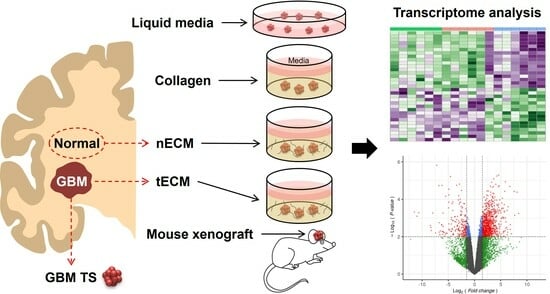
Comparison of Glioblastoma Cell Culture Platforms Based on Transcriptional Similarity with Paired Tissue
Abstract
1. Introduction
2. Results
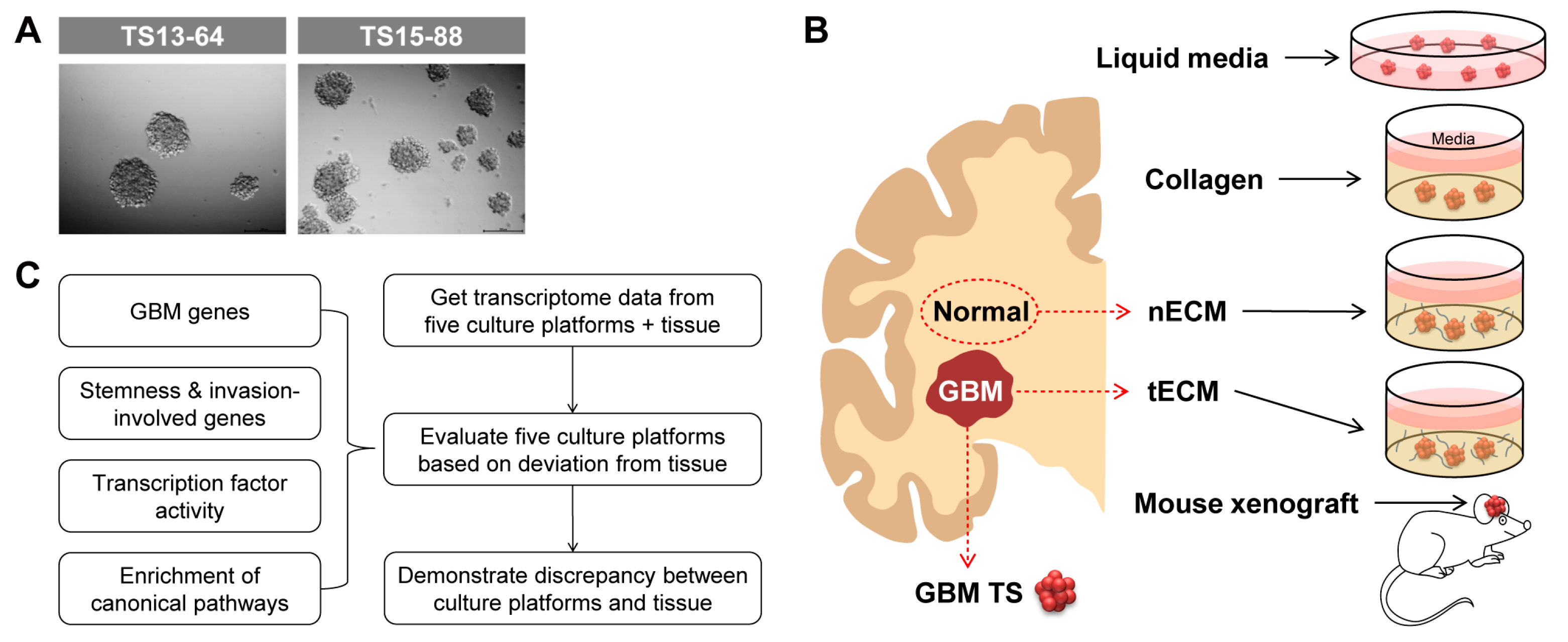
2.1. Five Culture Platforms for GBM TSs
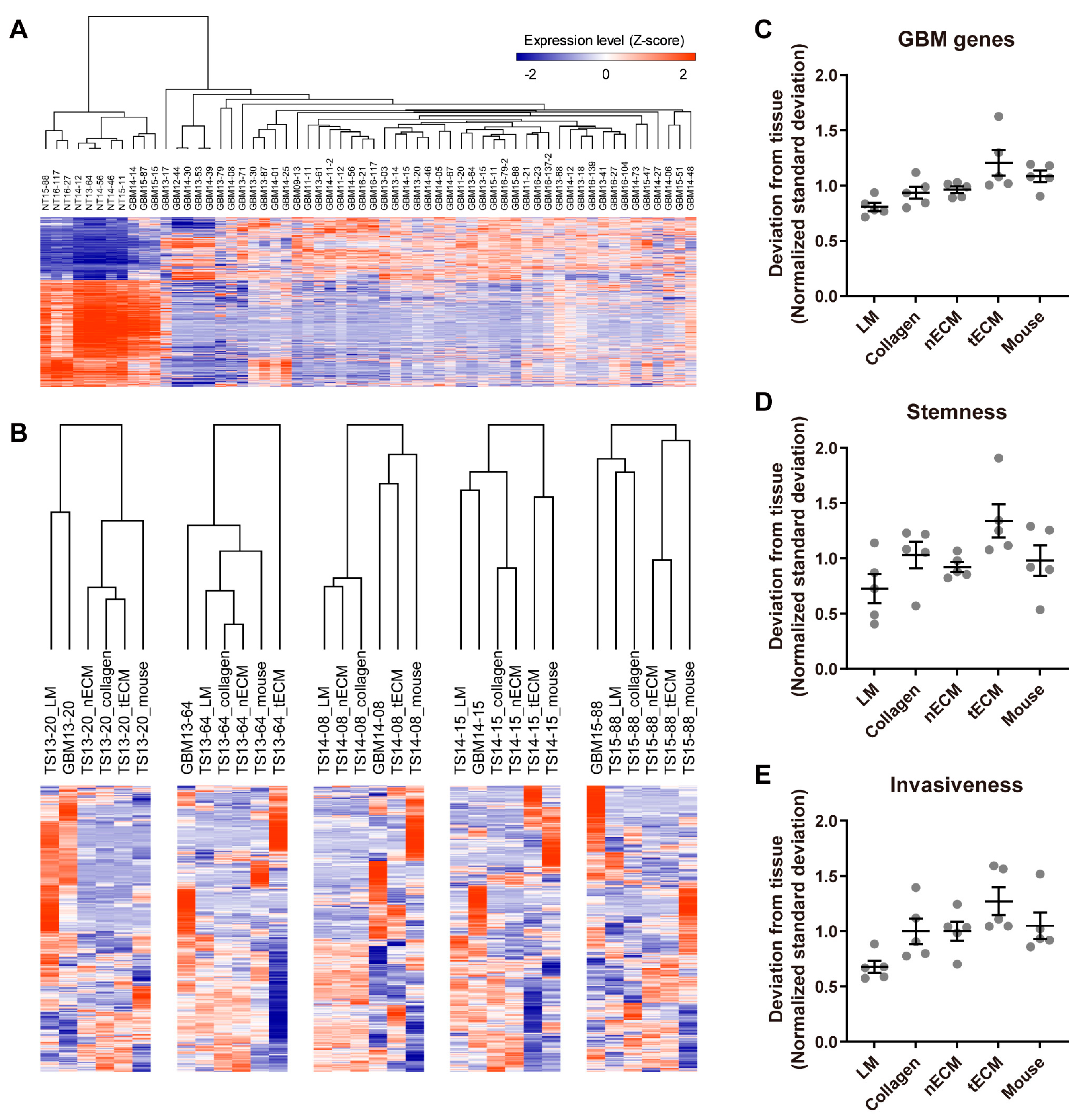
2.2. Differential Expression of GBM-Associated Genes among Culture Platforms
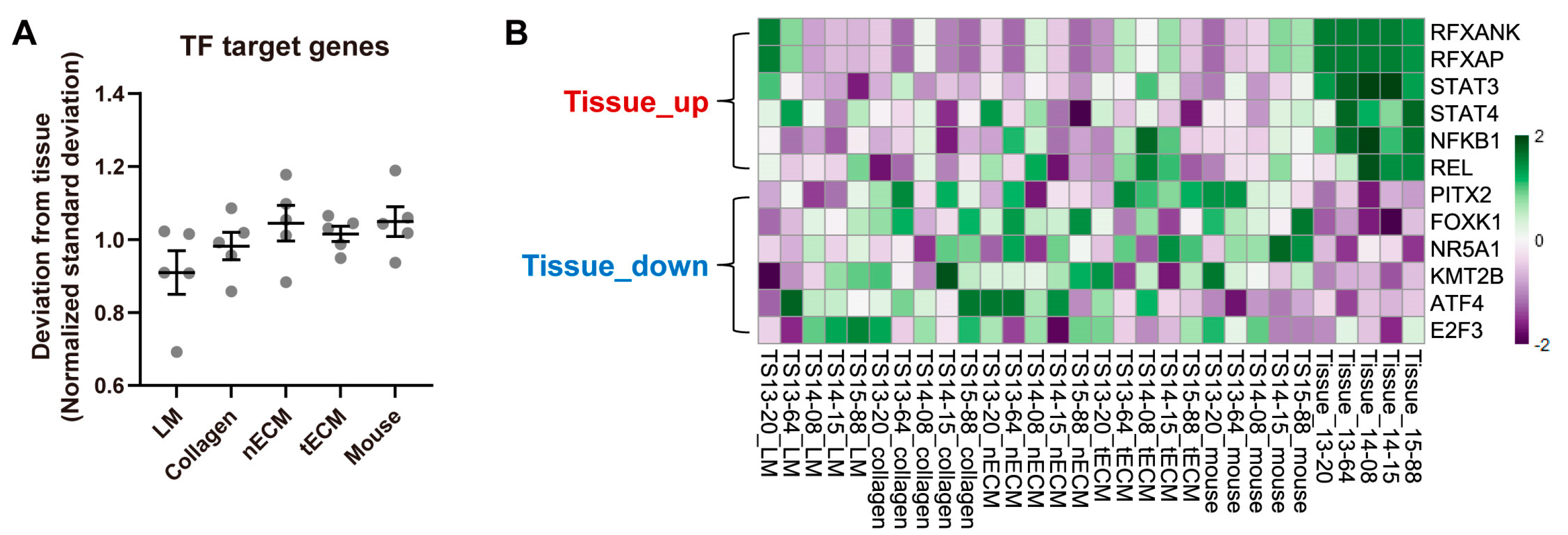
2.3. Differential Activity of TFs among Culture Platforms
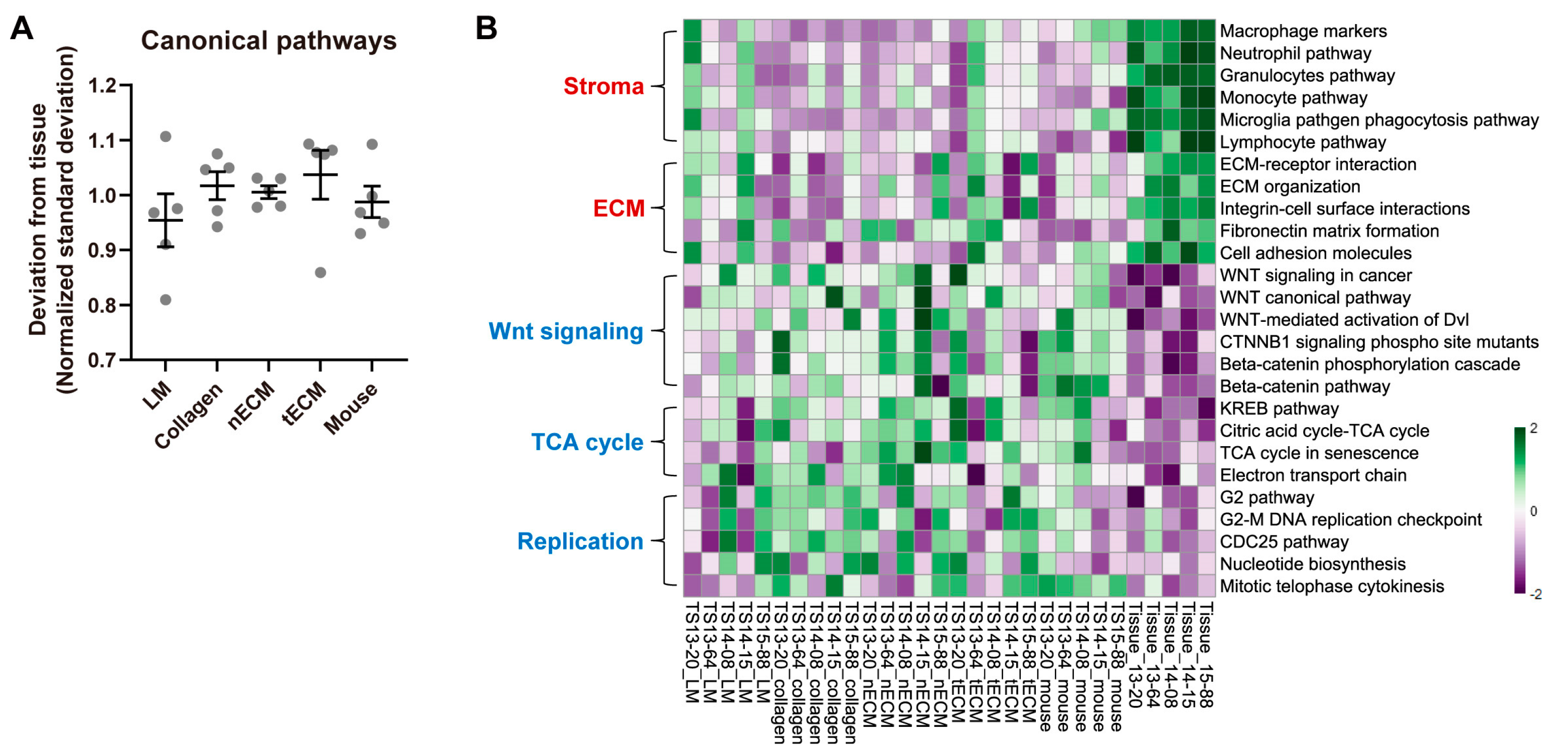
2.4. Differential Activity of Canonical Signaling Pathways among Culture Platforms
3. Discussion
4. Materials and Methods
4.1. Patient Information and Isolation of GBM TSs
4.2. Preparation of Patient-Derived ECM
4.3. Protein Extraction from Patient-Derived ECM
4.4. Nano LC–ESI-MS/MS Analysis
4.5. Proteome Data Analysis
4.6. Analysis of Gene Expression Profile
4.7. Mouse Orthotopic Xenograft Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoshide, R.; Jandial, R. 2016 World Health Organization Classification of Central Nervous System Tumors: An Era of Molecular Biology. World Neurosurg. 2016, 94, 561–562. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shim, J.K.; Yoon, S.J.; Kim, S.H.; Chang, J.H.; Kang, S.G. Transcriptome profiling-based identification of prognostic subtypes and multi-omics signatures of glioblastoma. Sci. Rep. 2019, 9, 10555. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e46. [Google Scholar] [CrossRef]
- Riedel, N.C.; de Faria, F.W.; Alfert, A.; Bruder, J.M.; Kerl, K. Three-Dimensional Cell Culture Systems in Pediatric and Adult Brain Tumor Precision Medicine. Cancers 2022, 14, 5972. [Google Scholar] [CrossRef] [PubMed]
- Bagley, S.J.; Kothari, S.; Rahman, R.; Lee, E.Q.; Dunn, G.P.; Galanis, E.; Chang, S.M.; Nabors, L.B.; Ahluwalia, M.S.; Stupp, R.; et al. Glioblastoma Clinical Trials: Current Landscape and Opportunities for Improvement. Clin. Cancer Res. 2022, 28, 594–602. [Google Scholar] [CrossRef] [PubMed]
- LeSavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-generation cancer organoids. Nat. Mater. 2022, 21, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Wanigasekara, J.; Cullen, P.J.; Bourke, P.; Tiwari, B.; Curtin, J.F. Advances in 3D culture systems for therapeutic discovery and development in brain cancer. Drug Discov. Today 2023, 28, 103426. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef]
- Alzeeb, G.; Metges, J.P.; Corcos, L.; Le Jossic-Corcos, C. Three-Dimensional Culture Systems in Gastric Cancer Research. Cancers 2020, 12, 2800. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, B.; Spencer, D.; Pytel, P.; Ahmed, A.U.; Lesniak, M.S. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev. Neurother. 2015, 15, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Hassiotou, F.; Nowak, A. Glioblastoma stem-like cells: At the root of tumor recurrence and a therapeutic target. Carcinogenesis 2015, 36, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.G.; Cheong, J.H.; Huh, Y.M.; Kim, E.H.; Kim, S.H.; Chang, J.H. Potential use of glioblastoma tumorsphere: Clinical credentialing. Arch. Pharm. Res. 2015, 38, 402–407. [Google Scholar] [CrossRef]
- Patrizii, M.; Bartucci, M.; Pine, S.R.; Sabaawy, H.E. Utility of Glioblastoma Patient-Derived Orthotopic Xenografts in Drug Discovery and Personalized Therapy. Front. Oncol. 2018, 8, 23. [Google Scholar] [CrossRef]
- Park, J.; Shim, J.K.; Kang, J.H.; Choi, J.; Chang, J.H.; Kim, S.Y.; Kang, S.G. Regulation of bioenergetics through dual inhibition of aldehyde dehydrogenase and mitochondrial complex I suppresses glioblastoma tumorspheres. Neuro. Oncol. 2018, 20, 954–965. [Google Scholar] [CrossRef]
- Mohiuddin, E.; Wakimoto, H. Extracellular matrix in glioblastoma: Opportunities for emerging therapeutic approaches. Am. J. Cancer Res. 2021, 11, 3742–3754. [Google Scholar] [PubMed]
- Damania, A.; Jain, E.; Kumar, A. Advancements in in vitro hepatic models: Application for drug screening and therapeutics. Hepatol. Int. 2014, 8, 23–38. [Google Scholar] [CrossRef]
- Edmonds, M.E.; Woodruff, T.K. Testicular organoid formation is a property of immature somatic cells, which self-assemble and exhibit long-term hormone-responsive endocrine function. Biofabrication 2020, 12, 045002. [Google Scholar] [CrossRef]
- Warschkau, D.; Delgado-Betancourt, E.; Holthaus, D.; Muller, A.; Kliem, G.; Krug, S.M.; Schulzke, J.D.; Aebischer, T.; Klotz, C.; Seeber, F. From 3D to 2D: Harmonization of Protocols for Two-dimensional Cultures on Cell Culture Inserts of Intestinal Organoids from Various Species. Bio. Protoc. 2022, 12, e4295. [Google Scholar] [CrossRef]
- Soldatow, V.Y.; Lecluyse, E.L.; Griffith, L.G.; Rusyn, I. In vitro models for liver toxicity testing. Toxicol. Res. 2013, 2, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Bissell, D.M.; Caron, J.M.; Babiss, L.E.; Friedman, J.M. Transcriptional regulation of the albumin gene in cultured rat hepatocytes. Role of basement-membrane matrix. Mol. Biol. Med. 1990, 7, 187–197. [Google Scholar]
- Bissell, D.M.; Arenson, D.M.; Maher, J.J.; Roll, F.J. Support of cultured hepatocytes by a laminin-rich gel. Evidence for a functionally significant subendothelial matrix in normal rat liver. J. Clin. Investig. 1987, 79, 801–812. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Balis, U.J.; Yarmush, M.L.; Toner, M. Effect of cell-cell interactions in preservation of cellular phenotype: Cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999, 13, 1883–1900. [Google Scholar] [CrossRef]
- Du, Y.; Chia, S.M.; Han, R.; Chang, S.; Tang, H.; Yu, H. 3D hepatocyte monolayer on hybrid RGD/galactose substratum. Biomaterials 2006, 27, 5669–5680. [Google Scholar] [CrossRef]
- Nyberg, S.L.; Hardin, J.; Amiot, B.; Argikar, U.A.; Remmel, R.P.; Rinaldo, P. Rapid, large-scale formation of porcine hepatocyte spheroids in a novel spheroid reservoir bioartificial liver. Liver Transpl. 2005, 11, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Barakat, O.; Abbasi, S.; Rodriguez, G.; Rios, J.; Wood, R.P.; Ozaki, C.; Holley, L.S.; Gauthier, P.K. Use of decellularized porcine liver for engineering humanized liver organ. J. Surg. Res. 2012, 173, e11–e25. [Google Scholar] [CrossRef]
- Zhou, P.; Lessa, N.; Estrada, D.C.; Severson, E.B.; Lingala, S.; Zern, M.A.; Nolta, J.A.; Wu, J. Decellularized liver matrix as a carrier for the transplantation of human fetal and primary hepatocytes in mice. Liver Transpl. 2011, 17, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.M.; Siddiqui, M.M.; Lozier, G.; Rodriguez, S.R.; Atala, A.; Soker, S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 2011, 53, 604–617. [Google Scholar] [CrossRef]
- Danova, K.; Klapetkova, A.; Kayserova, J.; Sediva, A.; Spisek, R.; Jelinkova, L.P. NF-kappaB, p38 MAPK, ERK1/2, mTOR, STAT3 and increased glycolysis regulate stability of paricalcitol/dexamethasone-generated tolerogenic dendritic cells in the inflammatory environment. Oncotarget 2015, 6, 14123–14138. [Google Scholar] [CrossRef]
- Park, J.; Shim, J.K.; Lee, M.; Kim, D.; Yoon, S.J.; Moon, J.H.; Kim, E.H.; Park, J.Y.; Chang, J.H.; Kang, S.G. Classification of IDH wild-type glioblastoma tumorspheres into low- and high-invasion groups based on their transcriptional program. Br. J. Cancer 2023, 129, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Leone, G.; DeGregori, J.; Yan, Z.; Jakoi, L.; Ishida, S.; Williams, R.S.; Nevins, J.R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998, 12, 2120–2130. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Tian, H.; Gao, X.; Wang, W.; Wang, X.; Zhang, Z. KMT2D deficiency disturbs the proliferation and cell cycle activity of dental epithelial cell line (LS8) partially via Wnt signaling. Biosci. Rep. 2021, 41, BSR20211148. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Poornima, K.; Francis, A.P.; Hoda, M.; Eladl, M.A.; Subramanian, S.; Veeraraghavan, V.P.; El-Sherbiny, M.; Asseri, S.M.; Hussamuldin, A.B.A.; Surapaneni, K.M.; et al. Implications of Three-Dimensional Cell Culture in Cancer Therapeutic Research. Front. Oncol. 2022, 12, 891673. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.; Choi, C. Evaluation of drug-targetable genes by defining modes of abnormality in gene expression. Sci. Rep. 2015, 5, 13576. [Google Scholar] [CrossRef] [PubMed]
- Koh, I.; Cha, J.; Park, J.; Choi, J.; Kang, S.G.; Kim, P. The mode and dynamics of glioblastoma cell invasion into a decellularized tissue-derived extracellular matrix-based three-dimensional tumor model. Sci. Rep. 2018, 8, 4608. [Google Scholar] [CrossRef]
- Du, P.; Kibbe, W.A.; Lin, S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Khokhlova, O.N.; Borozdina, N.A.; Sadovnikova, E.S.; Pakhomova, I.A.; Rudenko, P.A.; Korolkova, Y.V.; Kozlov, S.A.; Dyachenko, I.A. Comparative Study of the Aftereffect of CO(2) Inhalation or Tiletamine-Zolazepam-Xylazine Anesthesia on Laboratory Outbred Rats and Mice. Biomedicines 2022, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; Lacroix, M.; Tofilon, P.; Fuller, G.N.; Sawaya, R.; Lang, F.F. An implantable guide-screw system for brain tumor studies in small animals. J. Neurosurg. 2000, 92, 326–333. [Google Scholar] [CrossRef] [PubMed]
| Case | Sex | Age | IDH1 Mutation | MGMT Promoter | 1p/19q Deletion |
|---|---|---|---|---|---|
| 13-20 | M | 61 | Wild-type | Methylated | Intact |
| 13-64 | F | 56 | Wild-type | Unmethylated | Intact |
| 14-08 | F | 57 | Wild-type | Unmethylated | Intact |
| 14-15 | M | 67 | Wild-type | Methylated | Intact |
| 15-88 | M | 61 | Wild-type | Unmethylated | Intact |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Koh, I.; Cha, J.; Oh, Y.; Shim, J.-K.; Kim, H.; Moon, J.H.; Kim, E.H.; Chang, J.H.; Kim, P.; et al. Comparison of Glioblastoma Cell Culture Platforms Based on Transcriptional Similarity with Paired Tissue. Pharmaceuticals 2024, 17, 529. https://doi.org/10.3390/ph17040529
Park J, Koh I, Cha J, Oh Y, Shim J-K, Kim H, Moon JH, Kim EH, Chang JH, Kim P, et al. Comparison of Glioblastoma Cell Culture Platforms Based on Transcriptional Similarity with Paired Tissue. Pharmaceuticals. 2024; 17(4):529. https://doi.org/10.3390/ph17040529
Chicago/Turabian StylePark, Junseong, Ilkyoo Koh, Junghwa Cha, Yoojung Oh, Jin-Kyoung Shim, Hyejin Kim, Ju Hyung Moon, Eui Hyun Kim, Jong Hee Chang, Pilnam Kim, and et al. 2024. "Comparison of Glioblastoma Cell Culture Platforms Based on Transcriptional Similarity with Paired Tissue" Pharmaceuticals 17, no. 4: 529. https://doi.org/10.3390/ph17040529
APA StylePark, J., Koh, I., Cha, J., Oh, Y., Shim, J.-K., Kim, H., Moon, J. H., Kim, E. H., Chang, J. H., Kim, P., & Kang, S.-G. (2024). Comparison of Glioblastoma Cell Culture Platforms Based on Transcriptional Similarity with Paired Tissue. Pharmaceuticals, 17(4), 529. https://doi.org/10.3390/ph17040529