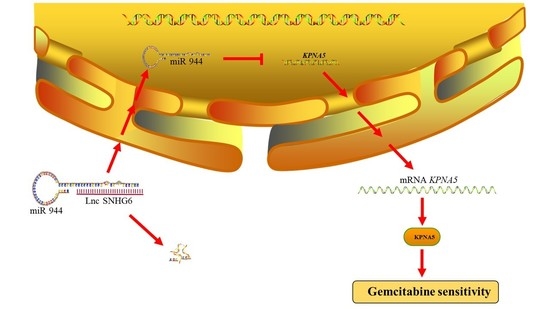
LncRNA SNHG6 Upregulates KPNA5 to Overcome Gemcitabine Resistance in Pancreatic Cancer via Sponging miR-944
Abstract
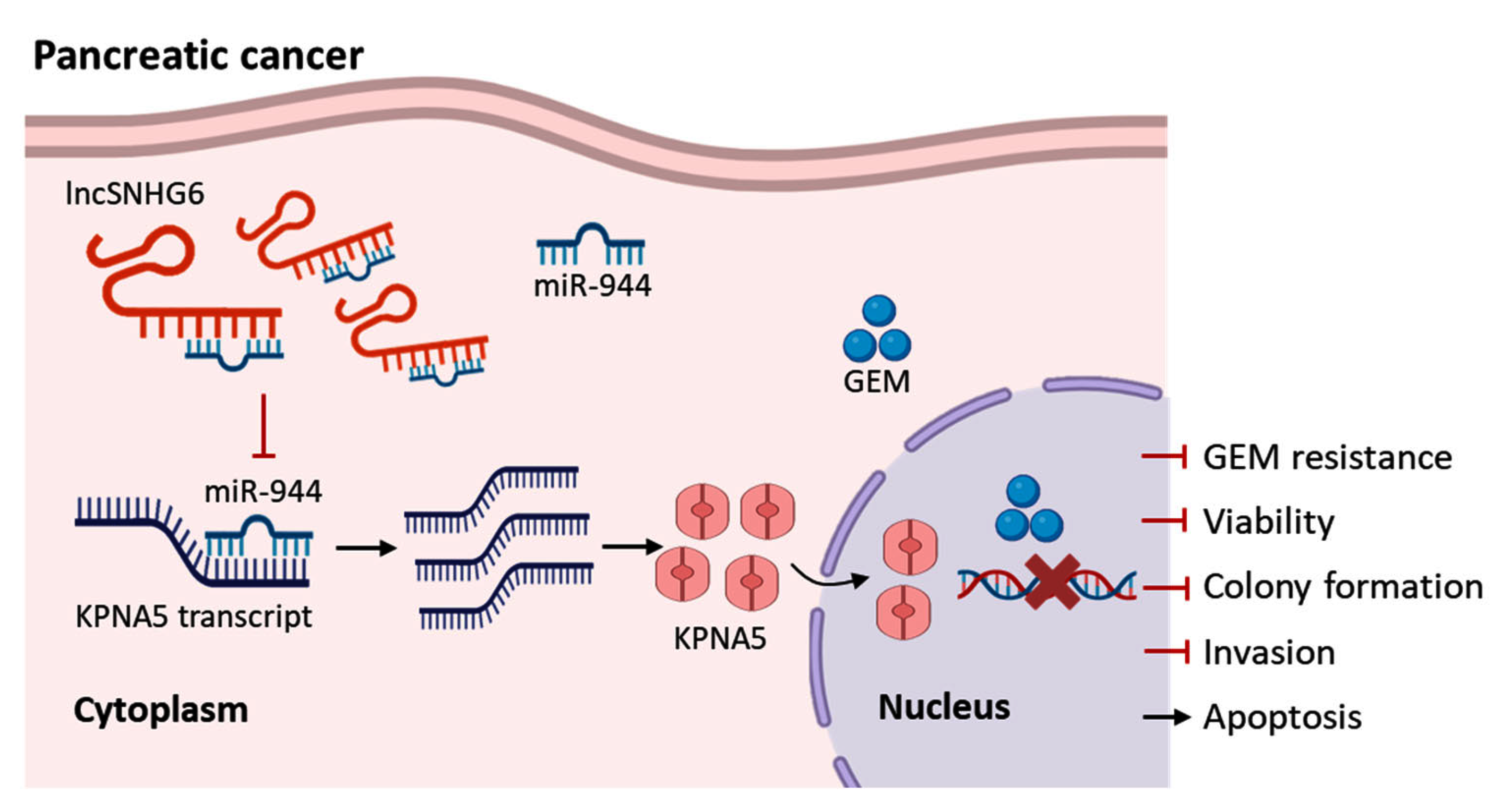
1. Introduction
2. Results and Discussion
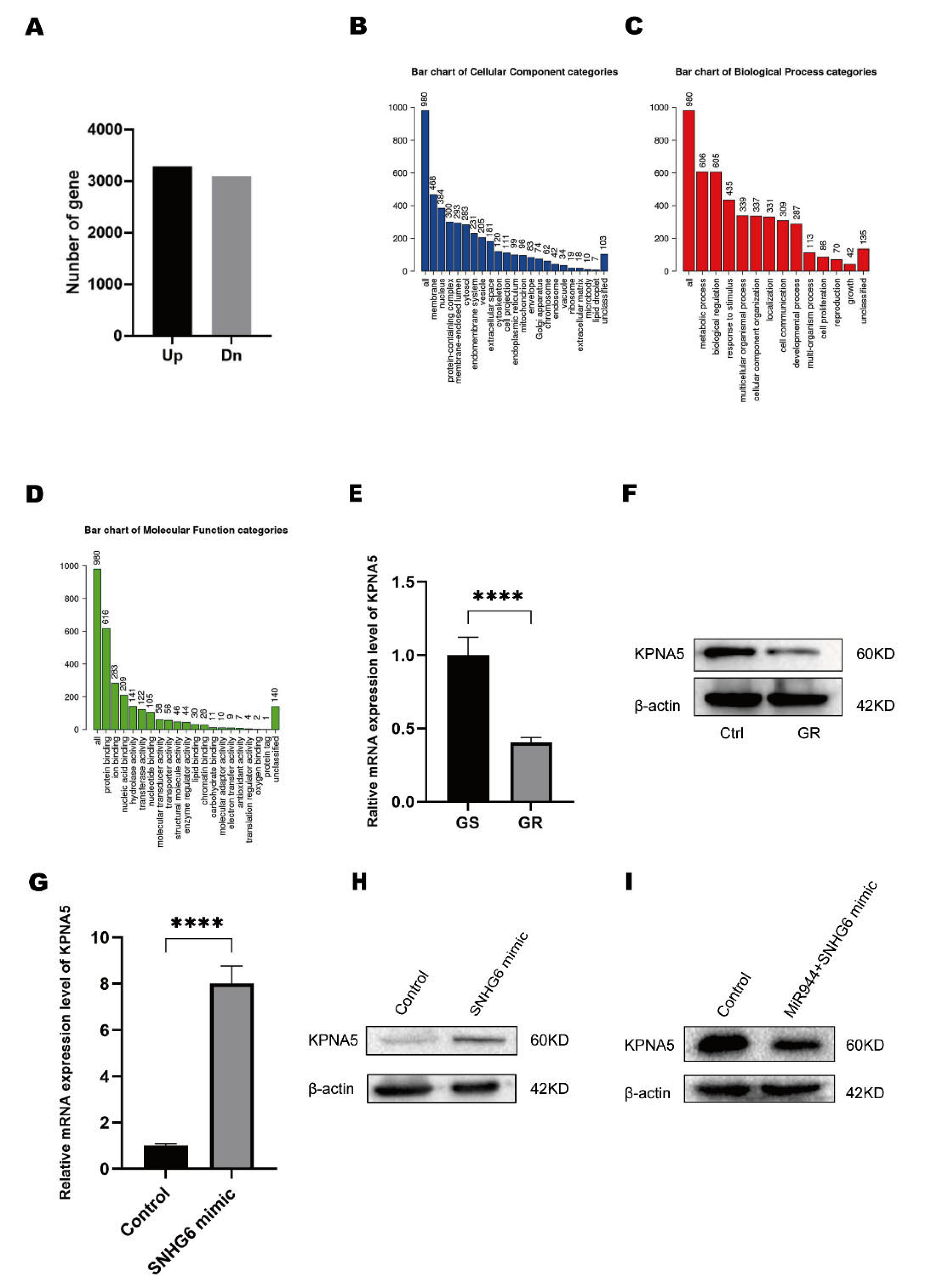
2.1. LncRNA SNHG6 Is Downregulated in GEM-Resistant PC
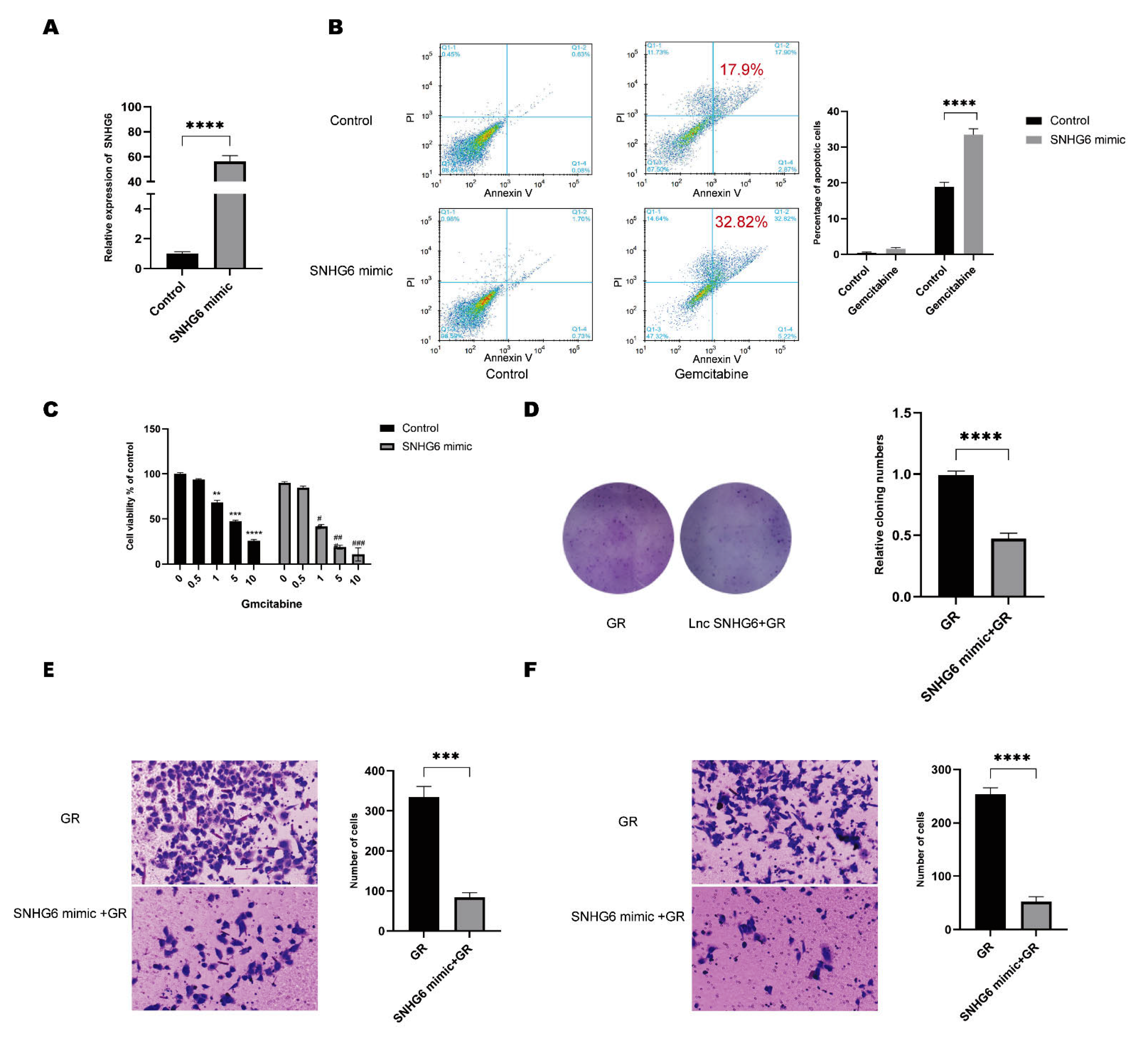
2.2. Upregulation of LncRNA SNHG6 Increases the Response of PC Cells to GEM
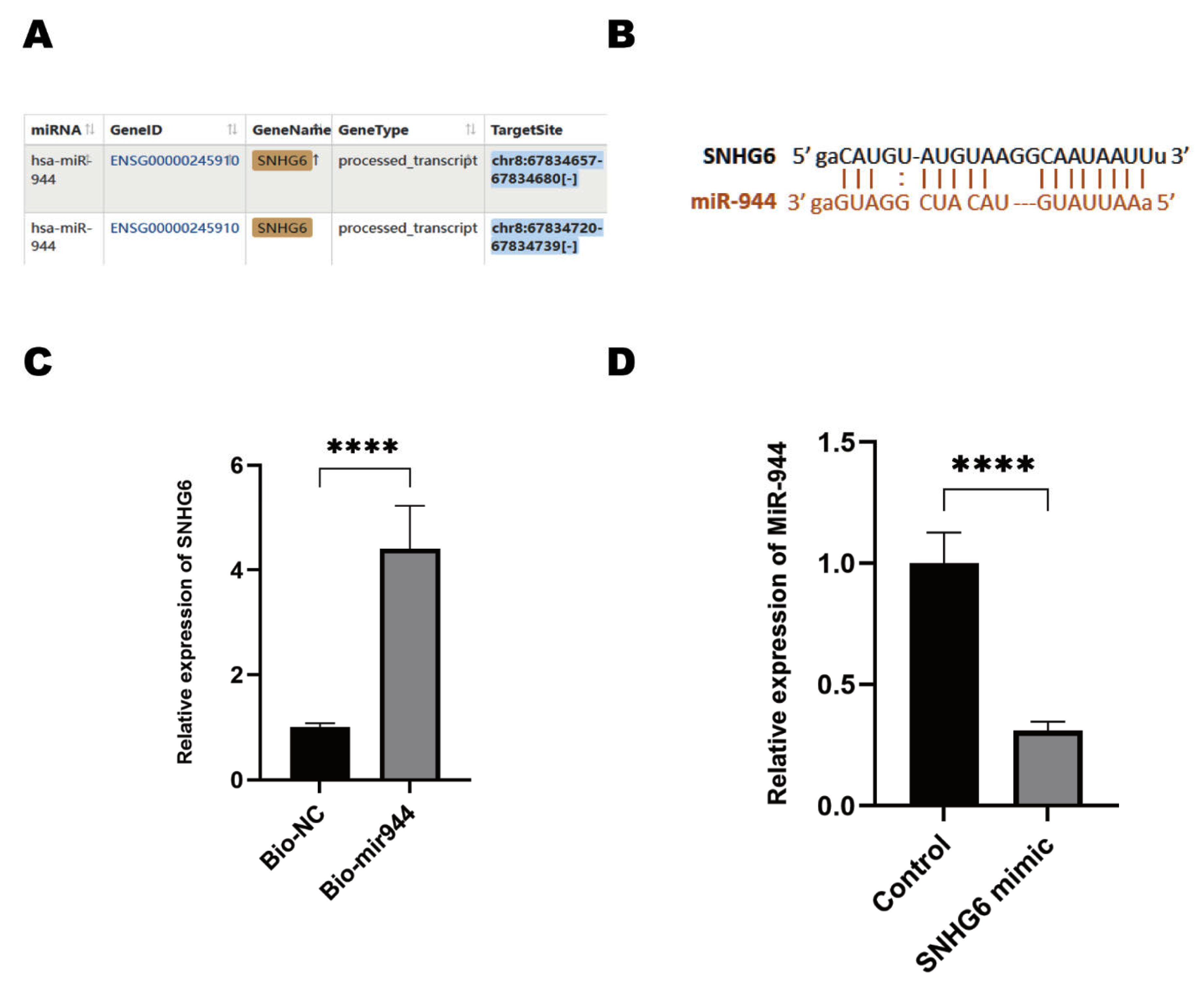
2.3. LncRNA SNHG6 Upregulates KPNA5 Expression by Acting as a Sponge of miR-944
2.4. LncRNA SNHG6/miR-944 Regulates the Expression of KPNA5
2.5. KPNA5 Helps Overcome GEM Resistance in Patients with PC
3. Materials and Methods
3.1. Biological Samples
3.2. Cell Culture
3.3. Transfection
3.4. Bioinformatic Analysis
3.5. Quantitative Polymerase Chain Reaction (qPCR)
3.6. Western Blotting
3.7. RNA Pulldown
3.8. Colony Formation
3.9. CCK8-Based Cell Viability Assay
3.10. Flow Cytometry Analysis
3.11. Transwell-Based Cell Invasion Assay
3.12. Immunohistochemistry Staining
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grossberg, A.J.; Chu, L.C.; Deig, C.R.; Fishman, E.K.; Hwang, W.L.; Maitra, A.; Marks, D.L.; Mehta, A.; Nabavizadeh, N.; Simeone, D.M.; et al. Multidisciplinary Standards of Care and Recent Progress in Pancreatic Ductal Adenocarcinoma. CA Cancer J. Clin. 2020, 70, 375–403. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yang, G.; Yang, J.; Ren, B.; Wang, H.; Chen, G.; Zhao, F.; You, L.; Wang, W.; Zhao, Y. Metabolism of Pancreatic Cancer: Paving the Way to Better Anticancer Strategies. Mol. Cancer 2020, 19, 50. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic Cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Cui Zhou, D.; Jayasinghe, R.G.; Chen, S.; Herndon, J.M.; Iglesia, M.D.; Navale, P.; Wendl, M.C.; Caravan, W.; Sato, K.; Storrs, E.; et al. Spatially Restricted Drivers and Transitional Cell Populations Cooperate with the Microenvironment in Untreated and Chemo-Resistant Pancreatic Cancer. Nat. Genet. 2022, 54, 1390–1405. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sanomachi, T.; Suzuki, S.; Uchida, H.; Yonezawa, H.; Higa, N.; Takajo, T.; Yamada, Y.; Sugai, A.; Togashi, K.; et al. Roles for HENT1 and DCK in Gemcitabine Sensitivity and Malignancy of Meningioma. Neuro-Oncology 2021, 23, 945–954. [Google Scholar] [CrossRef]
- Mini, E.; Nobili, S.; Caciagli, B.; Landini, I.; Mazzei, T. Cellular Pharmacology of Gemcitabine. Ann. Oncol. 2006, 17, v7–v12. [Google Scholar] [CrossRef]
- Zhou, C.; Yi, C.; Yi, Y.; Qin, W.; Yan, Y.; Dong, X.; Zhang, X.; Huang, Y.; Zhang, R.; Wei, J.; et al. LncRNA PVT1 Promotes Gemcitabine Resistance of Pancreatic Cancer via Activating Wnt/β-Catenin and Autophagy Pathway through Modulating the MiR-619-5p/Pygo2 and MiR-619-5p/ATG14 Axes. Mol. Cancer 2020, 19, 118. [Google Scholar] [CrossRef]
- Hu, C.; Xia, R.; Zhang, X.; Li, T.; Ye, Y.; Li, G.; He, R.; Li, Z.; Lin, Q.; Zheng, S.; et al. CircFARP1 Enables Cancer-Associated Fibroblasts to Promote Gemcitabine Resistance in Pancreatic Cancer via the LIF/STAT3 Axis. Mol. Cancer 2022, 21, 24. [Google Scholar] [CrossRef]
- Xu, S.; Jiang, C.; Lin, R.; Wang, X.; Hu, X.; Chen, W.; Chen, X.; Chen, T. Epigenetic Activation of the Elongator Complex Sensitizes Gallbladder Cancer to Gemcitabine Therapy. J. Exp. Clin. Cancer Res. 2021, 40, 373. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Zhang, B.; Tan, Z.; Meng, Q.; Hua, J.; Liu, J.; Wang, W.; Shi, S.; Yu, X.; et al. Ferroptosis: At the Crossroad of Gemcitabine Resistance and Tumorigenesis in Pancreatic Cancer. Int. J. Mol. Sci. 2021, 22, 10944. [Google Scholar] [CrossRef]
- Koltai, T.; Reshkin, S.J.; Carvalho, T.M.A.; Di Molfetta, D.; Greco, M.R.; Alfarouk, K.O.; Cardone, R.A. Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma: A Physiopathologic and Pharmacologic Review. Cancers 2022, 14, 2486. [Google Scholar] [CrossRef]
- Sun, M.; Kraus, W.L. From Discovery to Function: The Expanding Roles of Long NonCoding RNAs in Physiology and Disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-Coding RNAs as Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Yu, S.; Li, Y.; Liao, Z.; Wang, Z.; Wang, Z.; Li, Y.; Qian, L.; Zhao, J.; Zong, H.; Kang, B.; et al. Plasma Extracellular Vesicle Long RNA Profiling Identifies a Diagnostic Signature for the Detection of Pancreatic Ductal Adenocarcinoma. Gut 2020, 69, 540–550. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Li, W.; Han, S.; Hu, P.; Chen, D.; Zeng, Z.; Hu, Y.; Xu, F.; Tang, J.; Wang, F.; Zhao, Y.; et al. LncRNA ZNFTR Functions as an Inhibitor in Pancreatic Cancer by Modulating ATF3/ZNF24/VEGFA Pathway. Cell Death Dis. 2021, 12, 830. [Google Scholar] [CrossRef]
- Chen, K.; Hou, Y.; Liao, R.; Li, Y.; Yang, H.; Gong, J. LncRNA SNHG6 Promotes G1/S-Phase Transition in Hepatocellular Carcinoma by Impairing MiR-204-5p-Mediated Inhibition of E2F1. Oncogene 2021, 40, 3217–3230. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Xu, X.; Pan, B.; Xu, T.; Sun, L.; He, B.; et al. LncRNA SNHG6 Regulates EZH2 Expression by Sponging MiR-26a/b and MiR-214 in Colorectal Cancer. J. Hematol. Oncol. 2019, 12, 3. [Google Scholar] [CrossRef]
- Wang, H.; Ma, P.; Liu, P.; Guo, D.; Liu, Z.; Zhang, Z. LncRNA SNHG6 Promotes Hepatocellular Carcinoma Progression by Interacting with HNRNPL/PTBP1 to Facilitate SETD7/LZTFL1 MRNA Destabilization. Cancer Lett. 2021, 520, 121–131. [Google Scholar] [CrossRef]
- Yao, X.; Lan, Z.; Lai, Q.; Li, A.; Liu, S.; Wang, X. LncRNA SNHG6 Plays an Oncogenic Role in Colorectal Cancer and Can Be Used as a Prognostic Biomarker for Solid Tumors. J. Cell. Physiol. 2020, 235, 7620–7634. [Google Scholar] [CrossRef]
- Cao, C.; Zhang, T.; Zhang, D.; Xie, L.; Zou, X.; Lei, L.; Wu, D.; Liu, L. The Long Non-Coding RNA, SNHG6-003, Functions as a Competing Endogenous RNA to Promote the Progression of Hepatocellular Carcinoma. Oncogene 2017, 36, 1112–1122. [Google Scholar] [CrossRef]
- Biagioni, A.; Tavakol, S.; Ahmadirad, N.; Zahmatkeshan, M.; Magnelli, L.; Mandegary, A.; Samareh Fekri, H.; Asadi, M.H.; Mohammadinejad, R.; Ahn, K.S. Small Nucleolar RNA Host Genes Promoting Epithelial–Mesenchymal Transition Lead Cancer Progression and Metastasis. IUBMB Life 2021, 73, 825–842. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Jian, Z.; Yan, X.; Li, J.; Zhang, R. LncRNA SNHG6 Inhibits Cell Proliferation and Metastasis by Targeting ETS1 via the PI3K/AKT/MTOR Pathway in Colorectal Cancer. Mol. Med. Rep. 2019, 20, 2541–2548. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, D.; Zhao, M.; Huang, S.; Zhang, Q.; Lin, H.; Wang, W.; Li, K.; Li, Z.; Huang, W.; et al. Long Noncoding RNA SNHG6 Regulates P21 Expression via Activation of the JNK Pathway and Regulation of EZH2 in Gastric Cancer Cells. Life Sci. 2018, 208, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, Q.; Liang, C.; Su, X.; Ke, Y.; Mao, Y.; Fang, J.; Duan, S. Novel Insights into MiR-944 in Cancer. Cancers 2022, 14, 4232. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The First Metal Based Anticancer Drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Köhler, M.; Ansieau, S.; Prehn, S.; Leutz, A.; Haller, H.; Hartmann, E. Cloning of Two Novel Human Importin-α Subunits and Analysis of the Expression Pattern of the Importin-α Protein Family. FEBS Lett. 1997, 417, 104–108. [Google Scholar] [CrossRef]
- Lan, X.; Zhao, L.; Zhang, J.; Shao, Y.; Qv, Y.; Huang, J.; Cai, L. Comprehensive Analysis of Karyopherin Alpha Family Expression in Lung Adenocarcinoma: Association with Prognostic Value and Immune Homeostasis. Front. Genet. 2022, 13, 956314. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Yamada, K.; Yoneda, Y. Importin α: A Key Molecule in Nuclear Transport and Non-Transport Functions. J. Biochem. 2016, 160, 69–75. [Google Scholar] [CrossRef]
- Kim, N.-H.; Yoshimaru, T.; Chen, Y.-A.; Matsuo, T.; Komatsu, M.; Miyoshi, Y.; Tanaka, E.; Sasa, M.; Mizuguchi, K.; Katagiri, T. BIG3 Inhibits the Estrogen-Dependent Nuclear Translocation of PHB2 via Multiple Karyopherin-Alpha Proteins in Breast Cancer Cells. PLoS ONE 2015, 10, e0127707. [Google Scholar] [CrossRef]
- Mehmood, R.; Jibiki, K.; Shibazaki, N.; Yasuhara, N. Molecular Profiling of Nucleocytoplasmic Transport Factor Genes in Breast Cancer. Heliyon 2021, 7, e06039. [Google Scholar] [CrossRef]
- Ma, L.; Zhao, W.; Zhu, F.; Yuan, F.; Xie, N.; Li, T.; Wang, P.; Tong, T. Global Characteristics of CSIG-Associated Gene Expression Changes in Human HEK293 Cells and the Implications for CSIG Regulating Cell Proliferation and Senescence. Front. Endocrinol. 2015, 6, 69. [Google Scholar] [CrossRef]
- Pu, F.-F.; Shi, D.-Y.; Chen, T.; Liu, Y.-X.; Zhong, B.-L.; Zhang, Z.-C.; Liu, W.-J.; Wu, Q.; Wang, B.-C.; Shao, Z.-W.; et al. SP1-Induced Long Non-Coding RNA SNHG6 Facilitates the Carcinogenesis of Chondrosarcoma through Inhibiting KLF6 by Recruiting EZH2. Cell Death Dis. 2021, 12, 59. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Lee, J.H.; Jin, S.; Kim, J.H.; Kim, S.H. Primate-Specific MiR-944 Activates P53-Dependent Tumor Suppression in Human Colorectal Cancers. Cancer Lett. 2019, 440–441, 168–179. [Google Scholar] [CrossRef]
- Lv, W.; Tan, Y.; Xiong, M.; Zhao, C.; Wang, Y.; Wu, M.; Wu, Y.; Zhang, Q. Analysis and Validation of M6A Regulatory Network: A Novel CircBACH2/Has-MiR-944/HNRNPC Axis in Breast Cancer Progression. J. Transl. Med. 2021, 19, 527. [Google Scholar] [CrossRef]
- Lv, P.; Qiu, X.; Gu, Y.; Yang, X.; Xu, X.; Yang, Y. Long Non-Coding RNA SNHG6 Enhances Cell Proliferation, Migration and Invasion by Regulating MiR-26a-5p/MAPK6 in Breast Cancer. Biomed. Pharmacother. 2019, 110, 294–301. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Xue, Y.; Ouyang, K.; Huang, J.; Zhou, Y.; Ouyang, H.; Li, H.; Wang, G.; Wu, Q.; Wei, C.; Bi, Y.; et al. Direct Conversion of Fibroblasts to Neurons by Reprogramming PTB-Regulated MicroRNA Circuits. Cell 2013, 152, 82–96. [Google Scholar] [CrossRef]
- Balakrishnan, I.; Yang, X.; Brown, J.; Ramakrishnan, A.; Torok-Storb, B.; Kabos, P.; Hesselberth, J.R.; Pillai, M.M. Genome-Wide Analysis of MiRNA-MRNA Interactions in Marrow Stromal Cells. Stem Cells 2014, 32, 662–673. [Google Scholar] [CrossRef]
- Liu, B.; Wang, T.; Wang, H.; Zhang, L.; Xu, F.; Fang, R.; Li, L.; Cai, X.; Wu, Y.; Zhang, W.; et al. Oncoprotein HBXIP Enhances HOXB13 Acetylation and Co-Activates HOXB13 to Confer Tamoxifen Resistance in Breast Cancer. J. Hematol. Oncol. 2018, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, Y.; Zheng, X.; Dai, F.; Lu, Y.; Dai, L.; Niu, M.; Guo, H.; Li, W.; Xue, X.; et al. Circular RNA CircCORO1C Promotes Laryngeal Squamous Cell Carcinoma Progression by Modulating the Let-7c-5p/PBX3 Axis. Mol. Cancer 2020, 19, 99. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, X.; Zhang, C.; Zhou, X. Microplastics Induce Immune Suppression via S100A8 Downregulation. Ecotoxicol. Environ. Saf. 2022, 242, 113905. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhu, J.; Chen, H.; Qian, J.; Zhang, L.; Wan, Z.; Chen, F.; Sun, S.; Li, W.; Luo, C. A Novel LncRNA-LINC01116 Regulates Tumorigenesis of Glioma by Targeting VEGFA. Int. J. Cancer 2020, 146, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yang, Y.; Kang, M.; Wang, Y.; Wang, Y.; Bi, Y.; He, S.; Shimamoto, F. M6A Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol. Cancer 2020, 19, 3. [Google Scholar] [CrossRef]


| GENE | Primer Sequence |
|---|---|
| SNHG6 | Forward 5’-3’ TGCCAGCAGTGACAGCAGCA |
| Reverse 5’-3’ TACGGAGGTGGAGTGCCAT | |
| miR-944 | Forward 5’-3’ GCACTCCTAAAATTATTGTACATCG |
| Reverse 5’-3’ TATGGTTGTTCACGACTCCTTCAC | |
| KPNA5 | Forward 5’-3’ TCAGGAACAGGCTGTTTGGG |
| Reverse 5’-3’ TGGGGTCATCTTCTTCTACAC | |
| 18S rRNA | Forward 5’-3’ TGTGCCGCTAGAGGTGAAATT |
| Reverse 5’-3’ TGGCAAATGCTTTCGCTTT | |
| U6 | Forward 5’-3’ CTCGCTTCGGCAGCACATATACT Reverse 5’-3’ACGCTTCACGAATTTGCGTGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, G.; Li, X.; Wu, H.; Huang, L.-l.; Lin, Y.-x.; Huo, Z.; Xiang, Z.-y.; Zhou, X. LncRNA SNHG6 Upregulates KPNA5 to Overcome Gemcitabine Resistance in Pancreatic Cancer via Sponging miR-944. Pharmaceuticals 2023, 16, 184. https://doi.org/10.3390/ph16020184
Gao G, Li X, Wu H, Huang L-l, Lin Y-x, Huo Z, Xiang Z-y, Zhou X. LncRNA SNHG6 Upregulates KPNA5 to Overcome Gemcitabine Resistance in Pancreatic Cancer via Sponging miR-944. Pharmaceuticals. 2023; 16(2):184. https://doi.org/10.3390/ph16020184
Chicago/Turabian StyleGao, Ge, Xin Li, Hui Wu, Ling-li Huang, Yu-xin Lin, Zhi Huo, Zhong-yuan Xiang, and Xiao Zhou. 2023. "LncRNA SNHG6 Upregulates KPNA5 to Overcome Gemcitabine Resistance in Pancreatic Cancer via Sponging miR-944" Pharmaceuticals 16, no. 2: 184. https://doi.org/10.3390/ph16020184
APA StyleGao, G., Li, X., Wu, H., Huang, L.-l., Lin, Y.-x., Huo, Z., Xiang, Z.-y., & Zhou, X. (2023). LncRNA SNHG6 Upregulates KPNA5 to Overcome Gemcitabine Resistance in Pancreatic Cancer via Sponging miR-944. Pharmaceuticals, 16(2), 184. https://doi.org/10.3390/ph16020184