Construction and Validation of an Oxaliplatin-Resistant Gene Signature in Colorectal Cancer Patients Who Underwent Chemotherapy
Abstract
:1. Introduction
2. Results
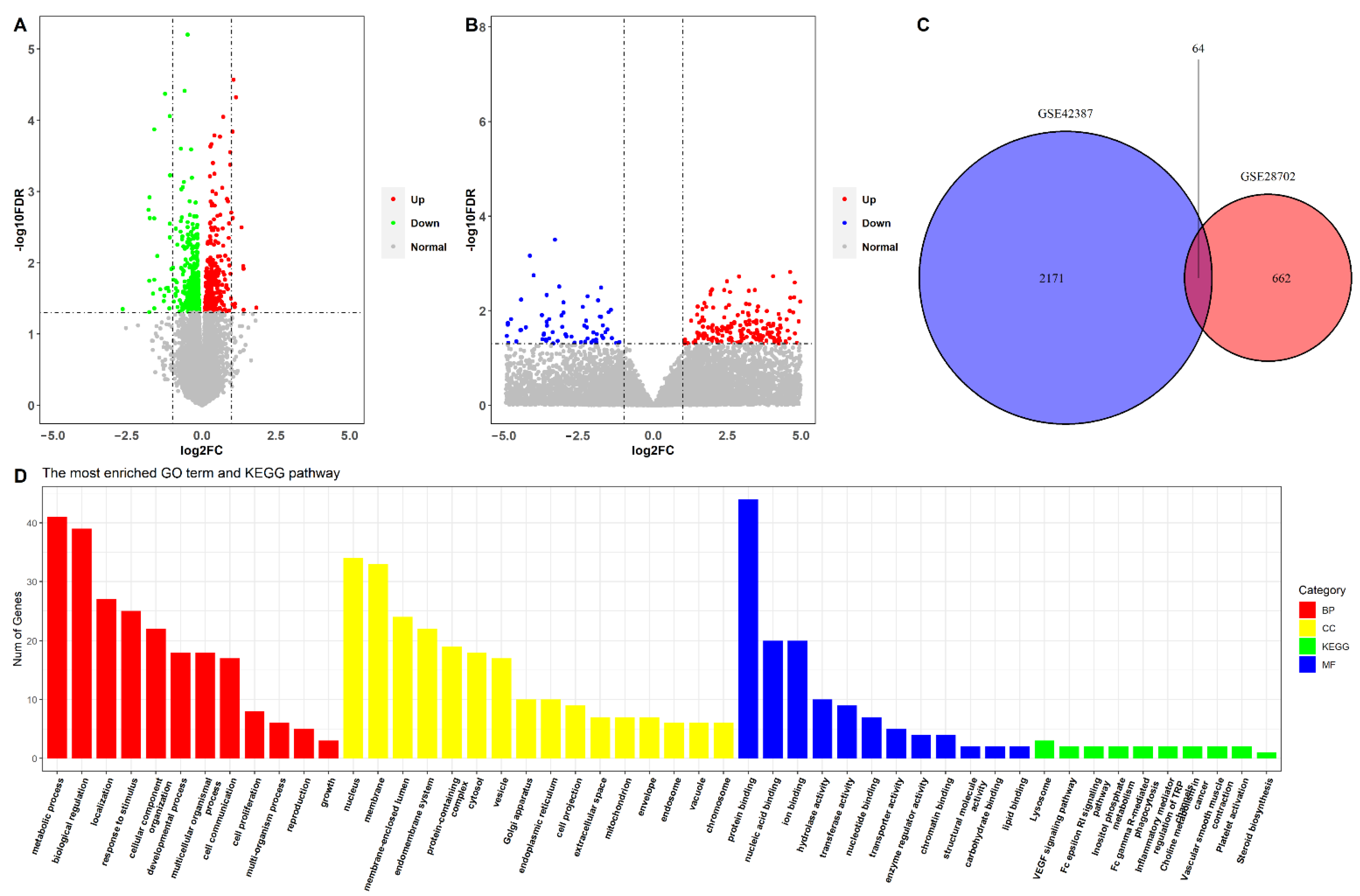
2.1. Identification of Oxaliplatin Resistance-Related DEGs in CRC Cells and Patients Who Underwent Oxaliplatin-Based Chemotherapy
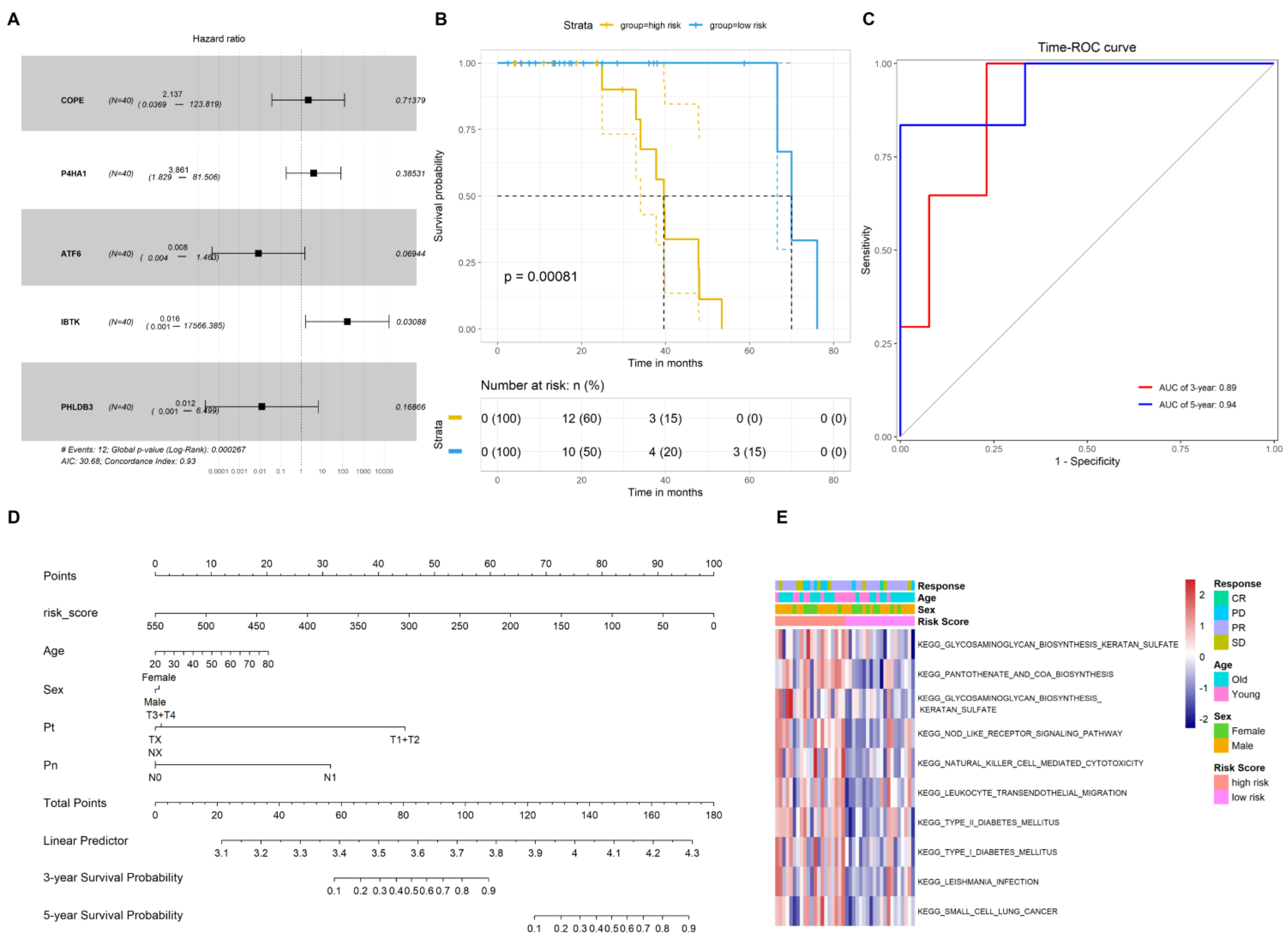
2.2. Construction of the Gene Signature to Predict the Prognosis of CRC Patients Who Underwent Chemotherapy
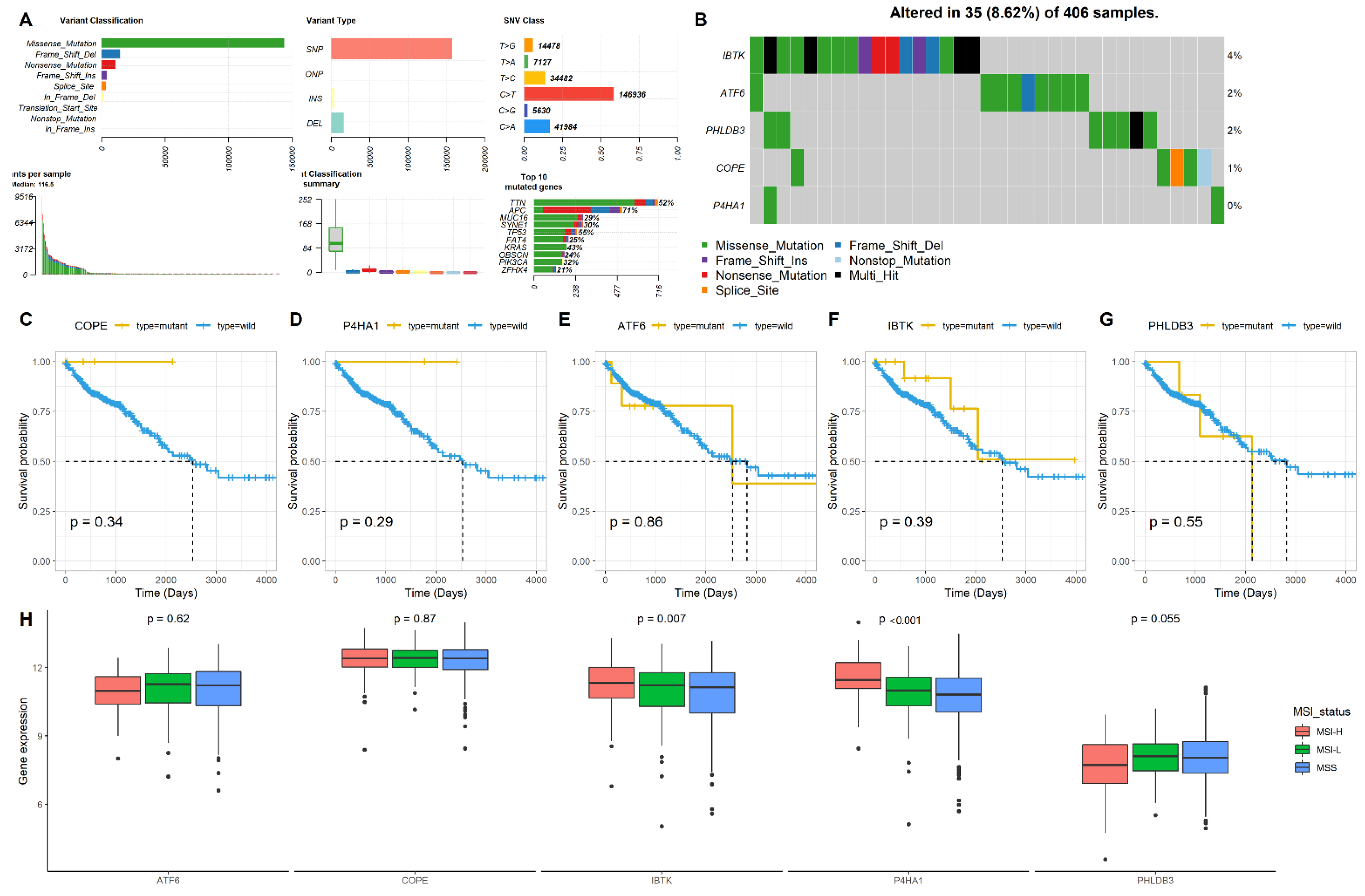
2.3. Mutation and MSI Status of the Five Genes in CRC
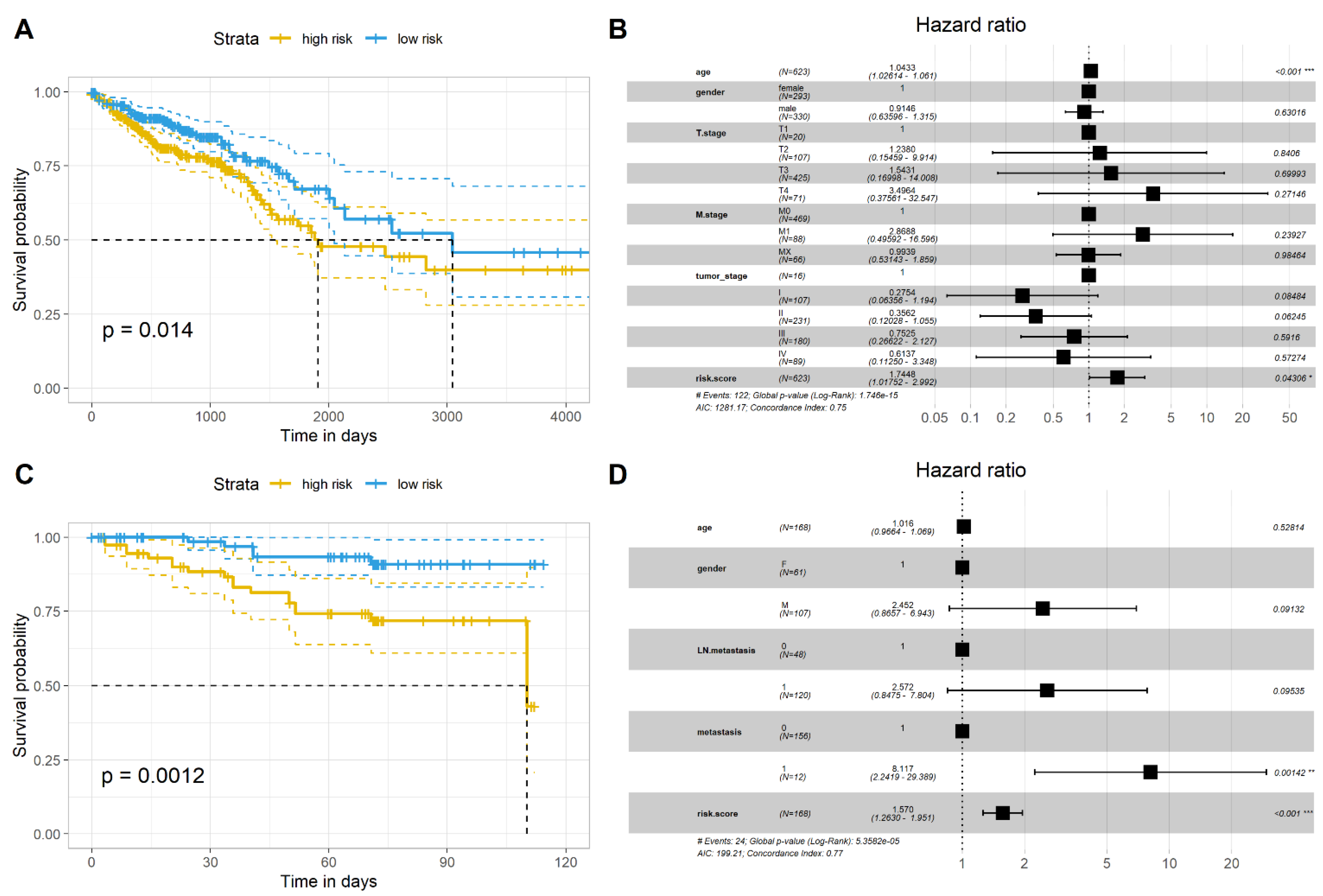
2.4. Validation of the Gene Signature in Two Independent Datasets
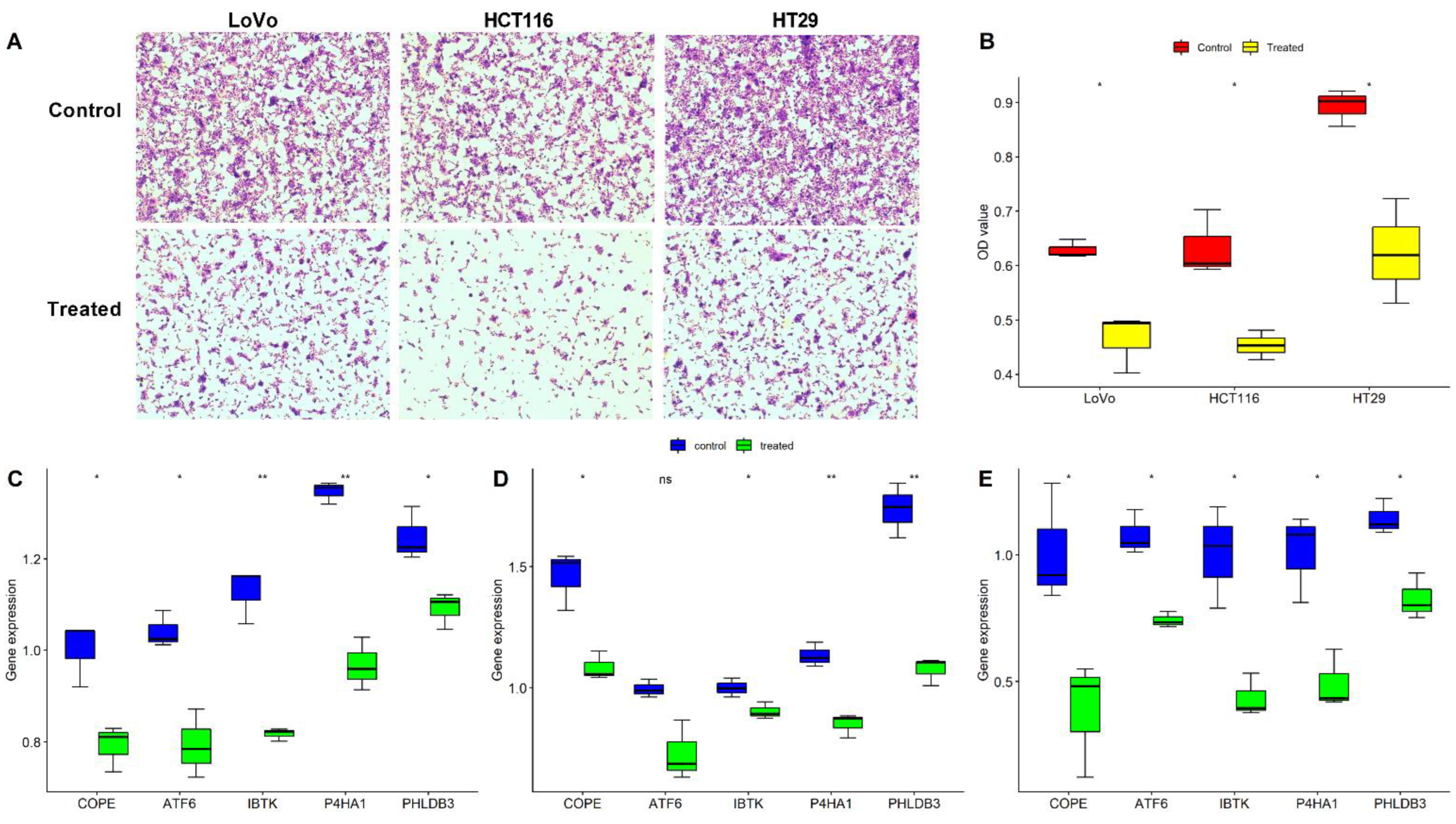
2.5. Validation of the Five Oxaliplatin-Resistant Genes in Three CRC Cells
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Processing
4.2. Gene Ontology Analysis and Gene Set Variation Analysis (GSVA)
4.3. Construction of an Oxaliplatin Resistance-Related Gene-Based Prognostic Signature
4.4. Cell Culture and Real-Time Polymerase Chain Reaction (RT-PCR) Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B., 3rd; Venook, A.P.; Cederquist, L.; Chan, E.; Chen, Y.J.; Cooper, H.S.; Deming, D.; Engstrom, P.F.; Enzinger, P.C.; Fichera, A.; et al. Colon Cancer, Version 1. 2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2017, 15, 370–398. [Google Scholar] [CrossRef] [PubMed]
- Escalante, P.I.; Quinones, L.A.; Contreras, H.R. Epithelial-Mesenchymal Transition and MicroRNAs in Colorectal Cancer Chemoresistance to FOLFOX. Pharmaceutics 2021, 13, 75. [Google Scholar] [CrossRef] [PubMed]
- Temraz, S.; Mukherji, D.; Alameddine, R.; Shamseddine, A. Methods of overcoming treatment resistance in colorectal cancer. Crit. Rev. Oncol. Hematol. 2014, 89, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Li, A.J.; Han, Y.; Yin, L.; Lin, M.B. Inhibition of Girdin enhances chemosensitivity of colorectal cancer cells to oxaliplatin. World J. Gastroenterol. 2014, 20, 8229–8236. [Google Scholar] [CrossRef]
- Lin, Q.; Luo, L.; Wang, H. A New Oxaliplatin Resistance-Related Gene Signature With Strong Predicting Ability in Colon Cancer Identified by Comprehensive Profiling. Front. Oncol. 2021, 11, 644956. [Google Scholar] [CrossRef]
- Rottenberg, S.; Disler, C.; Perego, P. The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer 2021, 21, 37–50. [Google Scholar] [CrossRef]
- Morii, Y.; Tsubaki, M.; Takeda, T.; Otubo, R.; Seki, S.; Yamatomo, Y.; Imano, M.; Satou, T.; Shimomura, K.; Nishida, S. Perifosine enhances the potential antitumor effect of 5-fluorourasil and oxaliplatin in colon cancer cells harboring the PIK3CA mutation. Eur. J. Pharmacol. 2021, 898, 173957. [Google Scholar] [CrossRef]
- Harada, K.; Okamoto, W.; Mimaki, S.; Kawamoto, Y.; Bando, H.; Yamashita, R.; Yuki, S.; Yoshino, T.; Komatsu, Y.; Ohtsu, A.; et al. Comparative sequence analysis of patient-matched primary colorectal cancer, metastatic, and recurrent metastatic tumors after adjuvant FOLFOX chemotherapy. BMC Cancer 2019, 19, 255. [Google Scholar] [CrossRef]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Sudan, S.K.; Deshmukh, S.K.; Poosarla, T.; Holliday, N.P.; Dyess, D.L.; Singh, A.P.; Singh, S. Resistin: An inflammatory cytokine with multi-faceted roles in cancer. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188419. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Z.; Cai, S.; Yu, L.; Hu, H.; Zeng, S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert Opin. Drug Metab. Toxicol. 2021, 17, 291–306. [Google Scholar] [CrossRef]
- Zhang, Y.; Devocelle, A.; Desterke, C.; de Souza, L.E.B.; Hadadi, E.; Acloque, H.; Foudi, A.; Xiang, Y.; Ballesta, A.; Chang, Y.; et al. BMAL1 Knockdown Leans Epithelial-Mesenchymal Balance toward Epithelial Properties and Decreases the Chemoresistance of Colon Carcinoma Cells. Int. J. Mol. Sci. 2021, 22, 5247. [Google Scholar] [CrossRef]
- Zhang, Q.; Yin, Y.; Zhao, H.; Shi, Y.; Zhang, W.; Yang, Z.; Liu, T.; Huang, Y.; Yu, Z. P4HA1 regulates human colorectal cancer cells through HIF1alpha-mediated Wnt signaling. Oncol. Lett. 2021, 21, 145. [Google Scholar] [CrossRef]
- Agarwal, S.; Behring, M.; Kim, H.G.; Bajpai, P.; Chakravarthi, B.; Gupta, N.; Elkholy, A.; Diffalha, S.A.; Varambally, S.; Manne, U. Corrigendum to ‘Targeting P4HA1 with a Small Molecule Inhibitor in a Colorectal Cancer PDX Model’ [Translational Oncology 13 (2020) 100754]. Transl. Oncol. 2021, 14, 101142. [Google Scholar] [CrossRef]
- Li, Y.; Ge, Y.Z.; Qian, Y.; Chen, K.; Zhao, F.; Qin, Z.; Zhou, L.; Xu, L.; Xu, Z.; Dou, Q.; et al. The Role of P4HA1 in Multiple Cancer Types and its Potential as a Target in Renal Cell Carcinoma. Front. Genet. 2022, 13, 848456. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, J. P4HA1, a Prognostic Biomarker that Correlates With Immune Infiltrates in Lung Adenocarcinoma and Pan-Cancer. Front. Cell Dev. Biol. 2021, 9, 754580. [Google Scholar] [CrossRef]
- Spaan, C.N.; Smit, W.L.; van Lidth de Jeude, J.F.; Meijer, B.J.; Muncan, V.; van den Brink, G.R.; Heijmans, J. Expression of UPR effector proteins ATF6 and XBP1 reduce colorectal cancer cell proliferation and stemness by activating PERK signaling. Cell Death Dis. 2019, 10, 490. [Google Scholar] [CrossRef]
- Chao, T.; Zhou, X.; Cao, B.; Liao, P.; Liu, H.; Chen, Y.; Park, H.W.; Zeng, S.X.; Lu, H. Pleckstrin homology domain-containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53. Nat. Commun. 2016, 7, 13755. [Google Scholar] [CrossRef]
- Vecchio, E.; Golino, G.; Pisano, A.; Albano, F.; Falcone, C.; Ceglia, S.; Iaccino, E.; Mimmi, S.; Fiume, G.; Giurato, G.; et al. IBTK contributes to B-cell lymphomagenesis in Emu-myc transgenic mice conferring resistance to apoptosis. Cell Death Dis. 2019, 10, 320. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, G.; Li, M.; Du, J.; Wang, M. COPB2 gene silencing inhibits colorectal cancer cell proliferation and induces apoptosis via the JNK/c-Jun signaling pathway. PLoS ONE 2020, 15, e0240106. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Taieb, J.; Fiskum, J.; Yothers, G.; Goldberg, R.; Yoshino, T.; Alberts, S.; Allegra, C.; de Gramont, A.; Seitz, J.F.; et al. Microsatellite Instability in Patients With Stage III Colon Cancer Receiving Fluoropyrimidine With or Without Oxaliplatin: An ACCENT Pooled Analysis of 12 Adjuvant Trials. J. Clin. Oncol. 2021, 39, 642–651. [Google Scholar] [CrossRef]
- Liu, J.; Wang, B.; Fang, W. Microsatellite instability and sensitivity to fluoropyrimidine and oxaliplatin containing first-line chemotherapy in metastatic colorectal cancer. Eur. J. Hosp. Pharm. 2020, 27, 267–270. [Google Scholar] [CrossRef]
- Del Rio, M.; Mollevi, C.; Bibeau, F.; Vie, N.; Selves, J.; Emile, J.F.; Roger, P.; Gongora, C.; Robert, J.; Tubiana-Mathieu, N.; et al. Molecular subtypes of metastatic colorectal cancer are associated with patient response to irinotecan-based therapies. Eur. J. Cancer 2017, 76, 68–75. [Google Scholar] [CrossRef]
- Jensen, N.F.; Stenvang, J.; Beck, M.K.; Hanakova, B.; Belling, K.C.; Do, K.N.; Viuff, B.; Nygard, S.B.; Gupta, R.; Rasmussen, M.H.; et al. Establishment and characterization of models of chemotherapy resistance in colorectal cancer: Towards a predictive signature of chemoresistance. Mol. Oncol. 2015, 9, 1169–1185. [Google Scholar] [CrossRef]
- Tsuji, S.; Midorikawa, Y.; Takahashi, T.; Yagi, K.; Takayama, T.; Yoshida, K.; Sugiyama, Y.; Aburatani, H. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br. J. Cancer 2012, 106, 126–132. [Google Scholar] [CrossRef]
- Hu, Y.; Gaedcke, J.; Emons, G.; Beissbarth, T.; Grade, M.; Jo, P.; Yeager, M.; Chanock, S.J.; Wolff, H.; Camps, J.; et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer 2018, 57, 140–149. [Google Scholar] [CrossRef]
- Chen, H.; Chen, C.; Yuan, X.; Xu, W.; Yang, M.Q.; Li, Q.; Shen, Z.; Yin, L. Identification of Immune Cell Landscape and Construction of a Novel Diagnostic Nomogram for Crohn’s Disease. Front. Genet. 2020, 11, 423. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Mo, Z.; Yu, L.; Cao, Z.; Hu, H.; Luo, S.; Zhang, S. Identification of a Hypoxia-Associated Signature for Lung Adenocarcinoma. Front. Genet. 2020, 11, 647. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Y.; Li, S.; Liang, X.; Li, K.; Xie, M.; Hu, B. Construction and Validation of an Oxaliplatin-Resistant Gene Signature in Colorectal Cancer Patients Who Underwent Chemotherapy. Pharmaceuticals 2022, 15, 1139. https://doi.org/10.3390/ph15091139
Yin Y, Li S, Liang X, Li K, Xie M, Hu B. Construction and Validation of an Oxaliplatin-Resistant Gene Signature in Colorectal Cancer Patients Who Underwent Chemotherapy. Pharmaceuticals. 2022; 15(9):1139. https://doi.org/10.3390/ph15091139
Chicago/Turabian StyleYin, Yixin, Siqi Li, Xinqiang Liang, Kezhi Li, Mingzhi Xie, and Bangli Hu. 2022. "Construction and Validation of an Oxaliplatin-Resistant Gene Signature in Colorectal Cancer Patients Who Underwent Chemotherapy" Pharmaceuticals 15, no. 9: 1139. https://doi.org/10.3390/ph15091139
APA StyleYin, Y., Li, S., Liang, X., Li, K., Xie, M., & Hu, B. (2022). Construction and Validation of an Oxaliplatin-Resistant Gene Signature in Colorectal Cancer Patients Who Underwent Chemotherapy. Pharmaceuticals, 15(9), 1139. https://doi.org/10.3390/ph15091139




