Correlating Basal Gene Expression across Chemical Sensitivity Data to Screen for Novel Synergistic Interactors of HDAC Inhibitors in Pancreatic Carcinoma
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Data Preparation
3.2. Synergy Predictions
3.3. Cell Culture
3.4. Quantitative Analysis of Drug Synergy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Koltai, T.; Reshkin, S.J.; Carvalho, T.M.A.; Di Molfetta, D.; Greco, M.R.; Alfarouk, K.O.; Cardone, R.A. Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma: A Physiopathologic and Pharmacologic Review. Cancers 2022, 14, 2486. [Google Scholar] [CrossRef]
- Rahib, L.; Wehner, M.R.; Matrisian, L.M.; Nead, K.T. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw. Open 2021, 4, e214708. [Google Scholar] [CrossRef] [PubMed]
- Labori, K.J.; Katz, M.H.; Tzeng, C.W.; Bjørnbeth, B.A.; Cvancarova, M.; Edwin, B.; Kure, E.H.; Eide, T.J.; Dueland, S.; Buanes, T.; et al. Impact of Early Disease Progression and Surgical Complications on Adjuvant Chemotherapy Completion Rates and Survival in Patients Undergoing the Surgery First Approach for Resectable Pancreatic Ductal Adenocarcinoma—A Population-Based Cohort Study. Acta Oncol. 2016, 55, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic Ductal Adenocarcinoma: Biological Hallmarks, Current Status, and Future Perspectives of Combined Modality Treatment Approaches. Radiat. Oncol. 2019, 14, 141. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, C.; Xie, K.-P. Therapeutic Resistance of Pancreatic Cancer: Roadmap to Its Reversal. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188461. [Google Scholar] [CrossRef] [PubMed]
- Cancer Facts & Figures. 2022. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2022/2022-cancer-facts-and-figures.pdf (accessed on 9 October 2022).
- Xiang, X.-S.; Li, P.-C.; Wang, W.-Q.; Liu, L. Histone Deacetylases: A Novel Class of Therapeutic Targets for Pancreatic Cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer 2022, 1877, 188676. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Zhou, L.; Zhu, W.; Wang, T.; Wang, J.; Shu, Y.; Liu, P. Effects of Treatment with Histone Deacetylase Inhibitors in Solid Tumors: A Review Based on 30 Clinical Trials. Future Oncol. 2013, 9, 255–269. [Google Scholar] [CrossRef]
- Laschanzky, R.S.; Humphrey, L.E.; Ma, J.; Smith, L.M.; Enke, T.J.; Shukla, S.K.; Dasgupta, A.; Singh, P.K.; Howell, G.M.; Brattain, M.G.; et al. Selective Inhibition of Histone Deacetylases 1/2/6 in Combination with Gemcitabine: A Promising Combination for Pancreatic Cancer Therapy. Cancers 2019, 11, 1327. [Google Scholar] [CrossRef] [PubMed]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating Chemical Sensitivity and Basal Gene Expression Reveals Mechanism of Action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef]
- Yang, H.; Sun, B.; Xu, K.; He, Y.; Zhang, T.; Hall, S.R.R.; Tan, S.T.; Schmid, R.A.; Peng, R.-W.; Hu, G.; et al. Pharmaco-Transcriptomic Correlation Analysis Reveals Novel Responsive Signatures to HDAC Inhibitors and Identifies Dasatinib as a Synergistic Interactor in Small-Cell Lung Cancer. eBioMedicine 2021, 69, 103457. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A Resource for Therapeutic Biomarker Discovery in Cancer Cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modeling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Susanto, J.M.; Colvin, E.K.; Pinese, M.; Chang, D.K.; Pajic, M.; Mawson, A.; Caldon, C.E.; Musgrove, E.A.; Henshall, S.M.; Sutherland, R.L.; et al. The Epigenetic Agents Suberoylanilide Hydroxamic Acid and 5-AZA-2′ Deoxycytidine Decrease Cell Proliferation, Induce Cell Death and Delay the Growth of MiaPaCa2 Pancreatic Cancer Cells in Vivo. Int. J. Oncol. 2015, 46, 2223–2230. [Google Scholar] [CrossRef]
- Poklepovic, A.S.; Fields, E.C.; Bandyopadhyay, D.; Tombes, M.B.; Kmieciak, M.; McGuire, W.P.; Gordon, S.W.; Kaplan, B.J.; Myers, J.L.; Matin, K.; et al. A Phase 1 Study of Neoadjuvant Chemotherapy Followed by Concurrent Chemoradiation with Gemcitabine, Sorafenib, and Vorinostat in Pancreatic Cancer. J. Clin. Oncol. 2021, 39, e16268. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, X.; Yang, H.; Xu, X.; Hu, Z.; Yan, J.; Zheng, X.; Wei, R.; Zhang, Z.; Tang, S.; et al. CUDC-907 Displays Potent Antitumor Activity against Human Pancreatic Adenocarcinoma in Vitro and in Vivo through Inhibition of HDAC6 to Downregulate c-Myc Expression. Acta Pharm. Sin. 2019, 40, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Biermann, M.; Quintero, C.; Ferguson, P.; Rajbhandari, N.; Park, D.E.; Patel, H.; Reya, T. Repurposing HDAC and MTOR Inhibitors for Pancreatic Cancer. J. Clin. Oncol. 2022, 40, e16234. [Google Scholar] [CrossRef]
- Zhang, X.; Zegar, T.; Weiser, T.; Hamdan, F.H.; Berger, B.-T.; Lucas, R.; Balourdas, D.-I.; Ladigan, S.; Cheung, P.F.; Liffers, S.-T.; et al. Characterization of a Dual BET/HDAC Inhibitor for Treatment of Pancreatic Ductal Adenocarcinoma. Int. J. Cancer 2020, 147, 2847–2861. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- KEGG PATHWAY: Sphingolipid Signaling Pathway–Homo Sapiens (Human). Available online: https://www.genome.jp/pathway/hsa04071 (accessed on 9 October 2022).
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A Multifaceted Pathway Database Bridging Metabolomics to Other Omics Research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the Sphingolipid Rheostat: Evolving Concepts in Cancer Therapy. Exp. Cell Res. 2015, 333, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Speirs, M.M.P.; Swensen, A.C.; Chan, T.Y.; Jones, P.M.; Holman, J.C.; Harris, M.B.; Maschek, J.A.; Cox, J.E.; Carson, R.H.; Hill, J.T.; et al. Imbalanced Sphingolipid Signaling Is Maintained as a Core Proponent of a Cancerous Phenotype in Spite of Metabolic Pressure and Epigenetic Drift. Oncotarget 2019, 10, 449–479. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-X.; Ma, Y.-J.; Han, L.; Wang, Y.-J.; Han, J.-A.; Zhu, Y. Role of Sphingosine 1-Phosphate in Human Pancreatic Cancer Cells Proliferation and Migration. Int. J. Clin. Exp. Med. 2015, 8, 20349–20354. [Google Scholar] [PubMed]
- Lankadasari, M.B.; Aparna, J.S.; Mohammed, S.; James, S.; Aoki, K.; Binu, V.S.; Nair, S.; Harikumar, K.B. Targeting S1PR1/STAT3 Loop Abrogates Desmoplasia and Chemosensitizes Pancreatic Cancer to Gemcitabine. Theranostics 2018, 8, 3824–3840. [Google Scholar] [CrossRef]
- Lewis, C.S.; Voelkel-Johnson, C.; Smith, C.D. Targeting Sphingosine Kinases for the Treatment of Cancer. Adv. Cancer Res. 2018, 140, 295–325. [Google Scholar] [CrossRef]
- Yuza, K.; Nakajima, M.; Nagahashi, M.; Tsuchida, J.; Hirose, Y.; Miura, K.; Tajima, Y.; Abe, M.; Sakimura, K.; Takabe, K.; et al. Different Roles of Sphingosine Kinase 1 and 2 in Pancreatic Cancer Progression. J. Surg. Res. 2018, 232, 186–194. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Mrakovcic, M.; Kleinheinz, J.; Fröhlich, L.F. P53 at the Crossroads between Different Types of HDAC Inhibitor-Mediated Cancer Cell Death. Int. J. Mol. Sci. 2019, 20, 2415. [Google Scholar] [CrossRef]
- Heffernan-Stroud, L.A.; Obeid, L.M. P53 and Regulation of Bioactive Sphingolipids. Adv. Enzym. Regul. 2011, 51, 219–228. [Google Scholar] [CrossRef]
- Ruzic, D.; Ellinger, B.; Djokovic, N.; Santibanez, J.F.; Gul, S.; Beljkas, M.; Djuric, A.; Ganesan, A.; Pavic, A.; Srdic-Rajic, T.; et al. Discovery of 1-Benzhydryl-Piperazine-Based HDAC Inhibitors with Anti-Breast Cancer Activity: Synthesis, Molecular Modeling, In Vitro and In Vivo Biological Evaluation. Pharmaceutics 2022, 14, 2600. [Google Scholar] [CrossRef]
- Vennin, C.; Rath, N.; Pajic, M.; Olson, M.F.; Timpson, P. Targeting ROCK Activity to Disrupt and Prime Pancreatic Cancer for Chemotherapy. Small GTPases 2020, 11, 45–52. [Google Scholar] [CrossRef]
- Rath, N.; Munro, J.; Cutiongco, M.F.; Jagiełło, A.; Gadegaard, N.; McGarry, L.; Unbekandt, M.; Michalopoulou, E.; Kamphorst, J.J.; Sumpton, D.; et al. Rho Kinase Inhibition by AT13148 Blocks Pancreatic Ductal Adenocarcinoma Invasion and Tumor Growth. Cancer Res. 2018, 78, 3321–3336. [Google Scholar] [CrossRef] [PubMed]
- Lepley, D.; Paik, J.-H.; Hla, T.; Ferrer, F. The G Protein-Coupled Receptor S1P2 Regulates Rho/Rho Kinase Pathway to Inhibit Tumor Cell Migration. Cancer Res. 2005, 65, 3788–3795. [Google Scholar] [CrossRef] [PubMed]
- Huwiler, A.; Zangemeister-Wittke, U. The Sphingosine 1-Phosphate Receptor Modulator Fingolimod as a Therapeutic Agent: Recent Findings and New Perspectives. Pharmacol. Ther. 2018, 185, 34–49. [Google Scholar] [CrossRef]
- Hait, N.C.; Wise, L.E.; Allegood, J.C.; O’Brien, M.; Avni, D.; Reeves, T.M.; Knapp, P.E.; Lu, J.; Luo, C.; Miles, M.F.; et al. Active, Phosphorylated Fingolimod Inhibits Histone Deacetylases and Facilitates Fear Extinction Memory. Nat. Neurosci. 2014, 17, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Sriram, K.; Salmerón, C.; Wiley, S.Z.; Insel, P.A. GPCRs in Pancreatic Adenocarcinoma: Contributors to Tumour Biology and Novel Therapeutic Targets. Br. J. Pharm. 2020, 177, 2434–2455. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Talalay, P. Quantitative Analysis of Dose-Effect Relationships: The Combined Effects of Multiple Drugs or Enzyme Inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X. CytoNCA: A Cytoscape Plugin for Centrality Analysis and Evaluation of Protein Interaction Networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
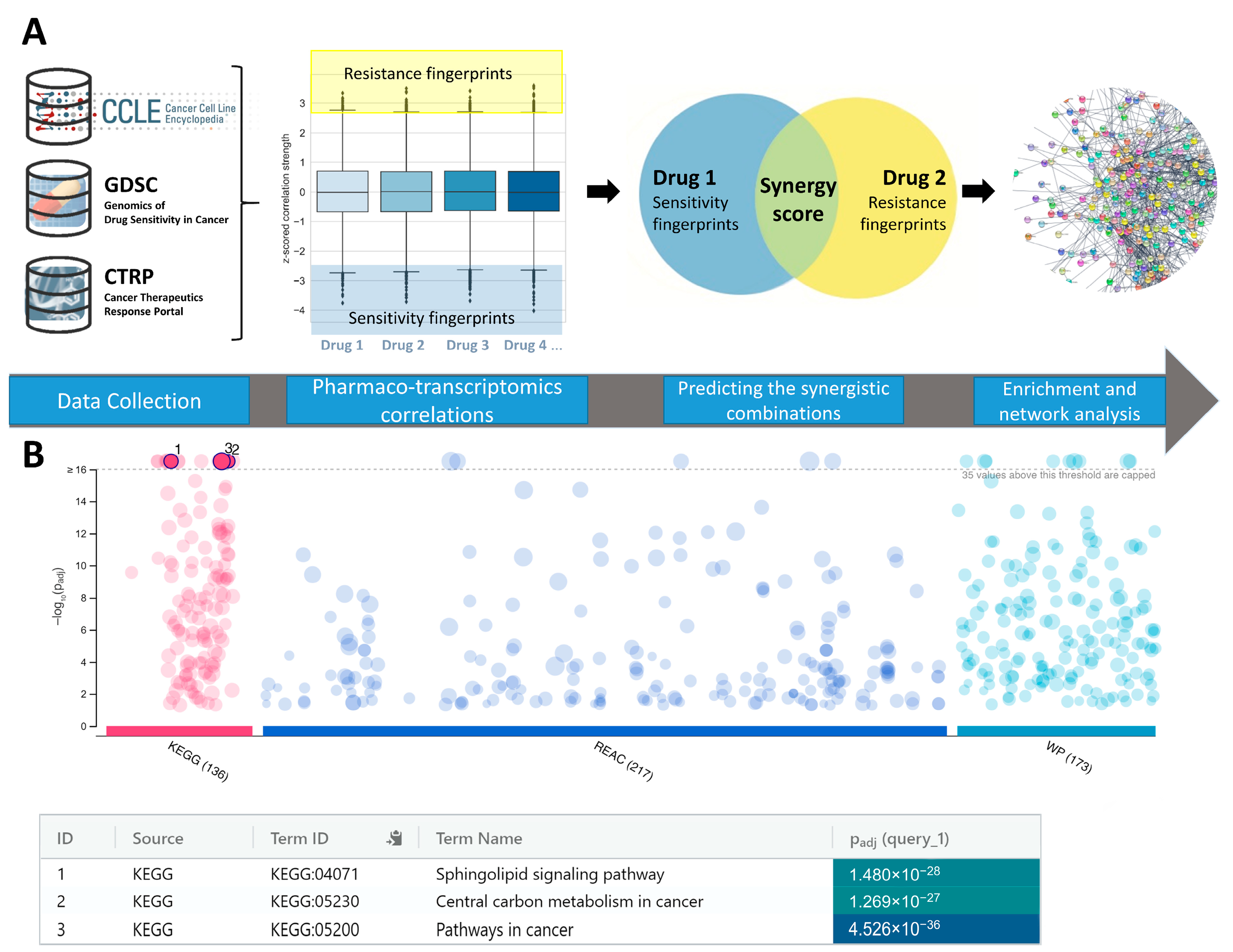
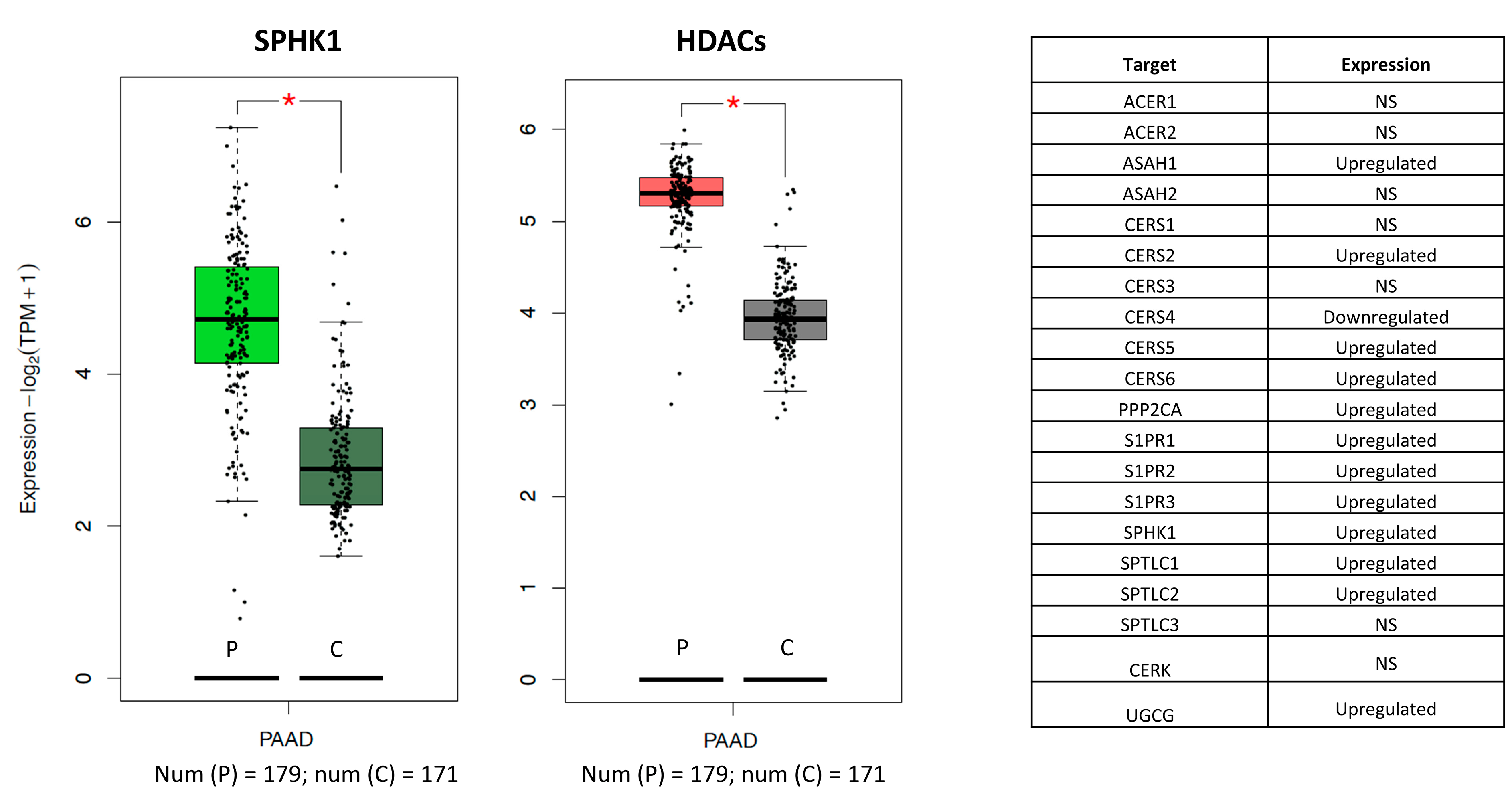
| Molecule 1 (HDACi) | Molecule 2 | Target Profile of Molecule 1 | Target Profile of Molecule 2 | Count +/− a | Count −/+ b | Final_Syn_Score c |
|---|---|---|---|---|---|---|
| tacedinaline | RG-108 | HDAC1; HDAC2; HDAC3; HDAC6; HDAC8 | DNMT1 | 37 | 403 | 440 (0.3%) |
| Merck60 | Sorafenib | HDAC1; HDAC2 | BRAF; FLT3; KDR; RAF1 | 14 | 61 | 75 (5%) |
| tacedinaline | PI-103 | HDAC1; HDAC2; HDAC3; HDAC6; HDAC8 | PI3Kalpha, DAPK3, CLK4, PIM3, HIPK2 | 23 | 243 | 266 (1%) |
| tacedinaline | KU-0063794 | HDAC1; HDAC2; HDAC3; HDAC6; HDAC8 | MTOR | 189 | 425 | 614 (0.1%) |
| BRD-K85133207 | AZD5153 | HDAC1 | BRD4 | 36 | 47 | 83 (4.6%) |
| PCI-34051 | gemcitabine | HDAC8, HDAC6, HDAC1 | CMPK1; RRM1; TYMS | 32 | 34 | 66 (5.7%) |
| PCI-34051 | GSK429286A | HDAC8, HDAC6, HDAC1 | ROCK1, ROCK2 | 65 | 79 | 144 (2.3%) |
| BRD-K51490254 | SKI-II | HDAC6; HDAC8 | SPHK1 | 12 | 63 | 75 (5%) |
| PCI-34051 | fingolimod | HDAC8, HDAC6, HDAC1 | S1PR1 | 10 | 17 | 27 (12.7%) |
| Gene ID | BC | Gene ID | BC |
|---|---|---|---|
| TP53 | 56,527 | APOA1 | 8858 |
| VWF | 19,076 | ETV1 | 8308 |
| CALR | 17,531 | PEBP1 | 7894 |
| NTRK1 | 16,901 | ITGA4 | 7747 |
| MMP9 | 16,341 | GNGT2 | 7426 |
| CYCS | 13,336 | SLC17A7 | 7102 |
| DNM1 | 11,804 | TPT1 | 6997 |
| FOXP3 | 10,884 | BUD13 | 6632 |
| H2AFX | 10,415 | COPS5 | 6290 |
| RCC1 | 9091 | DDB2 | 6156 |
| MIA PaCa-2 Cell Line | Panc-1 Cell Line | |||||||
|---|---|---|---|---|---|---|---|---|
| Compound Concentration a | Compound Concentration a | |||||||
| I | II | III | IV | I | II | III | IV | |
| HDACi (6b) | 48.60 ± 7.45 | 13.58 ± 2.33 | 4.80 ± 1.01 | 2.91 ± 0.87 | 23.61 ± 3.26 | 1.39 ± 0.55 | 0.47 ± 0.21 | 0.5 ± 0.2 |
| ROCKi (RKI-1447) | 64.18 ± 8.12 | 40.49 ± 5.55 | 37.49 ± 3.02 | 30.84 ± 7.33 | 62.76 ± 11.23 | 33.89 ± 8.92 | 22.02 ± 7.22 | 18.11 ± 5.90 |
| 6b + RKI-1447 | 83.28 ± 10.34 | 55.48 ± 7.51 | 37.69 ± 5.38 | 38.31 ± 4.56 | 69.79 ± 10.45 | 48.51 ± 6.34 | 24.98 ± 3.13 | 8.92 ± 4.21 |
| Combination Index (CI) | 0.445 | 0.981 | 1.253 | 0.605 | 0.992 | 1.050 | 1.334 | 2.117 |
| Interaction 6b + RKI-1447 b | + + + | + | − − | + + | ± | ± | − − | − − − |
| HDACi (8b) | 27.30 ± 4.44 | 1.83 ± 0.70 | 0.94 ± 0.45 | 0.57 ± 0.19 | 49.78 ± 6.77 | 38.21 ± 5.89 | 17.13 ± 2.11 | 0.1 ± 0.05 |
| ROCKi (RKI-1447) | 62.94 ± 10.11 | 42.93 ± 6.99 | 36.07 ± 7.51 | 35.46 ± 4.09 | 58.78 ± 5.55 | 30.77 ± 3.45 | 17.27 ± 2.28 | 7.41 ± 2.56 |
| 8b + RKI-1447 | 73.02 ± 7.44 | 45.32 ± 5.14 | 31.14 ± 5.21 | 28.91 ± 2.34 | 63.94 ± 6.33 | 42.96 ± 5.79 | 21.99 ± 3.45 | 6.79 ± 3.06 |
| Combination Index (CI) | 0.333 | 1.564 | 2.469 | 1.505 | 3.133 | 1.433 | 1.266 | 1.474 |
| Interaction 8b + RKI-1447 b | + + + | − − − | − − − | − − − | − − − | − − | − − | − − − |
| HDACi (9b) | 44.65 ± 3.39 | 18.25 ± 2.22 | 1.71 ± 0.67 | 0.46 ± 0.20 | 45.18 ± 4.55 | 23.14 ± 7.42 | 7.56 ± 3.71 | 1.12 ± 0.89 |
| ROCKi (RKI-1447) | 61.64 ± 7.78 | 43.28 ± 5.38 | 35.02 ± 3.25 | 29.16 ± 2.16 | 64.82 ± 6.41 | 37.92 ± 5.34 | 19.76 ± 2.30 | 9.41 ± 1.06 |
| 9b + RKI-1447 | 78.03 ± 7.18 | 64.37 ± 6.78 | 41.91 ± 5.98 | 33.32 ± 4.21 | 64.68 ± 5.01 | 51.26 ± 7.20 | 33.14 ± 6.10 | 18.01 ± 4.02 |
| Combination Index (CI) | 0.513 | 0.577 | 0.964 | 0.803 | 1.705 | 1.207 | 0.975 | 0.823 |
| Interaction 9b + RKI-1447 b | + + + | + + + | ± | + + | − − − | − − | ± | + |
| MIA PaCa-2 Cell Line | Panc-1 Cell Line | |||||||
|---|---|---|---|---|---|---|---|---|
| Compound Concentration a | Compound Concentration a | |||||||
| I | II | III | IV | I | II | III | IV | |
| HDACi (6b) | 71.49 ± 11.91 | 41.62 ± 11.91 | 23.06 ± 9.75 | 16.27 ± 5.34 | 63.68 ± 4.70 | 25.29 ± 6.71 | 23.93 ± 6.23 | 10.30 ± 1.96 |
| Fingolimod | 79.98 ± 10.36 | 48.31 ± 12.04 | 20.57 ± 4.85 | 20.33 ± 5.37 | 79.18 ± 1.45 | 40.09 ± 4.97 | 24.56 ± 1.24 | 18.99 ± 0.95 |
| 6b + Fingolimod | 89.28 ± 4.05 | 60.81 ± 20.19 | 34.11 ± 14.58 | 26.96 ± 7.59 | 84.96 ± 2.05 | 67.22 ± 5.98 | 37.87 ± 5.71 | 21.98 ± 0.78 |
| Combination Index (CI) | 0.780 | 1.422 | 1.648 | 1.070 | 0.847 | 0.945 | 1.232 | 1.141 |
| Interaction 6b + Fingolimod b | + + | − − | − − − | ± | + + | ± | − − | − |
| HDACi (8b) | 81.22 ± 0.96 | 68.72 ± 2.69 | 36.08 ± 3.29 | 16.72 ± 4.28 | 58.10 ± 0.24 | 23.86 ± 9.24 | 4.77 ± 2.69 | 4.68 ± 2.33 |
| Fingolimod | 87.22 ± 2.01 | 52.02 ± 2.47 | 23.73 ± 3.92 | 22.83 ± 1.89 | 68.24 ± 4.62 | 41.81 ± 5.37 | 19.44 ± 10.97 | 12.27 ± 8.75 |
| 8b + Fingolimod | 91.31 ± 10.08 | 65.06 ± 1.92 | 38.98 ± 5.26 | 26.55 ± 2.94 | 74.74 ± 5.61 | 30.91 ± 6.84 | 16.60 ± 7.22 | 9.42 ± 5.57 |
| Combination Index (CI) | 1.188 | 1.836 | 1.852 | 1.338 | 1.267 | 1.806 | 2.027 | 1.592 |
| Interaction 8b + Fingolimod b | − − − | − − − | − − − | − − | − − | − − − | − − − − | − − − |
| HDACi (9b) | 83.61 ± 0.27 | 69.42 ± 11.34 | 53.31 ± 17.80 | 44.84 ± 11.21 | 61.28 ± 6.91 | 50.93 ± 3.83 | 40.71 ± 3.74 | 24.03 ± 5.73 |
| Fingolimod | 76.58 ± 4.48 | 63.82 ± 6.81 | 43.78 ± 12.65 | 35.34 ± 6.99 | 62.10 ± 4.94 | 50.04 ± 3.20 | 31.21 ± 11.85 | 21.17 ± 3.33 |
| 9b + Fingolimod | 89.11 ± 1.57 | 73.59 ± 12.40 | 55.57 ± 18.16 | 47.22 ± 9.51 | 76.50 ± 5.14 | 66.12 ± 10.76 | 53.41 ± 18.12 | 33.34 ± 10.07 |
| Combination Index (CI) | 0.870 | 1.449 | 1.773 | 1.293 | 0.911 | 0.848 | 0.810 | 1.120 |
| Interaction 9b + Fingolimod b | + | − − | − − − | − − | ± | + + | + + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djokovic, N.; Djuric, A.; Ruzic, D.; Srdic-Rajic, T.; Nikolic, K. Correlating Basal Gene Expression across Chemical Sensitivity Data to Screen for Novel Synergistic Interactors of HDAC Inhibitors in Pancreatic Carcinoma. Pharmaceuticals 2023, 16, 294. https://doi.org/10.3390/ph16020294
Djokovic N, Djuric A, Ruzic D, Srdic-Rajic T, Nikolic K. Correlating Basal Gene Expression across Chemical Sensitivity Data to Screen for Novel Synergistic Interactors of HDAC Inhibitors in Pancreatic Carcinoma. Pharmaceuticals. 2023; 16(2):294. https://doi.org/10.3390/ph16020294
Chicago/Turabian StyleDjokovic, Nemanja, Ana Djuric, Dusan Ruzic, Tatjana Srdic-Rajic, and Katarina Nikolic. 2023. "Correlating Basal Gene Expression across Chemical Sensitivity Data to Screen for Novel Synergistic Interactors of HDAC Inhibitors in Pancreatic Carcinoma" Pharmaceuticals 16, no. 2: 294. https://doi.org/10.3390/ph16020294
APA StyleDjokovic, N., Djuric, A., Ruzic, D., Srdic-Rajic, T., & Nikolic, K. (2023). Correlating Basal Gene Expression across Chemical Sensitivity Data to Screen for Novel Synergistic Interactors of HDAC Inhibitors in Pancreatic Carcinoma. Pharmaceuticals, 16(2), 294. https://doi.org/10.3390/ph16020294









