondrial ABC transporter ABCB10 in apo and biliverdin-bound form. Nat. Commun. 2023, 14, 2030. [Google Scholar] [CrossRef] [PubMed]
- Shum, M.; Shintre, C.A.; Althoff, T.; Gutierrez, V.; Segawa, M.; Saxberg, A.D.; Martinez, M.; Adamson, R.; Young, M.R.; Faust, B.; et al. ABCB10 exports mitochondrial biliverdin, driving metabolic maladaptation in obesity. Sci. Transl. Med. 2021, 13, eabd1869. [Google Scholar] [CrossRef]
- Schaedler, T.A.; Faust, B.; Shintre, C.A.; Carpenter, E.P.; Srinivasan, V.; van Veen, H.W.; Balk, J. Structures and functions of mitochondrial ABC transporters. Biochem. Soc. Trans. 2015, 43, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Grixti, J.; O’Hagan, S.; Day, P.J.; Kell, D.B. Enhancing drug efficacy and therapeutic index through cheminformatics-based selection of small molecule binary weapons that improve transporter-mediated targeting: A cytotoxicity system based on gemcitabine. Front. Pharmacol. 2017, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. Supravital staining of mitochondria with phenosafranin dyes. Biochim. Biophys. Acta 1953, 11, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Jungwirth, H.; Kuchler, K. Yeast ABC transporters—A tale of sex, stress, drugs and aging. FEBS Lett. 2006, 580, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Zutz, A.; Gompf, S.; Schagger, H.; Tampé, R. Mitochondrial ABC proteins in health and disease. Biochim. Biophys. Acta 2009, 1787, 681–690. [Google Scholar] [CrossRef]
- Ryter, S.W. Significance of Heme and Heme Degradation in the Pathogenesis of Acute Lung and Inflammatory Disorders. Int. J. Mol. Sci. 2021, 22, 5509. [Google Scholar] [CrossRef]
- Sadler, J.C.; Currin, A.; Kell, D.B. Ultra-high-throughput functional enrichment of large monoamine oxidase (MAO-N) libraries by fluorescence activated cell sorting. Analyst 2018, 143, 4747–4755. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.; Green, E.M. Assessing Yeast Cell Survival Following Hydrogen Peroxide Exposure. Bio Protoc. 2019, 9, e3149. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B. Iron behaving badly: Inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Med. Genom. 2009, 2, 2. [Google Scholar]
- Kell, D.B. Towards a unifying, systems biology understanding of large-scale cellular death and destruction caused by poorly liganded iron: Parkinson’s, Huntington’s, Alzheimer’s, prions, bactericides, chemical toxicology and others as examples. Arch. Toxicol. 2010, 577, 825–889. [Google Scholar]
- Grixti, J.M.; Theron, C.W.; Salcedo-Sora, J.E.; Pretorius, E.; Kell, D.B. Automated microscopic measurement of fibrinaloid microclots and their degradation by nattokinase, the main natto protease. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kell, D.B.; Knowles, J.D. The role of modeling in systems biology. In System Modeling in Cellular Biology: From Concepts to Nuts and Bolts; Szallasi, Z., Stelling, J., Periwal, V., Eds.; MIT Press: Cambridge, UK, 2006; pp. 3–18. [Google Scholar]
- Oliver, S.G. Functional genomics: Lessons from yeast. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002, 357, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Grenson, M.; Mousset, M.; Wiame, J.M.; Bechet, J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta 1966, 127, 325–338. [Google Scholar] [CrossRef]
- Hoffmann, W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. A transmembrane protein without N-terminal hydrophobic signal sequence. J. Biol. Chem. 1985, 260, 11831–11837. [Google Scholar] [CrossRef] [PubMed]
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas system for yeast genome engineering: Advances and applications. FEMS Yeast Res. 2017, 17, fox030. [Google Scholar] [CrossRef]
- Wang, G.; Møller-Hansen, I.; Babaei, M.; D’Ambrosio, V.; Christensen, H.B.; Darbani, B.; Jensen, M.K.; Borodina, I. Transportome-wide engineering of Saccharomyces cerevisiae. Metab. Eng. 2021, 64, 52–63. [Google Scholar] [CrossRef]
- Tedeschi, H. The transport of cations in mitochondria. Biochim. Biophys. Acta 1981, 639, 157–196. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Smith, R.A.J. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 629–656. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. Targeting lipophilic cations to mitochondria. Biochim. Biophys. Acta 2008, 1777, 1028–1031. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.F.; Da Ros, T.; Blaikie, F.H.; Prime, T.A.; Porteous, C.M.; Severina, I.I.; Skulachev, V.P.; Kjaergaard, H.G.; Smith, R.A.J.; Murphy, M.P. Accumulation of lipophilic dications by mitochondria and cells. Biochem. J. 2006, 400, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Allikmets, R.; Gerrard, B.; Stewart, C.; Kistler, A.; Shafer, B.; Michaelis, S.; Strathern, J. Mapping and sequencing of two yeast genes belonging to the ATP-binding cassette superfamily. Yeast 1994, 10, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Melin, J.; Kilisch, M.; Neumann, P.; Lytovchenko, O.; Gomkale, R.; Schendzielorz, A.; Schmidt, B.; Liepold, T.; Ficner, R.; Jahn, O.; et al. A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim. Biophys. Acta 2015, 1853, 1850–1859. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Jung, S.J.; Kim, H.; Kim, H. Mode of membrane insertion of individual transmembrane segments in Mdl1 and Mdl2, multi-spanning mitochondrial ABC transporters. FEBS Lett. 2014, 588, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Miljkovic, M.; Seguin, A.; Jia, X.; Cox, J.E.; Catrow, J.L.; Bergonia, H.; Phillips, J.D.; Stephens, W.Z.; Ward, D.M. Loss of the mitochondrial protein Abcb10 results in altered arginine metabolism in MEL and K562 cells and nutrient stress signaling through ATF4. J. Biol. Chem. 2023, 299, 104877. [Google Scholar] [CrossRef]
- Casademont, J.; Garrabou, G.; Miro, O.; Lopez, S.; Pons, A.; Bernardo, M.; Cardellach, F. Neuroleptic treatment effect on mitochondrial electron transport chain: Peripheral blood mononuclear cells analysis in psychotic patients. J. Clin. Psychopharmacol. 2007, 27, 284–288. [Google Scholar] [CrossRef]
- Hillenmeyer, M.E.; Ericson, E.; Davis, R.W.; Nislow, C.; Koller, D.; Giaever, G. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010, 11, R30. [Google Scholar] [CrossRef] [PubMed]
- Horng, Y.C.; Cobine, P.A.; Maxfield, A.B.; Carr, H.S.; Winge, D.R. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 2004, 279, 35334–35340. [Google Scholar] [CrossRef] [PubMed]
- Abajian, C.; Yatsunyk, L.A.; Ramirez, B.E.; Rosenzweig, A.C. Yeast cox17 solution structure and Copper(I) binding. J. Biol. Chem. 2004, 279, 53584–53592. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Jiang, Y.; Yang, Y.; Peng, Y.; Li, C. Copper metabolism in Saccharomyces cerevisiae: An update. Biometals 2021, 34, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Voronova, A.; Meyer-Klaucke, W.; Meyer, T.; Rompel, A.; Krebs, B.; Kazantseva, J.; Sillard, R.; Palumaa, P. Oxidative switches in functioning of mammalian copper chaperone Cox17. Biochem. J. 2007, 408, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, Y.; Shi, H.; Peng, Y.; Fan, X.; Li, C. The molecular mechanisms of copper metabolism and its roles in human diseases. Pflugers Arch. 2020, 472, 1415–1429. [Google Scholar] [CrossRef] [PubMed]
- Salcedo-Sora, J.E.; Robison, A.T.R.; Zaengle-Barone, J.; Franz, K.J.; Kell, D.B. Membrane transporters involved in the antimicrobial activities of pyrithione in Escherichia coli. Molecules 2021, 26, 5826. [Google Scholar] [CrossRef] [PubMed]
- Baylay, A.J.; Ivens, A.; Piddock, L.J. A novel gene amplification causes upregulation of the PatAB ABC transporter and fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2015, 59, 3098–3108. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Locher, K.P. Structure and Mechanism of Human ABC Transporters. Annu. Rev. Biophys. 2023, 52, 275–300. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, T.; Zhang, L.; Zhu, P.; Deng, M.; Huang, C.; Hu, T.; Jiang, L.; Li, J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016, 370, 153–164. [Google Scholar] [CrossRef]
- Orelle, C.; Mathieu, K.; Jault, J.M. Multidrug ABC transporters in bacteria. Res. Microbiol. 2019, 170, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Aller, S.G.; Beis, K.; Carpenter, E.P.; Chang, G.; Chen, L.; Dassa, E.; Dean, M.; Duong Van Hoa, F.; Ekiert, D.; et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 2020, 594, 3767–3775. [Google Scholar] [CrossRef] [PubMed]
- Ter Beek, J.; Guskov, A.; Slotboom, D.J. Structural diversity of ABC transporters. J. Gen. Physiol. 2014, 143, 419–435. [Google Scholar] [CrossRef]
- Lewinson, O.; Livnat-Levanon, N. Mechanism of Action of ABC Importers: Conservation, Divergence, and Physiological Adaptations. J. Mol. Biol. 2017, 429, 606–619. [Google Scholar] [CrossRef]
- Mendes, P.; Girardi, E.; Superti-Furga, G.; Kell, D.B. Why most transporter mutations that cause antibiotic resistance are to efflux pumps rather than to import transporters. bioRxiv 2001. [Google Scholar] [CrossRef]
- Elmorsy, E.; Smith, P.A. Bioenergetic disruption of human micro-vascular endothelial cells by antipsychotics. Biochem. Biophys. Res. Commun. 2015, 460, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef]
- Stovicek, V.; Borja, G.M.; Forster, J.; Borodina, I. EasyClone 2.0: Expanded toolkit of integrative vectors for stable gene expression in industrial Saccharomyces cerevisiae strains. J. Ind. Microbiol. Biotechnol. 2015, 42, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Jessop-Fabre, M.M.; Jakociunas, T.; Stovicek, V.; Dai, Z.J.; Jensen, M.K.; Keasling, J.D.; Borodina, I. EasyClone-MarkerFree: A vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol. J. 2016, 11, 1110–1117. [Google Scholar] [CrossRef]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth rates made easy. Mol. Biol. Evol. 2014, 31, 232–238. [Google Scholar] [CrossRef]
- Vaas, L.A.; Sikorski, J.; Michael, V.; Göker, M.; Klenk, H.P. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 2012, 7, e34846. [Google Scholar] [CrossRef] [PubMed]
- Misonou, Y.; Kikuchi, M.; Sato, H.; Inai, T.; Kuroiwa, T.; Tanaka, K.; Miyakawa, I. Aldehyde dehydrogenase, Ald4p, is a major component of mitochondrial fluorescent inclusion bodies in the yeast Saccharomyces cerevisiae. Biol. Open 2014, 3, 387–396. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. MetMaxStruct: A Tversky-similarity-based strategy for analysing the (sub)structural similarities of drugs and endogenous metabolites. Front. Pharmacol. 2016, 7, 266. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. Structural similarities between some common fluorophores used in biology, marketed drugs, endogenous metabolites, and natural products. Marine Drugs 2020, 18, 582. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.D.; Kell, D.B. FragNet, a contrastive learning-based transformer model for clustering, interpreting, visualising and navigating chemical space. Molecules 2021, 26, 2065. [Google Scholar] [CrossRef] [PubMed]
- Åkerman, K.E.O.; Wikström, M.K.F. Safranine as a probe of the mitochondrial membrane potential. FEBS Lett. 1976, 68, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analysis. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [CrossRef]
- van der Hoek, S.A.; Borodina, I. Transporter engineering in microbial cell factories: The ins, the outs, and the in-betweens. Curr. Opin. Biotechnol. 2020, 66, 186–194. [Google Scholar] [CrossRef]
- Kell, D.B. Control of metabolite efflux in microbial cell factories: Current advances and future prospects. In Fermentation Microbiology and Biotechnology, 4th ed.; El-Mansi, E.M.T., Nielsen, J., Mousdale, D., Allman, T., Carlson, R., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 117–138. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theron, C.W.; Salcedo-Sora, J.E.; Grixti, J.M.; Møller-Hansen, I.; Borodina, I.; Kell, D.B. Evidence for the Role of the Mitochondrial ABC Transporter MDL1 in the Uptake of Clozapine and Related Molecules into the Yeast Saccharomyces cerevisiae. Pharmaceuticals 2024, 17, 938. https://doi.org/10.3390/ph17070938
Theron CW, Salcedo-Sora JE, Grixti JM, Møller-Hansen I, Borodina I, Kell DB. Evidence for the Role of the Mitochondrial ABC Transporter MDL1 in the Uptake of Clozapine and Related Molecules into the Yeast Saccharomyces cerevisiae. Pharmaceuticals. 2024; 17(7):938. https://doi.org/10.3390/ph17070938
Chicago/Turabian StyleTheron, Chrispian W., J. Enrique Salcedo-Sora, Justine M. Grixti, Iben Møller-Hansen, Irina Borodina, and Douglas B. Kell. 2024. "Evidence for the Role of the Mitochondrial ABC Transporter MDL1 in the Uptake of Clozapine and Related Molecules into the Yeast Saccharomyces cerevisiae" Pharmaceuticals 17, no. 7: 938. https://doi.org/10.3390/ph17070938
APA StyleTheron, C. W., Salcedo-Sora, J. E., Grixti, J. M., Møller-Hansen, I., Borodina, I., & Kell, D. B. (2024). Evidence for the Role of the Mitochondrial ABC Transporter MDL1 in the Uptake of Clozapine and Related Molecules into the Yeast Saccharomyces cerevisiae. Pharmaceuticals, 17(7), 938. https://doi.org/10.3390/ph17070938







