Cryptolepine Suppresses Colorectal Cancer Cell Proliferation, Stemness, and Metastatic Processes by Inhibiting WNT/β-Catenin Signaling
Abstract
1. Introduction
2. Results
2.1. Effects of Cryptolepine, XAV 939, and WNT3a on Cell Viability
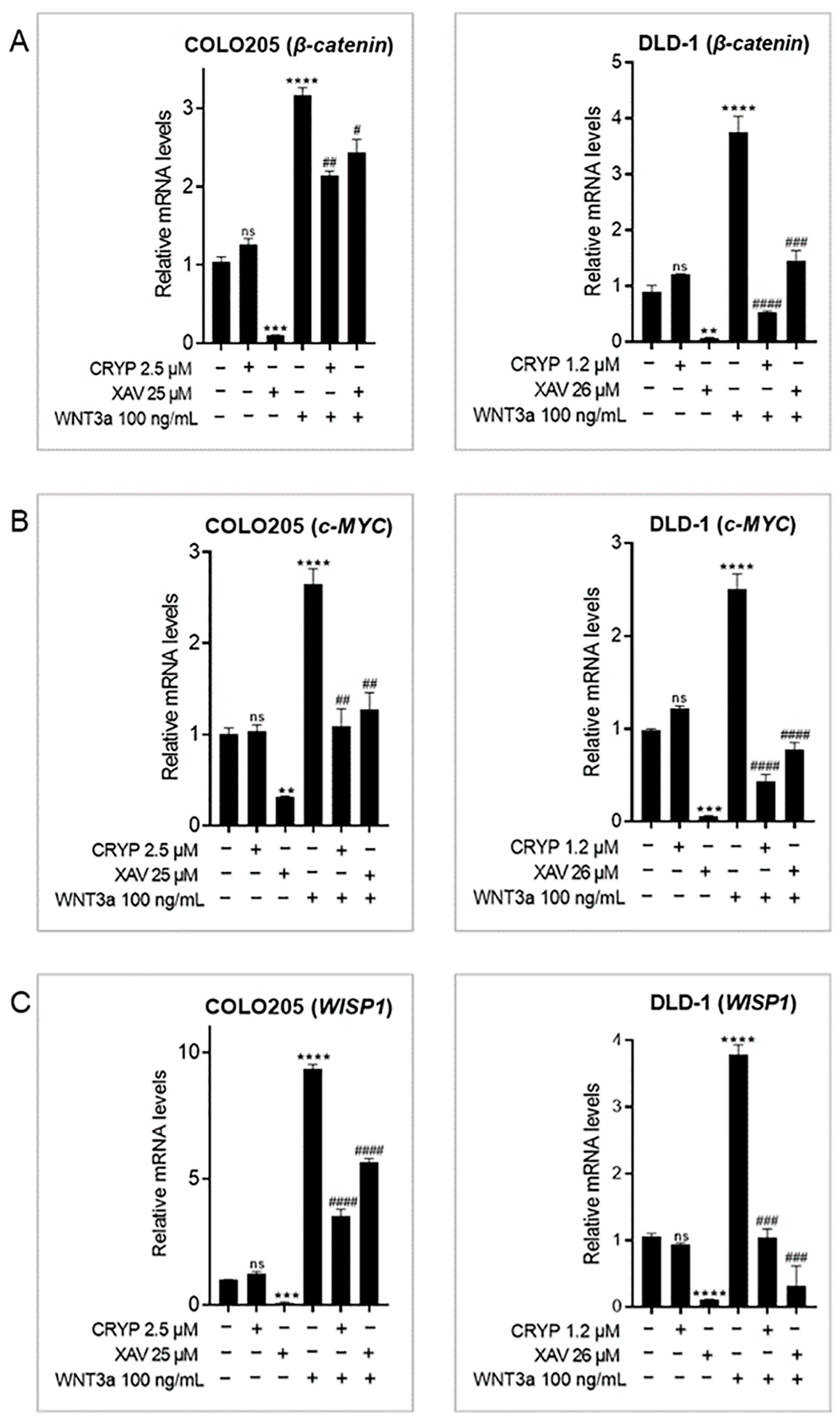
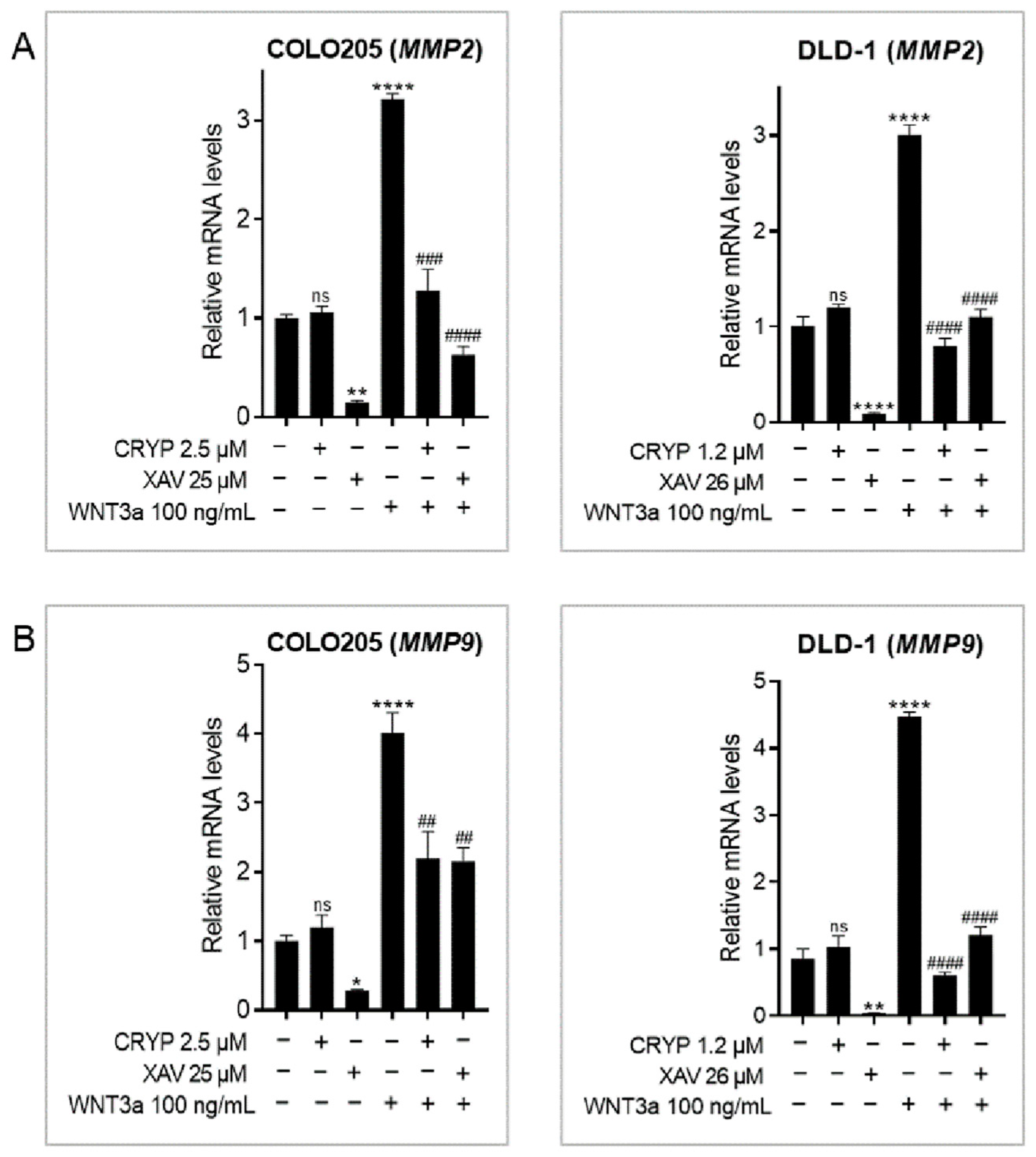
2.2. Cryptolepine Inhibits WNT/β-Catenin Signaling in CRC Cells
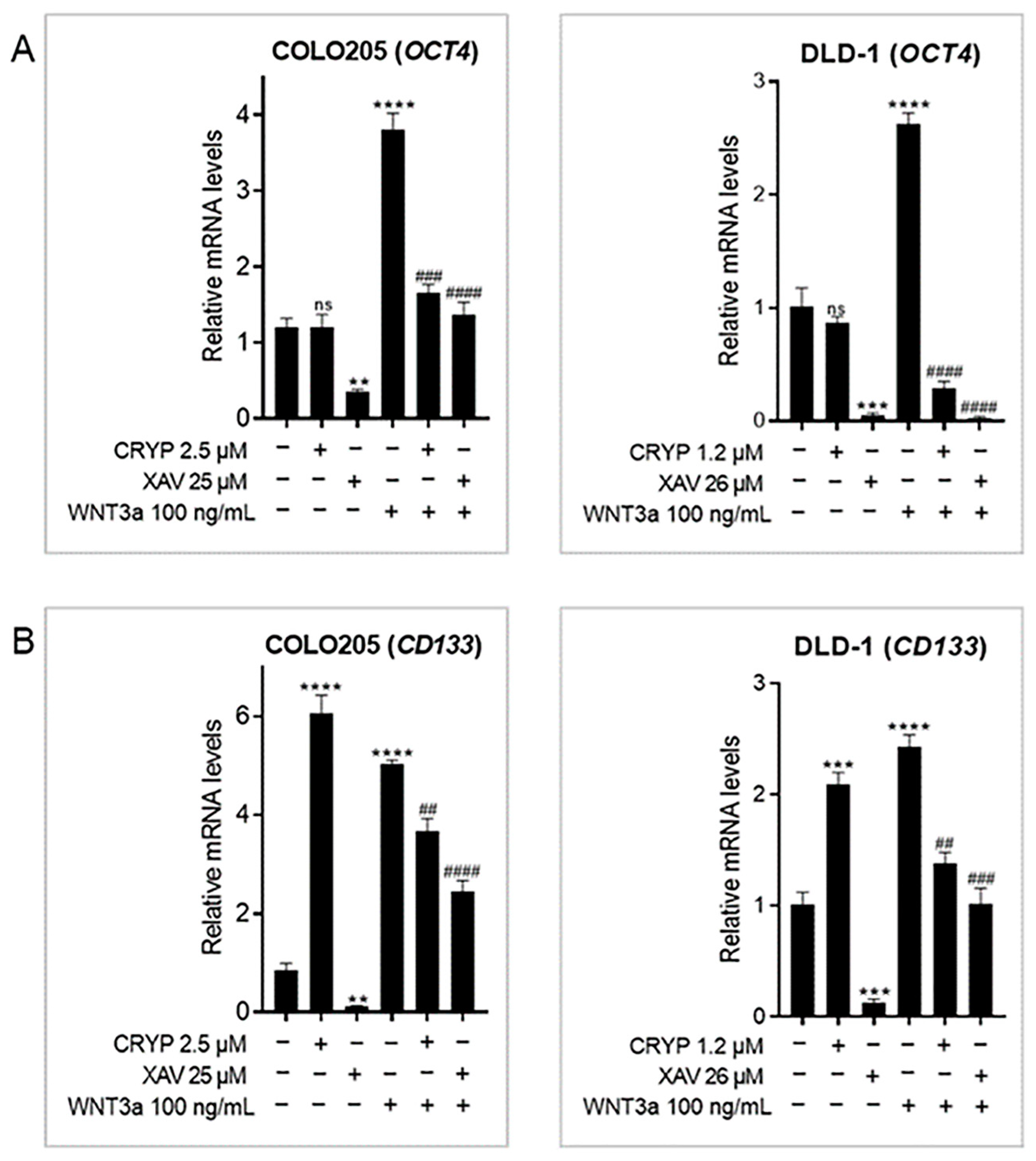
2.3. Cryptolepine Downregulates Stem Cell Markers in CRC
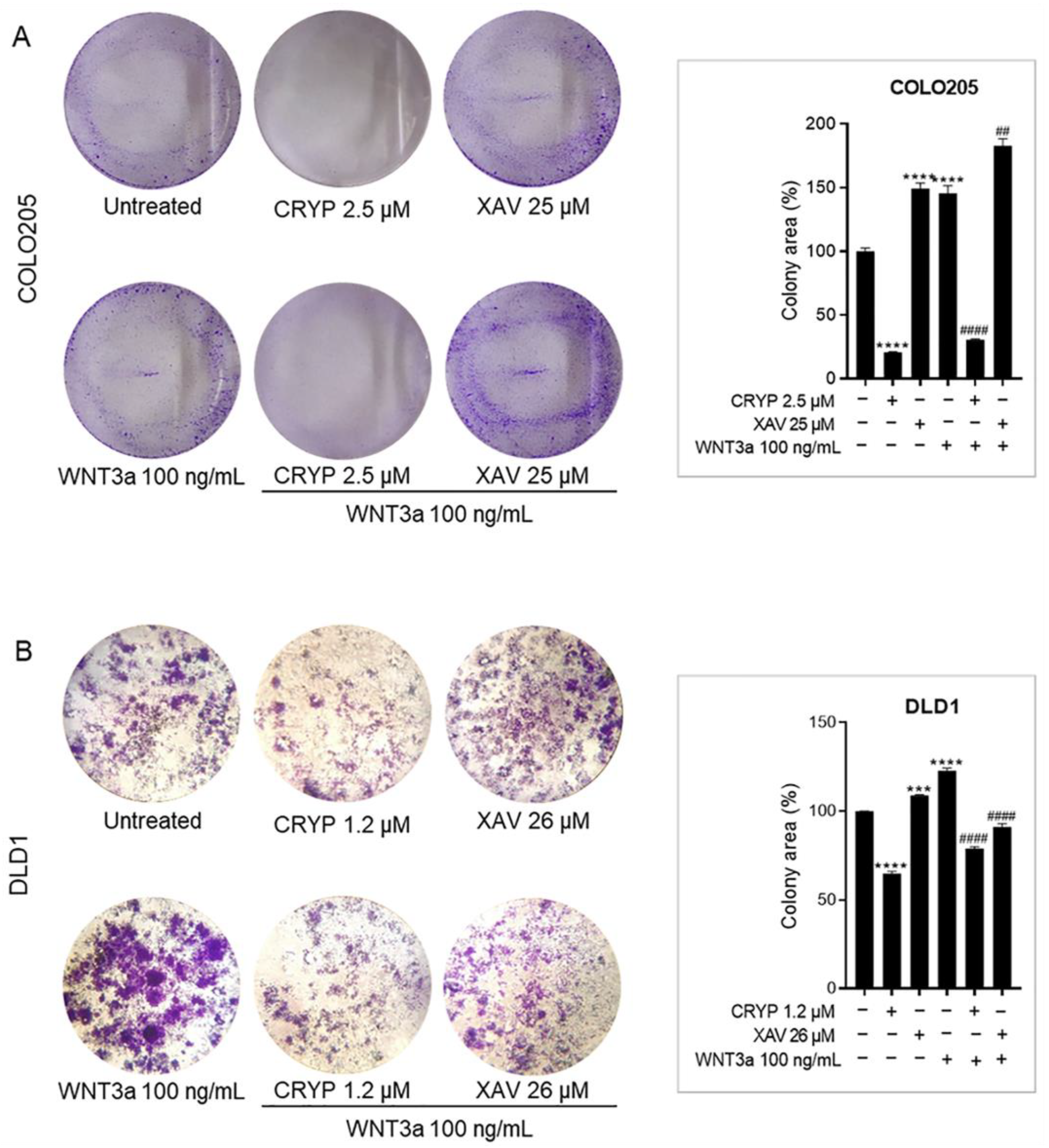
2.4. Cryptolepine Inhibits Clonogenicity in CRC
2.5. Cryptolepine Represses Epithelial-to-Mesenchymal Transition (EMT) in CRC
2.6. Cryptolepine Reduces CRC Cell Migration
2.7. Cryptolepine Reduces CRC Cell Invasiveness
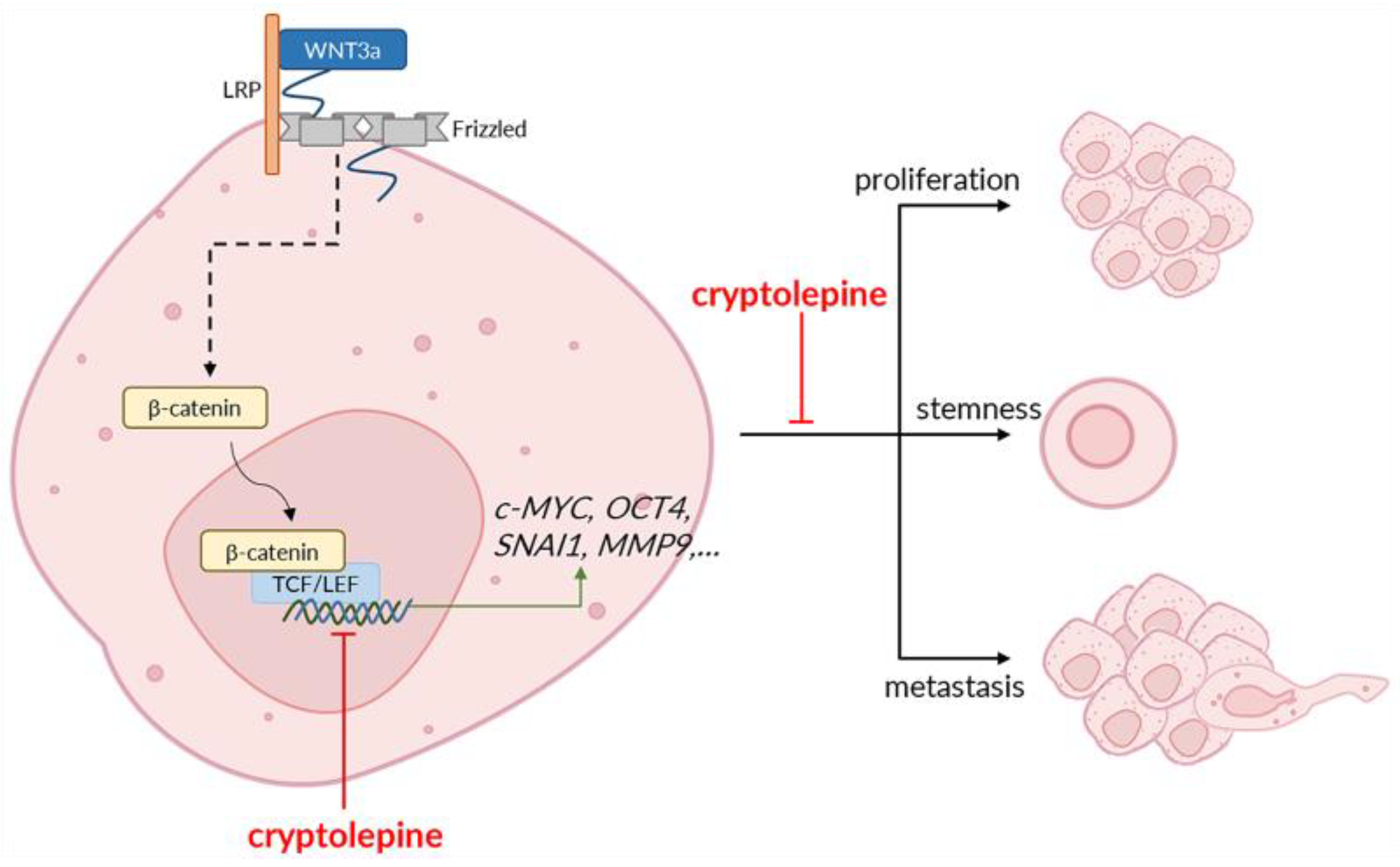
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Compounds/Drugs
4.3. Cell Viability Assay
4.4. Colony Formation/Clonogenic Assay
4.5. Wound Healing Assay
4.6. Reverse-Transcription Quantitative PCR (RT-qPCR)
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Duong, H. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Clinton, S.K.; Giovannucci, E.L.; Hursting, S.D. The World Cancer Research Fund/American Institute for Cancer Research Third Expert Report on Diet, Nutrition, Physical Activity, and Cancer: Impact and Future Directions. J. Nutr. 2020, 150, 663–671. [Google Scholar] [CrossRef]
- Munro, M.; Wickremesekera, S.K.; Peng, L.; Tan, S.T.; Itinteang, T. Cancer stem cells in colorectal cancer: A review. J. Clin. Pathol. 2018, 71, 110–116. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/β-Catenin Signaling in Development and Disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Liu, L.-J.; Xie, S.-X.; Chen, Y.-T.; Xue, J.-L.; Zhang, C.-J.; Zhu, F. Aberrant regulation of WNT signaling in hepatocellular carcinoma. World J. Gastroenterol. 2016, 22, 7486–7499. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.; Kuipers, J.; Kujala, P.; Van Den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Flanagan, D.J.; Pentinmikko, N.; Luopajärvi, K.; Willis, N.J.; Gilroy, K.; Raven, A.P.; Mcgarry, L.; Englund, J.I.; Webb, A.T.; Scharaw, S.; et al. NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature 2021, 594, 430–435. [Google Scholar] [CrossRef]
- Osafo, N.; Mensah, K.; Yeboah, O. Phytochemical and Pharmacological Review of Cryptolepis sanguinolenta (Lindl.) Schlechter. Adv. Pharmacol. Sci. 2017, 2017, 3026370. [Google Scholar] [CrossRef]
- Mensah, K.; Benneh, C.; Forkuo, A.; Ansah, C. Cryptolepine, the Main Alkaloid of the Antimalarial Cryptolepis sanguinolenta (Lindl.) Schlechter, Induces Malformations in Zebrafish Embryos. Biochem. Res. Int. 2019, 2019, 7076986. [Google Scholar] [CrossRef]
- Batiha, G.-S.; Beshbishy, A.; Alkazmi, L.; Nadwa, E.; Rashwan, E.; Yokoyama, N.; Igarashi, I. In vitro and in vivo growth inhibitory activities of cryptolepine hydrate against several Babesia species and Theileria equi. PLOS Negl. Trop. Dis. 2020, 14, e0008489. [Google Scholar] [CrossRef]
- Ansah, C.; Mensah, K. A review of the anticancer potential of the antimalarial herbal Cryptolepis sanguinolenta and its major alkaloid cryptolepine. Ghana Med. J. 2013, 47, 137–147. [Google Scholar]
- Pal, H.; Katiyar, S. Cryptolepine, a Plant Alkaloid, Inhibits the Growth of Non-Melanoma Skin Cancer Cells through Inhibition of Topoisomerase and Induction of DNA Damage. Molecules 2016, 21, 1758. [Google Scholar] [CrossRef]
- Domfeh, S.; Narkwa, P.; Quaye, O.; Kusi, K.; Rivera, O.; Danaah, M.M.; Musah, B.A.N.; Awandare, G.A.; Mensah, K.B.; Mutocheluh, M. Cryptolepine and Nibima inhibit hepatitis B virus replication. Sci. Afr. 2021, 13, e00942. [Google Scholar] [CrossRef]
- Bonjean, K.; De Pauw-Gillet, M.; Defresne, M.; Colson, P.; Houssier, C.; Dassonneville, L.; Bailly, C.; Greimers, R.; Wright, C.; Quetin-Leclercq, J.; et al. The DNA Intercalating Alkaloid Cryptolepine Interferes with Topoisomerase II and Inhibits Primarily DNA Synthesis in B16 Melanoma Cells. Biochemistry 1998, 37, 5136–5146. [Google Scholar] [CrossRef]
- Lisgarten, J.; Coll, M.; Portugal, J.; Wright, C.; Aymami, J. The antimalarial and cytotoxic drug cryptolepine intercalates into DNA at cytosine-cytosine sites. Nat. Struct. Biol. 2002, 9, 57–60. [Google Scholar] [CrossRef]
- Guittat, L.; Alberti, P.; Rosu, F.; Van Miert, S.; Thetiot, E.; Pieters, L.; Gabelica, V.; De Pauw, E.; Ottaviani, A.; Riou, J.-F.; et al. Interactions of cryptolepine and neocryptolepine with unusual DNA structures. Biochimie 2003, 85, 535–547. [Google Scholar] [CrossRef]
- Domfeh, S.; Narkwa, P.; Quaye, O.; Kusi, K.; Awandare, G.; Ansah, C.; Salam, A.; Mutocheluh, M. Cryptolepine inhibits hepatocellular carcinoma growth through inhibiting interleukin-6/STAT3 signalling. BMC Complement. Med. Ther. 2021, 21, 161. [Google Scholar] [CrossRef]
- Du, B.; Shim, J.S. Targeting Epithelial–Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 2016, 21, 965. [Google Scholar] [CrossRef]
- Nemoto, E.; Sakisaka, Y.; Tsuchiya, M.; Tamura, M.; Nakamura, T.; Kanaya, S.; Shimonishi, M.; Shimauchi, H. Wnt3a signaling induces murine dental follicle cells to differentiate into cementoblastic/osteoblastic cells via an osterix-dependent pathway. J. Periodontal Res. 2016, 51, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Osugui, L.; de Roo, J.J.; De Oliveira, V.C.; Sodré, A.C.P.; Staal, F.J.T.; Popi, A.F. B-1 cells and B-1 cell precursors prompt different responses to Wnt signaling. PLoS ONE 2018, 13, e0199332. [Google Scholar] [CrossRef] [PubMed]
- Martin-Orozco, E.; Sanchez-Fernandez, A.; Ortiz-Parra, I.; Nicolas, M.A.-S. WNT Signaling in Tumors: The Way to Evade Drugs and Immunity. Front. Immunol. 2019, 10, 2854. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, V.; Jain, M.V. In Vitro Tumorigenic Assay: Colony Forming Assay for Cancer Stem Cells. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2018; Volume 1692, pp. 89–95. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ren, J.; Tang, L. Genistein inhibits invasion and migration of colon cancer cells by recovering WIF1 expression. Mol. Med. Rep. 2018, 17, 7265–7273. [Google Scholar] [CrossRef]
- Stakheev, D.; Taborska, P.; Strizova, Z.; Podrazil, M.; Bartunkova, J.; Smrz, D. The WNT/β-catenin signaling inhibitor XAV939 enhances the elimination of LNCaP and PC-3 prostate cancer cells by prostate cancer patient lymphocytes in vitro. Sci. Rep. 2019, 9, 4761. [Google Scholar] [CrossRef]
- Reischmann, P.; Fiebeck, J.; von der Weiden, N.; Müller, O. Measured Effects of Wnt3a on Proliferation of HEK293T Cells Depend on the Applied Assay. Int. J. Cell Biol. 2015, 2015, 928502. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, R.; Zhang, Q.; Wang, H.; Kang, X.; Li, J.; Yang, S.; Zhang, J.; Liu, Z.; Yang, X. Survivin, a key component of the Wnt/β-catenin signaling pathway, contributes to traumatic brain injury-induced adult neurogenesis in the mouse dentate gyrus. Int. J. Mol. Med. 2013, 32, 867–875. [Google Scholar] [CrossRef]
- Li, C.; Zheng, X.; Han, Y.; Lv, Y.; Lan, F.; Zhao, J. XAV939 inhibits the proliferation and migration of lung adenocarcinoma A549 cells through the WNT pathway. Oncol. Lett. 2018, 15, 8973–8982. [Google Scholar] [CrossRef]
- Pal, H.C.; Prasad, R.; Katiyar, S.K. Cryptolepine inhibits melanoma cell growth through coordinated changes in mitochondrial biogenesis, dynamics and metabolic tumor suppressor AMPKα1/2-LKB1. Sci. Rep. 2017, 7, 1498. [Google Scholar] [CrossRef]
- Elbadawy, M.; Usui, T.; Yamawaki, H.; Sasaki, K. Emerging Roles of C-Myc in Cancer Stem Cell-Related Signaling and Resistance to Cancer Chemotherapy: A Potential Therapeutic Target Against Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 2340. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, B.-R.; Wu, T.-S.; Jeng, Y.-M.; Kuo, M.-L. Input of microenvironmental regulation on colorectal cancer: Role of the CCN family. World J. Gastroenterol. 2014, 20, 6826–6831. [Google Scholar] [CrossRef]
- Fujino, S.; Miyoshi, N. Oct4 Gene Expression in Primary Colorectal Cancer Promotes Liver Metastasis. Stem Cells Int. 2019, 2019, 7896524. [Google Scholar] [CrossRef]
- Dai, X.; Ge, J.; Wang, X.; Qian, X.; Li, X. OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol. Rep. 2012, 29, 155–160. [Google Scholar] [CrossRef]
- Chen, S.; Song, X.; Chen, Z.; Li, X.; Li, M.; Liu, H.; Li, J. CD133 Expression and the Prognosis of Colorectal Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2013, 8, e56380. [Google Scholar] [CrossRef]
- Saigusa, S.; Tanaka, K.; Toiyama, Y.; Yokoe, T.; Okugawa, Y.; Ioue, Y.; Miki, C.; Kusunoki, M. Correlation of CD133, OCT4, and SOX2 in Rectal Cancer and Their Association with Distant Recurrence After Chemoradiotherapy. Ann. Surg. Oncol. 2009, 16, 3488–3498. [Google Scholar] [CrossRef]
- You, H.; Ding, W.; Rountree, C.B. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-β. Hepatology 2010, 51, 1635–1644. [Google Scholar] [CrossRef]
- Kemper, K.; Versloot, M.; Cameron, K.; Colak, S.; Melo, F.D.S.E.; de Jong, J.H.; Bleackley, J.; Vermeulen, L.; Versteeg, R.; Koster, J.; et al. Mutations in the Ras–Raf Axis Underlie the Prognostic Value of CD133 in Colorectal Cancer. Clin. Cancer Res. 2012, 18, 3132–3141. [Google Scholar] [CrossRef]
- Park, E.K.; Lee, J.C.; Park, J.W.; Bang, S.Y.; Yi, S.A.; Kim, B.K.; Park, J.H.; Kwon, S.H.; You, J.S.; Nam, S.W.; et al. Transcriptional repression of cancer stem cell marker CD133 by tumor suppressor P53. Cell Death Dis. 2015, 6, e1964. [Google Scholar] [CrossRef]
- Ryu, H.; Nam, K.-Y.; Kim, H.J.; Song, J.-Y.; Hwang, S.-G.; Kim, J.S.; Kim, J.; Ahn, J. Discovery of a Novel Triazolopyridine Derivative as a Tankyrase Inhibitor. Int. J. Mol. Sci. 2021, 22, 7330. [Google Scholar] [CrossRef]
- Wang, W.; Liu, P.; Lavrijsen, M.; Li, S.; Zhang, R.; Li, S.; van de Geer, W.S.; van de Werken, H.J.G.; Peppelenbosch, M.P.; Smits, R. Evaluation of AXIN1 and AXIN2 as targets of tankyrase inhibition in hepatocellular carcinoma cell lines. Sci. Rep. 2021, 11, 7470. [Google Scholar] [CrossRef] [PubMed]
- Solomon, P.; Dong, Y.; Dogra, S.; Gupta, R. Interleukin 8 is a biomarker of telomerase inhibition in cancer cells. BMC Cancer 2018, 18, 730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Xie, Y.; Tai, Y.; Gao, Y.; Guo, W.; Yu, W.; Li, J.; Feng, X.; Hao, J.; Gao, Y.; et al. Bufalin Inhibits hTERT Expression and Colorectal Cancer Cell Growth by Targeting CPSF4. Cell. Physiol. Biochem. 2016, 40, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Ritch, S.J.; Brandhagen, B.N.; Goyeneche, A.A.; Telleria, C.M. Advanced assessment of migration and invasion of cancer cells in response to mifepristone therapy using double fluorescence cytochemical labeling. BMC Cancer 2019, 19, 376. [Google Scholar] [CrossRef] [PubMed]
- Yook, J.I.; Li, X.-Y.; Ota, I.; Fearon, E.R.; Weiss, S.J. Wnt-dependent Regulation of the E-cadherin Repressor Snail. J. Biol. Chem. 2005, 280, 11740–11748. [Google Scholar] [CrossRef] [PubMed]
- Yook, J.I.; Li, X.-Y.; Ota, I.; Hu, C.; Kim, H.S.; Kim, N.H.; Cha, S.Y.; Ryu, J.K.; Choi, Y.J.; Kim, J.; et al. A Wnt–Axin2–GSK3β cascade regulates Snail1 activity in breast cancer cells. Nature 2006, 8, 1398–1406. [Google Scholar] [CrossRef]
- Yang, H.-Y.; Shen, J.-X.; Wang, Y.; Liu, Y.; Shen, D.-Y.; Quan, S. Tankyrase Promotes Aerobic Glycolysis and Proliferation of Ovarian Cancer through Activation of Wnt/β-Catenin Signaling. BioMed. Res. Int. 2019, 2019, 2686340. [Google Scholar] [CrossRef]
- Zhu, M.; Gong, Z.; Wu, Q.; Shi, X.; Su, Q.; Zhang, Y. Sanguinarine suppresses migration and metastasis in colorectal carcinoma associated with the inversion of EMT through the Wnt/β-catenin signaling. Clin. Transl. Med. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Wu, X.; Luo, F.; Li, J.; Zhong, X.; Liu, K. Tankyrase 1 inhibitior XAV939 increases chemosensitivity in colon cancer cell lines via inhibition of the Wnt signaling pathway. Int. J. Oncol. 2016, 48, 1333–1340. [Google Scholar] [CrossRef]
- Wu, Z.-Q.; Li, X.-Y.; Hu, C.Y.; Ford, M.; Kleer, C.G.; Weiss, S.J. Canonical Wnt signaling regulates Slug activity and links epithelial–mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc. Natl. Acad. Sci. USA 2012, 109, 16654–16659. [Google Scholar] [CrossRef]
- Ren, H.; Du, P.; Ge, Z.; Jin, Y.; Ding, D.; Liu, X.; Zou, Q. TWIST1 and BMI1 in Cancer Metastasis and Chemoresistance. J. Cancer 2016, 7, 1074–1080. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, M.; Xia, W.; Xu, Y.; Mao, Q.; Wang, J.; Dong, G.; Xu, L.; Yang, X.; Yin, R. Glypican-5 suppresses Epithelial-Mesenchymal Transition of the lung adenocarcinoma by competitively binding to Wnt3a. Oncotarget 2016, 7, 79736. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Su, Y.-J.; Hsiao, M.; Wei, K.-C.; Lin, W.-H.; Liang, C.-J.; Chen, S.-C.; Lee, J.-L. Diverse Targets of β-Catenin during the Epithelial–Mesenchymal Transition Define Cancer Stem Cells and Predict Disease Relapse. Cancer Res. 2015, 75, 3398–3410. [Google Scholar] [CrossRef]
- Bao, R.; Christova, T.; Song, S.; Angers, S.; Yan, X.; Attisano, L. Inhibition of Tankyrases Induces Axin Stabilization and Blocks Wnt Signalling in Breast Cancer Cells. PLoS ONE 2012, 7, e48670. [Google Scholar] [CrossRef]
- Park, K.S.; Kim, S.J.; Kim, K.H.; Kim, J.C. Clinical characteristics of TIMP2, MMP2, and MMP9 gene polymorphisms in colorectal cancer. J. Gastroenterol. Hepatol. 2011, 26, 391–397. [Google Scholar] [CrossRef]
- Nandana, S.; Tripathi, M.; Duan, P.; Chu, C.-Y.; Mishra, R.; Liu, C.; Jin, R.; Yamashita, H.; Zayzafoon, M.; Bhowmick, N.A.; et al. Bone Metastasis of Prostate Cancer Can Be Therapeutically Targeted at the TBX2–WNT Signaling Axis. Cancer Res. 2017, 77, 1331–1344. [Google Scholar] [CrossRef]
- Lau, T.; Chan, E.; Callow, M.; Waaler, J.; Boggs, J.; Blake, R.A.; Magnuson, S.; Sambrone, A.; Schutten, M.; Firestein, R.; et al. A Novel Tankyrase Small-Molecule Inhibitor Suppresses APC Mutation–Driven Colorectal Tumor Growth. Cancer Res. 2013, 73, 3132–3144. [Google Scholar] [CrossRef]
- Guzmán, C.; Bagga, M.; Kaur, A.; Westermarck, J.; Abankwa, D. ColonyArea: An ImageJ Plugin to Automatically Quantify Colony Formation in Clonogenic Assays. PLoS ONE 2014, 9, e92444. [Google Scholar] [CrossRef]
- Suarez-Arnedo, A.; Figueroa, F.T.; Clavijo, C.; Arbeláez, P.; Cruz, J.C.; Muñoz-Camargo, C. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS ONE 2020, 15, e0232565. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
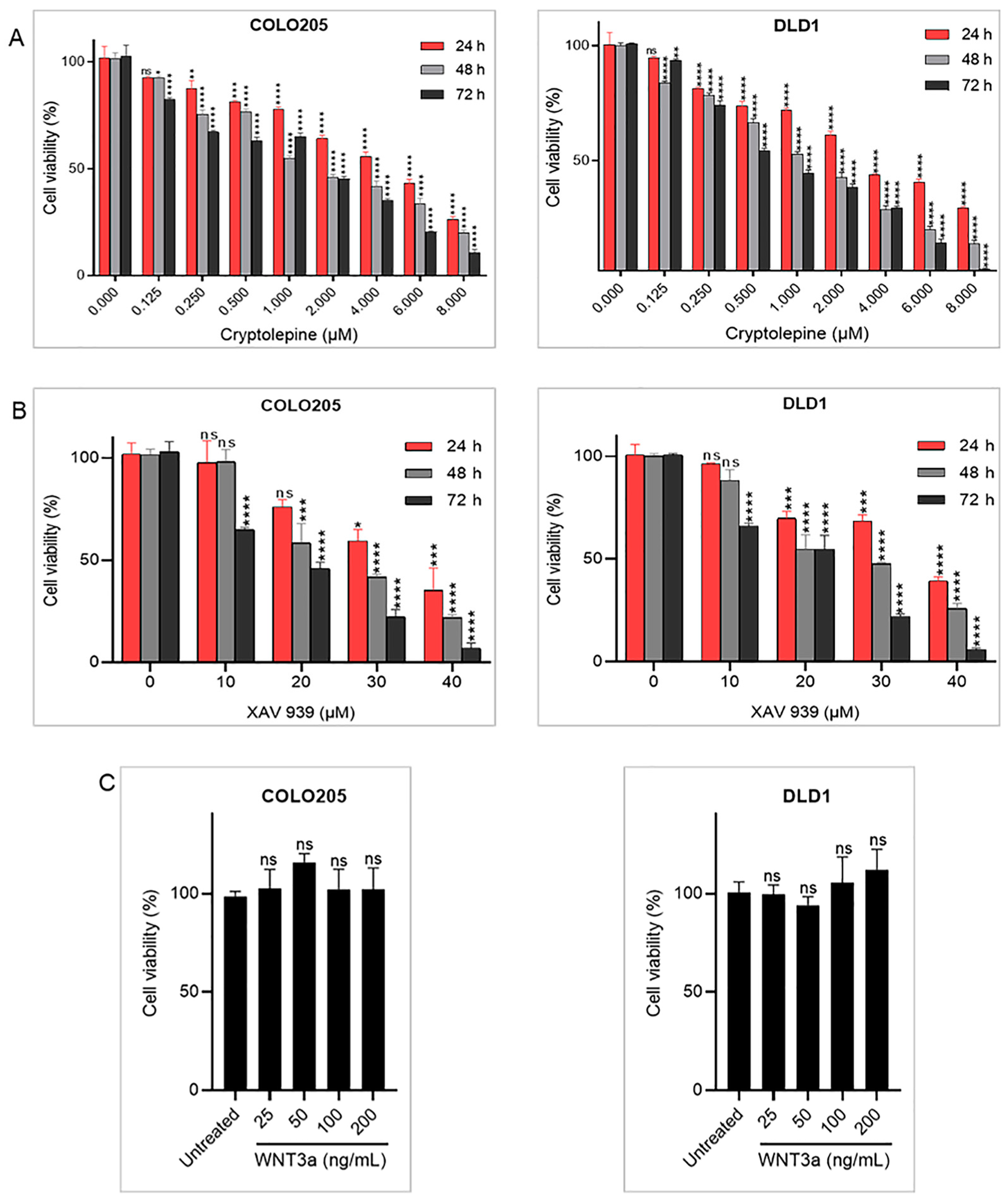


| Cell Lines | Time (h) | IC50 (µM) of Compounds | |
|---|---|---|---|
| Cryptolepine | XAV 939 | ||
| COLO205 | 24 | 5.01 ± 0.43 | 32.15 ± 3.19 |
| 48 | 2.45 ± 0.59 | 24.87 ± 2.45 | |
| 72 | 1.43 ± 0.15 | 16.51 ± 0.23 | |
| DLD1 | 24 | 3.51 ± 0.31 | 36.51 ± 1.04 |
| 48 | 1.16 ± 0.07 | 25.59 ± 3.22 | |
| 72 | 0.86 ± 0.02 | 19.19 ± 2.70 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quarshie, J.T.; Fosu, K.; Offei, N.A.; Sobo, A.K.; Quaye, O.; Aikins, A.R. Cryptolepine Suppresses Colorectal Cancer Cell Proliferation, Stemness, and Metastatic Processes by Inhibiting WNT/β-Catenin Signaling. Pharmaceuticals 2023, 16, 1026. https://doi.org/10.3390/ph16071026
Quarshie JT, Fosu K, Offei NA, Sobo AK, Quaye O, Aikins AR. Cryptolepine Suppresses Colorectal Cancer Cell Proliferation, Stemness, and Metastatic Processes by Inhibiting WNT/β-Catenin Signaling. Pharmaceuticals. 2023; 16(7):1026. https://doi.org/10.3390/ph16071026
Chicago/Turabian StyleQuarshie, Jude Tetteh, Kwadwo Fosu, Nicholas Awuku Offei, Augustine Kojo Sobo, Osbourne Quaye, and Anastasia Rosebud Aikins. 2023. "Cryptolepine Suppresses Colorectal Cancer Cell Proliferation, Stemness, and Metastatic Processes by Inhibiting WNT/β-Catenin Signaling" Pharmaceuticals 16, no. 7: 1026. https://doi.org/10.3390/ph16071026
APA StyleQuarshie, J. T., Fosu, K., Offei, N. A., Sobo, A. K., Quaye, O., & Aikins, A. R. (2023). Cryptolepine Suppresses Colorectal Cancer Cell Proliferation, Stemness, and Metastatic Processes by Inhibiting WNT/β-Catenin Signaling. Pharmaceuticals, 16(7), 1026. https://doi.org/10.3390/ph16071026






