Isolation and Characterization of Environmental Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Ouagadougou, Burkina Faso
Abstract
1. Introduction
2. Results
2.1. Prevalence of Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae in Environmental Samples
2.2. Resistance Profile of Environmental Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae Isolates
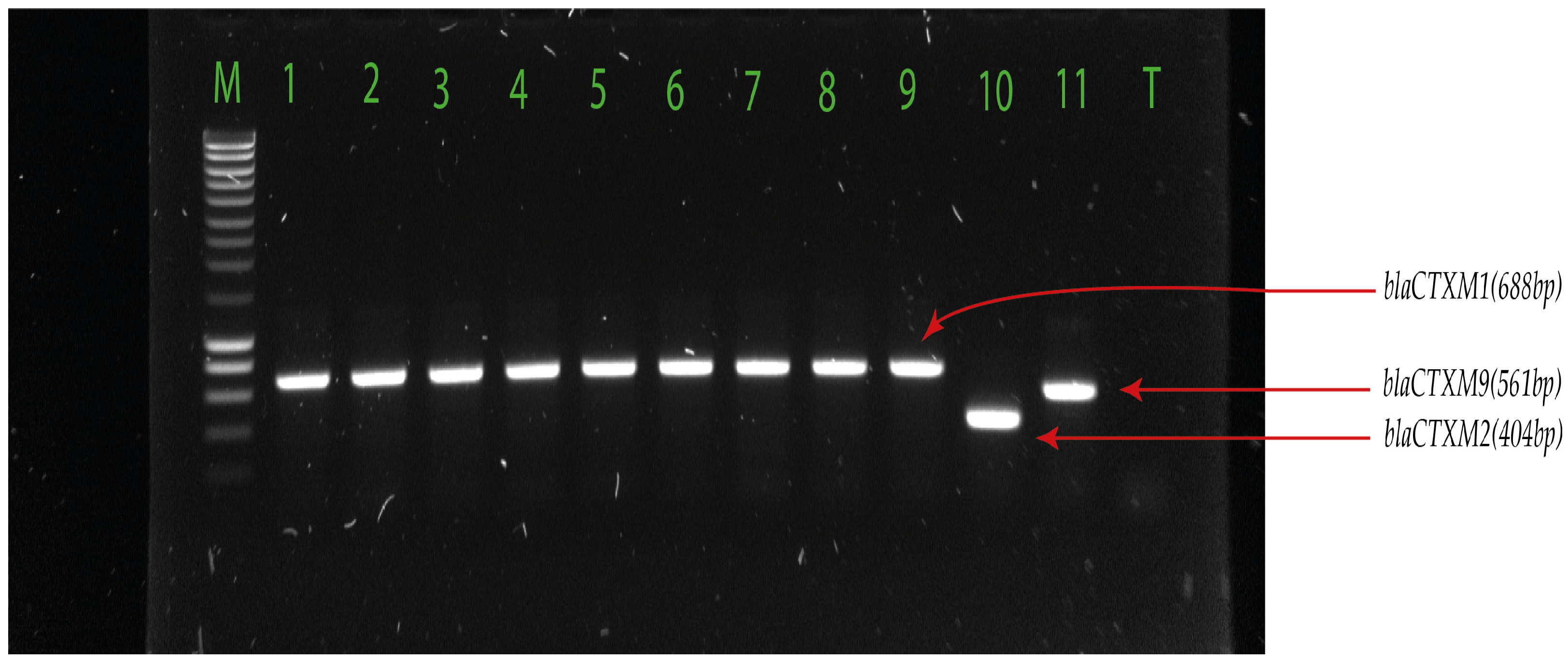
2.3. Detection of Extended-Spectrum β-Lactamase-Encoding Genes
3. Discussion
4. Materials and Methods
4.1. Study Design, Sites, and Sampling
4.2. Microbial Culturing and Identification
4.3. Antibiotic Susceptibility Testing
4.4. Detection of ESBL Genes
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GLASS. Whole-Genome Sequencing for Surveillance of Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/bitstream/handle/10665/334354/9789240011007-eng.pdf. (accessed on 27 July 2023).
- Jonas, O.; Irwin, A.; Berthe, F.; Le Gall, F.; Marquez, P. Drug-Resistant Infections: A Threat to Our Economic Future; World Bank: Washington, DC, USA, 2017; Available online: https://documents1.worldbank.org/curated/en/323311493396993758/pdf/final-report.pdf (accessed on 27 July 2023).
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Tasak, N. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. The burden of bacterial antimicrobial resistance in the WHO African region in 2019: A cross-country systematic analysis. Lancet Glob. Health 2024, 12, e201–e216. [Google Scholar] [CrossRef]
- Jasovský, D.; Littmann, J.; Zorzet, A.; Cars, O. Antimicrobial resistance-a threat to the world’s sustainable development. Upps. J. Med. Sci. 2016, 121, 159–164. [Google Scholar] [CrossRef]
- Bako, E. Caractérisation Moléculaire des Pathovars de Escherichia coli à Haut Potentiel Zoonotique Humaine Isolés des Effluents de 04 Marchés de Bétail de la Ville de Ouagadougou, Burkina Faso; Sciences Biologiques Appliquées; Université Joseph Ki-Zerbo: Ouagadougou, Burkina Faso, 2019; 111p. [Google Scholar]
- Ouédraogo, A.S.; Sanou, S.; Kissou, A.; Poda, A.; Aberkane, S.; Bouzinbi, N.; Nacro, B.; Ouédraogo, R.; Van De Perre, P.; Carriere, C.; et al. Fecal Carriage of Enterobacteriaceae Producing Extended-Spectrum Beta-Lactamases in Hospitalized Patients and Healthy Community Volunteers in Burkina Faso. Microb. Drug Resist. 2017, 23, 63–70. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (WOAH). Seventh Annual Report on Antimicrobial Agents Intended for Use in Animals; World Organisation for Animal Health: Paris, France, 2023; 137p. [Google Scholar]
- Organisation Internationnale de l’Environnement (OIE). Liste OIE des Agents Antimicrobiens Importants en Médecine Vétérinaire. 2021. Available online: https://www.oie.int/app/uploads/2021/06/f-oie-liste-antimicrobiens-juin2021.pdf (accessed on 15 September 2023).
- Dudkouski, A. L’épandage agricole des boues de stations d’épuration d’eaux usées urbaines. Courr. Environ. Inra 2000, 41, 134–135. [Google Scholar]
- Amos, G.C.A.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 2014, 69, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef]
- Garner, E.; Chen, C.; Xia, K.; Bowers, J.; Engelthaler, D.M.; McLain, J.; Edwards, M.A.; Pruden, A. Metagenomic characterization of antibiotic resistance genes in fullscale reclaimed water distribution systems and corresponding potable systems. Environ. Sci. Technol. 2018, 52, 6113–6125. [Google Scholar] [CrossRef]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Philippon, A.; Slama, P.; Dény, P.; Labia, R.A. Structure-Based Classification of Class A β-Lactamases, a Broadly Diverse Family of Enzymes. Clin. Microbiol. Rev. 2016, 29, 29–57. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of bêta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Ur Rahman, S.; Ahmad, S.; Khan, I.; Pakistan, P. Incidence of ESBL-producing-Escherichia coli in poultry farm environment and retail poultry meat. Pak. Vet. J. 2018, 39, 116–120. [Google Scholar] [CrossRef]
- Sirot, D. Extended-spectrum plasmid-mediated bêta-lactamases. J. Antimicrob. Chemother. 1995, 36, 19–34. [Google Scholar] [CrossRef]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- Bako, E.; Kagambèga, A.; Traor, K.A.; Bagre, T.S.; Ibrahim, H.B.; Bouda, S.C.; Bonkoungou, I.J.O.; Kaboré, S.; Zongo, C.; Traore, A.S.; et al. Characterization of Diarrheagenic Escherichia coli Isolated in Organic Waste Products (Cattle Fecal Matter, Manure and, Slurry) from Cattle’s Markets in Ouagadougou, Burkina Faso. Int. J. Environ. Res. Public Health 2017, 14, 1100. [Google Scholar] [CrossRef]
- Garba, Z.; Kaboré, B.; Bonkoungou, I.J.; Natama, M.H.; Rouamba, T.; Haukka, K.; Kirveskari, J.P.; Tinto, H.; Sangaré, L.; Barro, N.; et al. Phenotypic Detection of Carbapenemase and AmpC-β-Lactamase Production among Extended Spectrum β-Lactamase (ESBL)-Producing Escherichia coli and Klebsiella spp. Isolated from Clinical Specimens. Antibiotics 2023, 13, 31. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Brolund, A.; Sandegren, L. Infectious Diseases (London, England); Informa Healthcare: London, UK, 2016; Volume 48, pp. 18–25. [Google Scholar]
- Kaboré, B.; Ouédraogo, H.S.; Zongo, O.; Ouédraogo, G.A.; Tapsoba, F.; Bougma, S.; Zongo, K.J.; Zeba, B.; Traoré, Y.; Sanou, I.; et al. Emergence of New Delhi Metallo-β-Lactamase (NDM) Genes Detected from Clinical Strains of Escherichia coli Isolated in Ouagadougou, Burkina Faso. Int. J. Microbiol. 2023, 2023, 4813225. [Google Scholar] [CrossRef]
- Kpoda, D.S.; Ajayi, A.; Somda, M.; Traore, O.; Guessennd, N.; Ouattara, A.S.; Sangare, L.; Traore, A.S.; Dosso, M. Distribution of resistance genes encoding ESBLs in Enterobacteriaceae isolated from biological samples in health centers in Ouagadougou, Burkina Faso. BMC Res. Notes 2018, 11, 471. [Google Scholar] [CrossRef]
- Dembélé, R.; Konaté, A.; Traoré, O.; Kaboré, W.A.D.; Soulama, I.; Kagambèga, A.; Traoré, A.S.; Guessennd, N.K.; Aidara-Kane, A.; Gassama-Sow, A.; et al. Extended spectrum beta-lactamase and fluoroquinolone resistance genes among Escherichia coli and Salmonella isolates from children with diarrhea, Burkina Faso. BMC Pediatr. 2020, 20, 459. [Google Scholar] [CrossRef]
- Soré, S.; Sanou, S.; Sawadogo, Y.; Béogo, S.; Dakouo, S.N.P.; Djamalladine, M.D.; lboudo, K.S.; Ouoba, B.; Zoungrana, J.; Poda, A.; et al. Faecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in healthy volunteers and hospitalized patients in Ouagadougou, Burkina Faso: Prevalence, resistance profile, and associated risk factors. Afr. J. Clin. Exper. Microbiol. 2021, 22, 157–163. [Google Scholar] [CrossRef]
- Soré, S.; Sawadogo, Y.; Bonkoungou, J.I.; Kaboré, P.; Béogo, S.; Sawadogo, C.; Bationo, B.G.; Ky, H.; Madingar, P.D.M.; Ouédraogo, A.S.; et al. Detection, identification and characterization of extended-spectrum beta-lactamases producing Enterobacteriaceae in wastewater and salads marketed in Ouagadougou, Burkina Faso. Int. J. Biol. Chem. Sci. 2020, 14, 2746–2757. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum b-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 21, dlab092. [Google Scholar] [CrossRef]
- Kittinger, C.; Lipp, M.; Baumert, R.; Folli, B.; Koraimann, G.; Toplitsch, D.; Liebmann, A.; Grisold, A.J.; Farnleitner, A.H.; Kirschner, A.; et al. Antibiotic resistance patterns of Pseudomonas spp. isolated from the river Danube. Front. Microbiol. 2016, 7, 586. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Arnberger, E.; Inreiter, N.; Springer, B.; Allerberger, F.; Ruppitsch, W. Autria-wide survey on resistant, potentially pathogenic bacteria at Austrian bathing sites, 2017. J. Land Manag. Food Environ. 2019, 70, 81–88. [Google Scholar]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Soubelet, H.; Morel, G. Antibiorésistance et Environnement; SG/SPSSI/ATL2 Utilisant du Papier Issu de Forêts Durablement Gérées; Commissariat Général Au Développement Durable: La Défénse, France, 2017; 4p. [Google Scholar]
- Seiffert, S.N.; Hilty, M.; Perreten, V.; Endimiani, A. Extended-Spectrum Cephalosporin-Resistant Gram-Negative Organisms in Livestock: An Emerging Problem for Human Health? Drug Resist. Update 2013, 16, 22–45. [Google Scholar] [CrossRef]
- Kim, J.; Guk, J.; Mun, S.; An, J.; Song, H.; Kim, J.; Ryu, S.; Jeon, B.; Cho, S. Metagenomic Analysis of Isolation Methods of a Targeted Microbe, Campylobacter jejuni, from Chicken Feces with High Microbial Contamination. Microbiome 2019, 7, 67. [Google Scholar]
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of Extended Spectrum b-Lactamases, KPC-Type, and Mcr-1.2-Producing Enterobacteriaceae from Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 2017, 8, 2232. [Google Scholar] [CrossRef]
- Dorado-García, A.; Smid, J.H.; Pelt, V.W.; Mjm, B.; Veldman, K.T. Molecular Relatedness of Esbl/Ampc-Producing Escherichia coli From Humans, Animals, Food and the Environment: A Pooled Analysis. J. Antimicrob. Chemoth. 2018, 73, 339–347. [Google Scholar] [CrossRef]
- Sen, S.; Sarkar, K. Screening for ESBL Producing Bacterial Isolates of Agricultural Soil and Profiling for Multidrug Resistance. Ann. Agrar. Sci. 2018, 16, 272–280. [Google Scholar] [CrossRef]
- Al-Gheethi, A.A.; Efaq, A.N.; Bala, J.D.; Norli, I.; Abdel-Monem, M.O.; Kadir, M.O.A. Removal of pathogenic bacteria from sewage-treated effluent and biosolids for agricultural purposes. Appl. Water Sci. 2018, 8, 74. [Google Scholar] [CrossRef]
- Kesari, K.K.; Soni, R.; Jamal, Q.M.S.; Tripathi, P.; Lal, J.A.; Jha, N.K.; Siddiqui, M.H.; Kumar, P.; Tripathi, V.; Ruokolainen, J. Wastewater Treatment and Reuse: A Review of its Applications and Health Implications. Water Air Soil Pollut. 2021, 232, 208. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Jäger, T.; Hembach, N.; Elpers, C.; Wieland, A.; Alexander, J.; Hiller, C.; Krauter, G.; Schwartz, T. Reduction of Antibiotic Resistant Bacteria During Conventional and Advanced Wastewater Treatment, and the Disseminated Loads Released to the Environment. Front. Microbiol. 2018, 9, 2599. [Google Scholar] [CrossRef]
- Aubin, G.; Caron, F.; Cattoir, V.; Dubreuil, L.; Goutelle, S.; Jeannot, K.; Lepeule, R.; Lina, G.; Marchandin, H.; Merens, A.; et al. Comite de l’antibiogramme de la Societe Francaise de Microbiologie. Soc. Fr. Microbiol. 2021, 1–188. Available online: https://www.sfm-microbiologie.org/wp-content/uploads/2022/05/CASFM2022_V1.0.pdf (accessed on 25 October 2023).
- Dubois, V.; De Barbeyrac, B.; Rogues, A.M.; Arpin, C.; Coulange, L.; Andre, C.; M’zali, F.; Megraud, F.; Quentin, C. CTX-M-producing Escherichia coli in a maternity ward: A likely community importation and evidence of mother-to-neonate transmission. J. Antimicrob. Chemother. 2010, 65, 1368–1371. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]


| Sample Type | E. coli-Positive | K. pneumoniae-Positive | Negative Samples | Total | |
|---|---|---|---|---|---|
| Runoff water | 3 (5.2%) | 20 (34.5%) | 35 (60.3%) | 58 | |
| Wastewater | Untreated | 6 (14.6%) | 28 (68.3%) | 7 (17.1%) | 41 |
| Treated | 11 (26.8%) | 24 (58.5%) | 6 (14.6%) | 41 | |
| Manure | Ouaga-Inter livestock market | 30 (48.4%) | 11 (17.7%) | 21 (33.9%) | 62 |
| Tanghin livestock market | 24 (38.7%) | 12 (19.4%) | 26 (41.9%) | 62 | |
| Total | 74 (28.0%) | 95 (36.0%) | 95 (36.0%) | 264 | |
| Antibiotics | E. coli | K. pneumoniae |
|---|---|---|
| Ampicillin | 63/74 (85.1%) | 71/95 (74.7%) |
| Piperacillin | 63/74 (85.1%) | 71/95 (74.7%) |
| Cefoxitin | 1/74 (1.4%) | 7/95 (7.4%) |
| Ceftazidime | 49/74 (66.2%) | 67/95 (70.5%) |
| Ceftriaxone | 73/74 (98.6%) | 91/95 (95.8%) |
| Cefepime | 38/74 (51.4%) | 64/95 (67.4%) |
| Aztreonam | 50/74 (67.6%) | 62/95 (65.3%) |
| Ertapenem | 4/74 (5.4%) | 7/95 (7.4%) |
| Imipenem | 0 | 0 |
| Amoxicillin-Clavulanic acid | 6/74 (8.1%) | 17/95 (17.9%) |
| Piperacillin-Tazobactam | 1/74 (1.4%) | 1/95 (1.5%) |
| Gentamicin | 6/74 (8.1%) | 7/95 (7.4%) |
| Amikacin | 1/74 (1.4%) | 0 |
| Nalidixic acid | 32/74 (43.2%) | 9/95 (9.5%) |
| Ciprofloxacin | 24/74 (32.4%) | 33/95 (34.7%) |
| Sulfamethoxazole-Trimethoprim | 44/74 (59.5%) | 66/95 (69.5%) |
| Genes | E. coli | K. pneumoniae |
|---|---|---|
| blaTEM | 33/74 (44.6%) | 28/95 (29.5%) |
| blaSHV | 7/74 (9.5%) | 56/95 (58.9%) |
| blaOXA | 10/74 (13.5%) | 10/95 (10.5%) |
| blaCTX-M1 | 63/74 (85.1%) | 89/95 (93.7%) |
| blaCTX-M9 | 8/74 (10.8%) | 3/95 (3.2%) |
| blaCTX-M2 | 0 | 0 |
| Genes | E. coli | K. pneumoniae |
|---|---|---|
| blaCTX-M-blaTEM-blaSHV-blaOXA | 0 | 4/95 (4.2%) |
| blaCTX-M-blaTEM-blaSHV | 2/75 (2.7%) | 10/95 (10.5%) |
| blaCTX-M-blaTEM-blaOXA | 2/75 (2.7%) | 2/95 (2.1%) |
| blaCTX-M-blaSHV-blaOXA | 1/75 (1.3%) | 3/95 (3.2%) |
| blaCTX-M-blaSHV | 3/75 (4%) | 36/95 (37.9%) |
| blaCTX-M-blaTEM | 27/75 (36%) | 11/95 (11.6%) |
| blaCTX-M-blaOXA | 7/75 (9.3%) | 1/95 (1.1%) |
| blaTEM-blaSHV | 1/75 (1.33%) | 1/95 (1.1%) |
| blaCTX-M | 29/75 (38.7%) | 25/95 (26.3%) |
| blaSHV | 0 | 2/95 (2.1%) |
| blaTEM | 1/75 (1.3%) | 0 |
| No ESBL gene | 1/75 (1.3%) | 0 |
| ESBL Target Genes | Primer Sequences (5′-3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|
| TEM | F: CATTTCCGTGTCGCCCTTATTC | 800 | [49] |
| R: CGTTCATCCATAGTTGCCTGAC | [49] | ||
| SHV | F: AGCCGCTTGAGCAAATTAAAC | 713 | [49] |
| R: ATCCCGCAGATAAATCACCAC | [49] | ||
| OXA | F: GGCACCAGATTCAACTTTCAAG | 564 | [49] |
| R: GACCCCAAGTTTCCTGTAAGTG | [49] | ||
| CTX-M1 | F: TTAGGAARTGTGCCGCTGYA b | 688 | [49] |
| R: CGATATCGTTGGTGGTRCCAT b | [49] | ||
| CTX-M2 | F: CGTTAACGGCACGATGAC | 404 | [49] |
| R: CGATATCGTTGGTGGTRCCAT b | [49] | ||
| CTX-M9 | F: TCAAGCCTGCCGATCTGGT | 561 | [49] |
| R: TGATTCTCGCCGCTGAAG | [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kagambèga, A.B.; Dembélé, R.; Traoré, O.; Wane, A.A.; Mohamed, A.H.; Coulibaly, H.; Fall, C.; Bientz, L.; M’Zali, F.; Mayonnove, L.; et al. Isolation and Characterization of Environmental Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Ouagadougou, Burkina Faso. Pharmaceuticals 2024, 17, 305. https://doi.org/10.3390/ph17030305
Kagambèga AB, Dembélé R, Traoré O, Wane AA, Mohamed AH, Coulibaly H, Fall C, Bientz L, M’Zali F, Mayonnove L, et al. Isolation and Characterization of Environmental Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Ouagadougou, Burkina Faso. Pharmaceuticals. 2024; 17(3):305. https://doi.org/10.3390/ph17030305
Chicago/Turabian StyleKagambèga, Alix Bénédicte, René Dembélé, Oumar Traoré, Abdoul Aziz Wane, Alassane Halawen Mohamed, Hiliassa Coulibaly, Cheikh Fall, Léa Bientz, Fatima M’Zali, Laure Mayonnove, and et al. 2024. "Isolation and Characterization of Environmental Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Ouagadougou, Burkina Faso" Pharmaceuticals 17, no. 3: 305. https://doi.org/10.3390/ph17030305
APA StyleKagambèga, A. B., Dembélé, R., Traoré, O., Wane, A. A., Mohamed, A. H., Coulibaly, H., Fall, C., Bientz, L., M’Zali, F., Mayonnove, L., Barro, N., Dubois, V., & Dieye, Y. (2024). Isolation and Characterization of Environmental Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Ouagadougou, Burkina Faso. Pharmaceuticals, 17(3), 305. https://doi.org/10.3390/ph17030305







