Shedding Light on the Drug–Target Prediction of the Anti-Inflammatory Peptide TnP with Bioinformatics Tools
Abstract
:1. Introduction
2. Results and Discussion
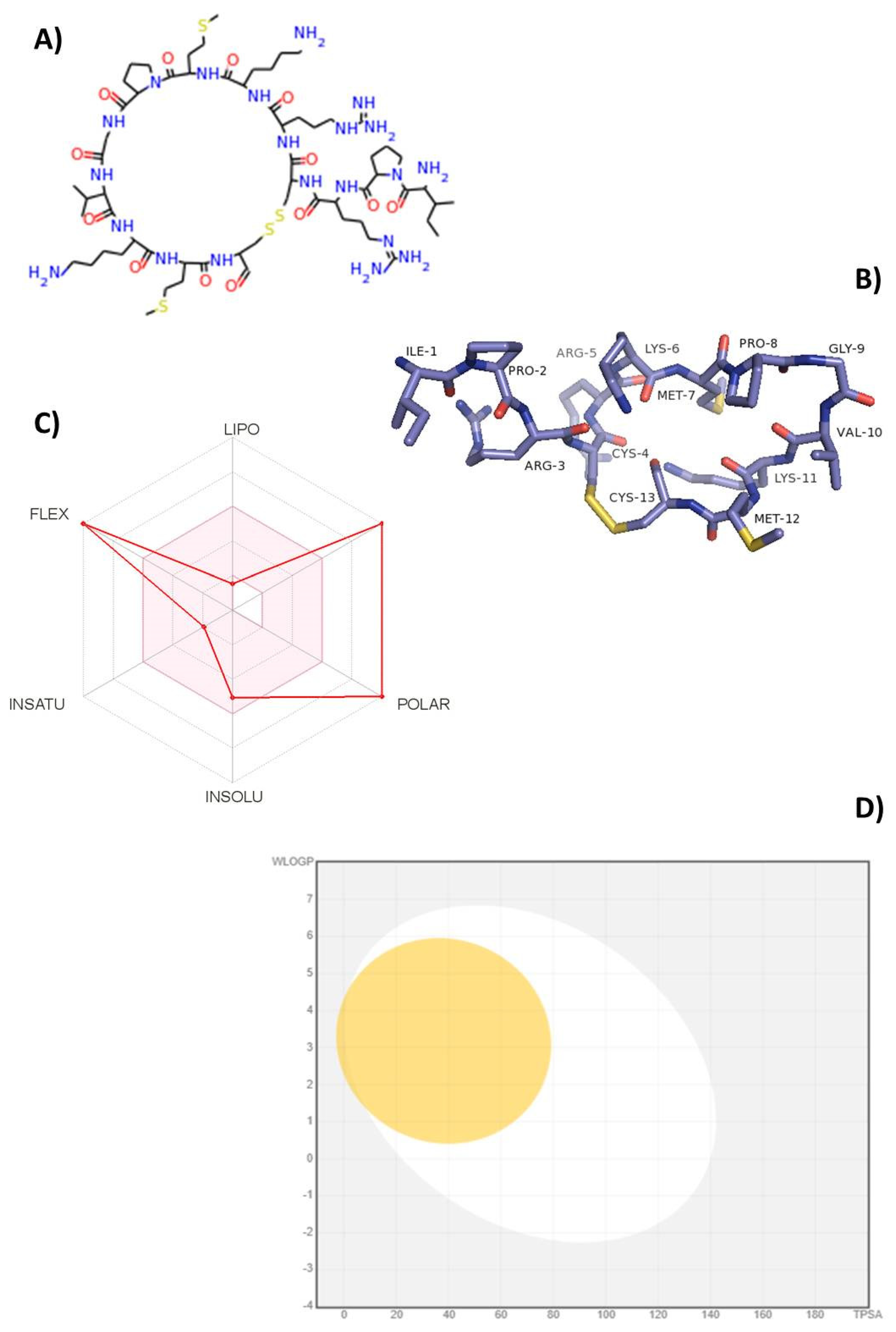
2.1. Bioinformatics and Computational Tools Predict Oral Bioavailability of TnP
2.2. In Silico TnP–Target Protein Interactions
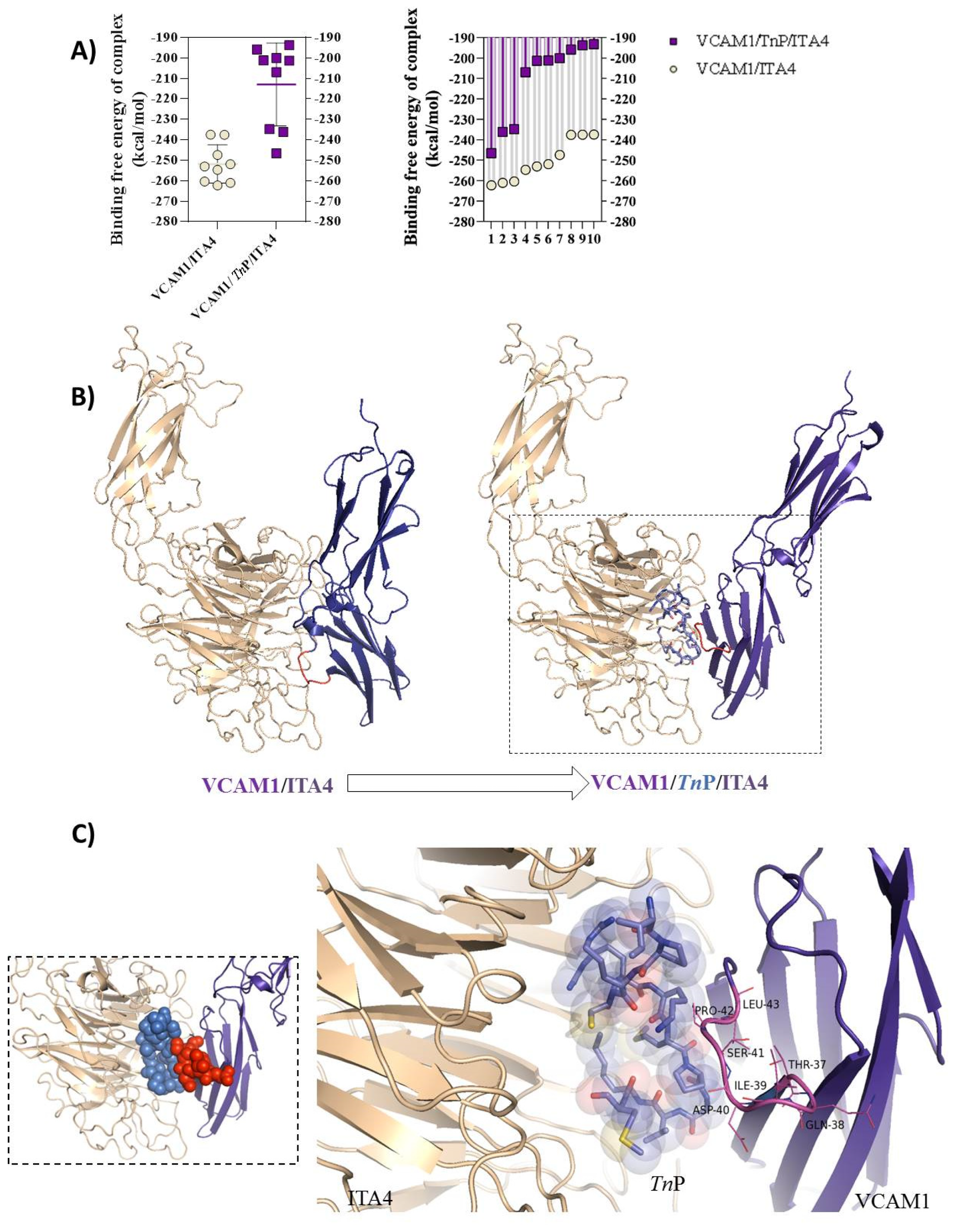
2.3. TnP Competition for the ITA4/VCAM-1 Complex Interaction
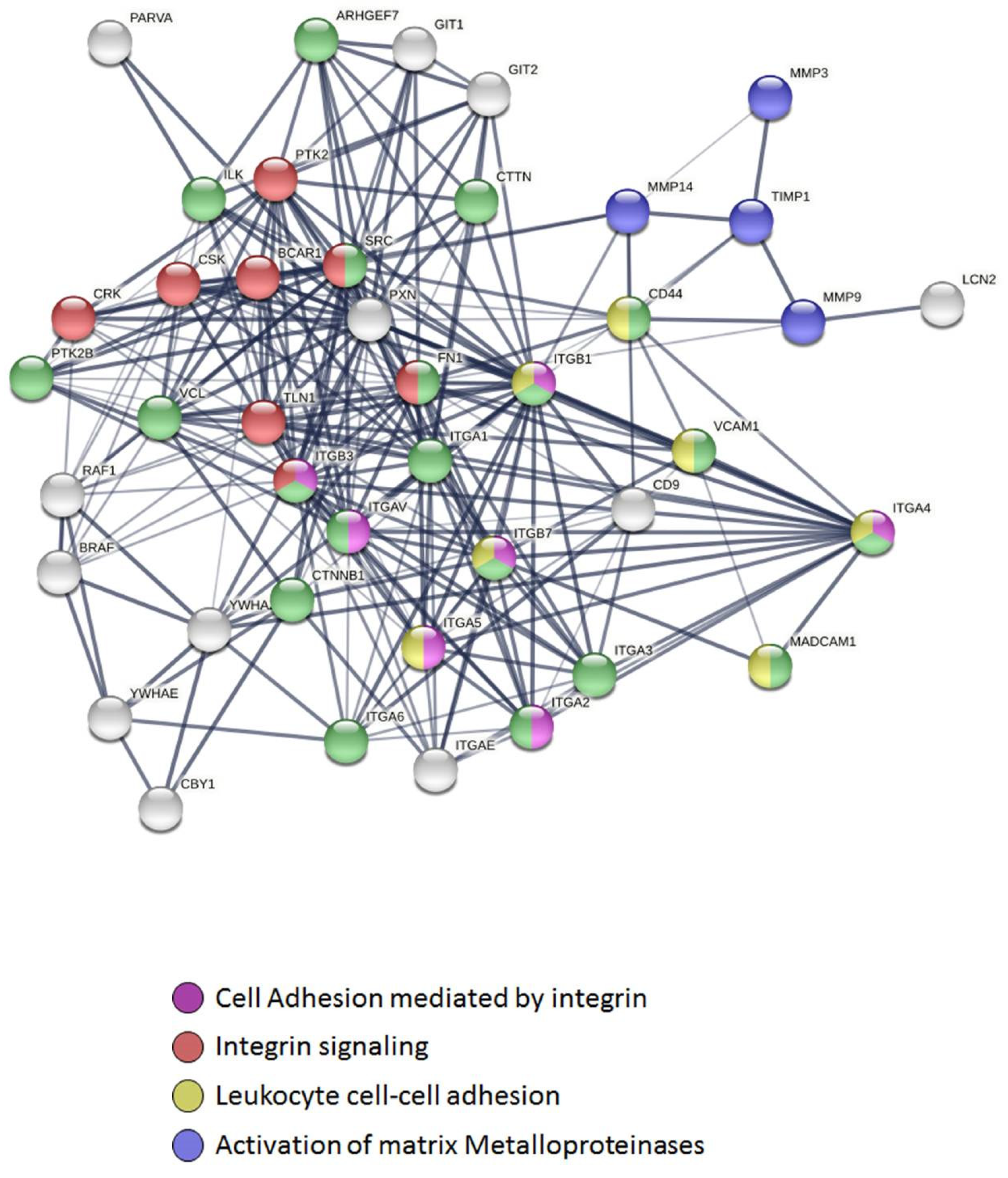
2.4. Connectivity and Interactions of Molecules as a Result of TnP Blockage
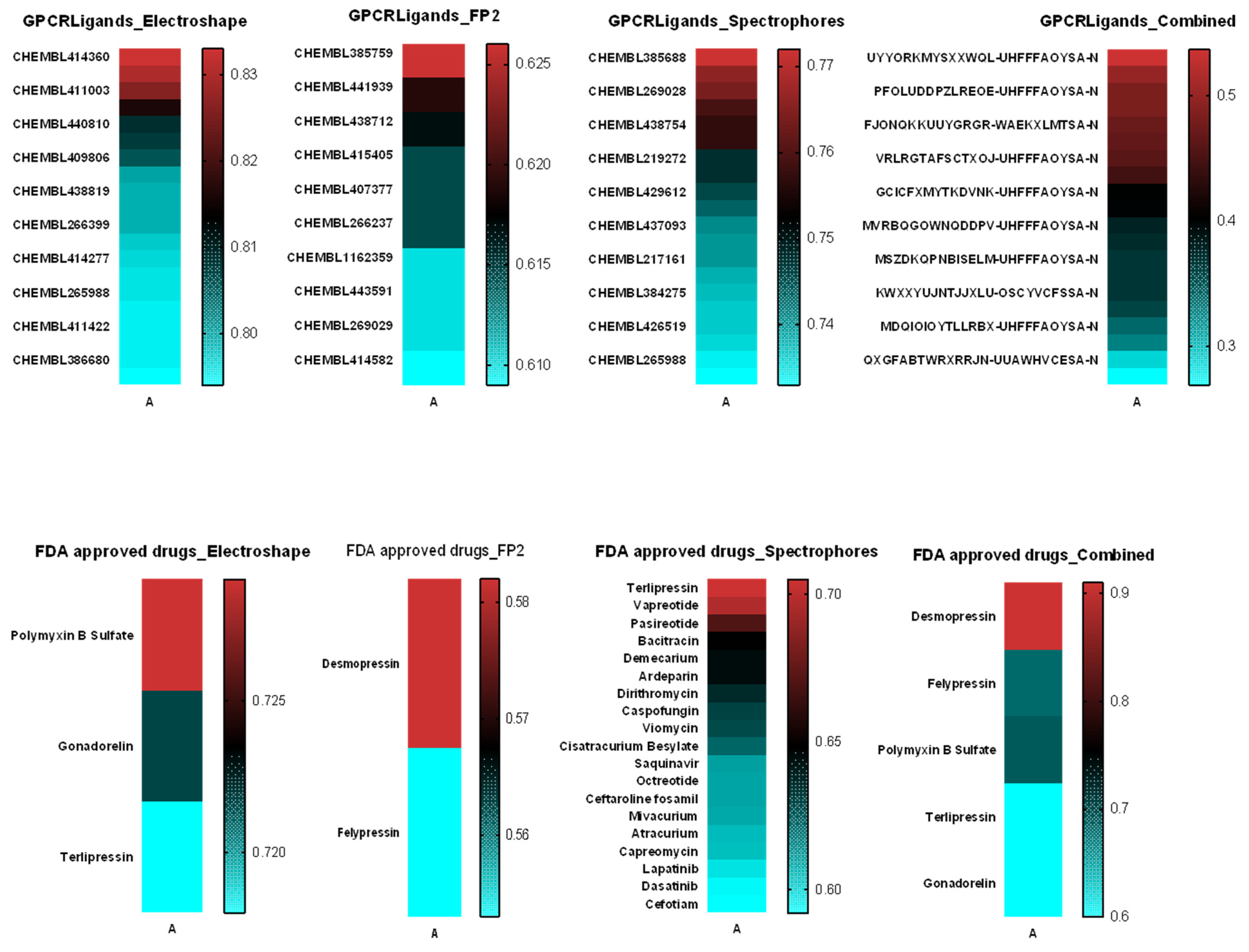
2.5. Similarity Analysis of TnP with Molecules from the Swiss Similarity Database
3. Methods
3.1. Virtual Construction of the TnP Peptide
3.2. ADMET In Silico
3.3. Estimation of Macromolecular Targets of TnP with Homo Sapiens Proteins
3.4. Computational Analysis of the Molecular Docking Interactions of TnP with the Human ITA4 Molecule
3.5. Functional and Pathway Enrichment Analysis
3.6. Similarity of TnP with Molecules from the Swiss Similarity Database
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Multiple sclerosis | MS |
| Central nervous system | CNS |
| Myelin oligodendrocyte glycoprotein | MOG |
| Dendritic cells | DC |
| Matrix metalloproteinase | MMP |
| Blood–brain barrier | BBB |
| T helper | Th |
| Integrin | ITG or IT |
| Integrin alpha 4 subunit | ITGA4, ITA4, or CD49d |
| Integrin beta 1 subunit | ITGB1, ITB1, or CD29 |
| Integrin alpha 4 subunit and integrin beta 1 subunit form | α4β1 dimer or very late antigen-4 (VLA-4) |
| Vascular cell adhesion protein | VCAM-1 |
| Simplified molecular-input line-entry system | SMILE |
| Absorption, distribution, metabolism, and excretion parameters | ADME |
| Topological polar surface area | TPSA |
| Molecular weight | MW |
| Split-intein circular ligation of peptides and proteins | SICLOPPS |
| G protein-coupled receptors | GPCRs |
| Post-translational modifications | PTMs |
| Melanocortin receptor family | MCR |
| Root-mean-square deviation | RMSD |
| Accessible surface area | SASA |
| IDSP motif | Ile-Asp-Ser-Pro |
| Drug–target interaction | DTI |
| Extracellular matrix | ECM |
| Tissue inhibitor of matrix metalloproteinase | TIMP |
| Vinculin | VCL |
| Protein tyrosine kinase 2 beta | PTK2B |
| Inside-out signaling pathway regulator of kinases protein | CRK |
| Cytosolic tyrosine kinase that negatively regulates Src family kinases | CSK |
| Adapter protein, which links cell–cell and cell–matrix adhesions to an extended signaling network | BCAR1 |
| Mammalian isoforms of talin | TLN1 |
| Src family protein tyrosine kinases member | SRC |
| Glycoprotein in the ECM fibronectin | FN1 |
| Addressin mucosal addressin cell adhesion molecule-1 | MAdCAM-1 |
| Individual database | ID |
| Protein Data Bank | PDB |
| Molecular dynamics | MD |
| Protein–protein interaction | PPI |
| Food and Drug Administration | FDA |
| Osteopontin | OPN |
Appendix A. Patent: WO2008009085A1

References
- Lopes-Ferreira, M.V.A.; Lima, C.; Pimenta, D.C.; Conceição, K.; Demasi, M.; Portaro, F.C.V. Anti-Inflammatory and Antiallergic Cyclic Peptides. WIPO Patent WO2008009085A1, 24 January 2008. [Google Scholar]
- Komegae, E.N.; Souza, T.A.M.; Grund, L.Z.; Lima, C.; Lopes-Ferreira, M. Multiple functional therapeutic effects of TnP: A small stable synthetic peptide derived from fish venom in a mouse model of multiple sclerosis. PLoS ONE 2017, 12, e0171796. [Google Scholar] [CrossRef] [PubMed]
- Batista-Filho, J.; Falcão, M.A.P.; Maleski, A.L.A.; Soares, A.B.S.; Balan-Lima, L.; Disner, G.R.; Lima, C.; Lopes-Ferreira, M. Early preclinical screening using zebrafish (Danio rerio) reveals the safety of the candidate anti-inflammatory therapeutic agent TnP. Toxicol. Rep. 2021, 8, 13–22. [Google Scholar] [CrossRef]
- Wooller, S.K.; Benstead-Hume, G.; Chen, X.; Ali, Y.; Pearl, F.M. Bioinformatics in translational drug discovery. Biosci. Rep. 2017, 37, BSR20160180. [Google Scholar] [CrossRef] [PubMed]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.; Maleski, A.L.A.; Bernardo, J.T.G.; Zelli, V.C.; Komegae, E.N.; Lopes-Ferreira, M. TnP Peptide Suppresses Experimental Autoimmune Encephalomyelitis (EAE) in a Preclinical Mouse Model. Front. Immunol. 2022, 13, 857692. [Google Scholar] [CrossRef] [PubMed]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Tesauro, D.; Accardo, A.; Diaferia, C.; Milano, V.; Guillon, J.; Ronga, L.; Rossi, F. Peptide-Based Drug-Delivery Systems in Biotechnological Applications: Recent Advances and Perspectives. Molecules 2019, 24, 351. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- McAllister, T.E.; Coleman, O.D.; Roper, G.; Kawamura, A. Structural diversity in de novo cyclic peptide ligands from genetically encoded library technologies. Pept. Sci. 2020, 113, e24204. [Google Scholar] [CrossRef]
- Falcao, M.A.P.; Walker, C.I.B.; Disner, G.R.; Batista-Filho, J.; Soares, A.B.S.; Balan-Lima, L.; Lima, C.; Lopes-Ferreira, M. Knockdown of miR-26a in zebrafish leads to impairment of the anti-inflammatory function of TnP in the control of neutrophilia. Fish Shellfish Immunol. 2021, 114, 301–310. [Google Scholar] [CrossRef]
- Harder, B.G.; Blomquist, M.R.; Wang, J.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A.; Loftus, J.C.; Tran, N.L. Developments in Blood-Brain Barrier Penetrance and Drug Repurposing for Improved Treatment of Glioblastoma. Front. Oncol. 2018, 8, 462. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, Y.; Araki, M.; Gutteridge, A.; Honda, W.; Kanehisa, M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2008, 24, i232–i240. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.A.; McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 2007, 450, 1001–1009. [Google Scholar] [CrossRef]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef]
- Fauman, E.; Rai, B.K.; Huang, E.S. Structure-based druggability assessment—identifying suitable targets for small molecule therapeutics. Curr. Opin. Chem. Biol. 2011, 15, 463–468. [Google Scholar] [CrossRef]
- Yu, Y.; Zhu, J.; Mi, L.-Z.; Walz, T.; Sun, H.; Chen, J.; Springer, T.A. Structural specializations of α4β7, an integrin that mediates rolling adhesion. J. Cell Biol. 2012, 196, 131–146. [Google Scholar] [CrossRef]
- Jawad, B.; Poudel, L.; Podgornik, R.; Steinmetz, N.F.; Ching, W.-Y. Molecular mechanism and binding free energy of doxorubicin intercalation in DNA. Phys. Chem. Chem. Phys. 2019, 21, 3877–3893. [Google Scholar] [CrossRef]
- Falchi, F.; Caporuscio, F.; Recanatini, M. Structure-based design of small-molecule protein–protein interaction modulators: The story so far. Futur. Med. Chem. 2014, 6, 343–357. [Google Scholar] [CrossRef]
- Clements, J.; Newham, P.; Shepherd, M.; Gilbert, R.; Dudgeon, T.; Needham, L.; Edwards, R.; Berry, L.; Brass, A.; Humphries, M. Identification of a key integrin-binding sequence in VCAM-1 homologous to the LDV active site in fibronectin. J. Cell Sci. 1994, 107, 2127–2135. [Google Scholar] [CrossRef]
- Wang, J.H.; Pepinsky, R.B.; Stehle, T.; Liu, J.H.; Karpusas, M.; Browning, B.; Osborn, L. The crystal structure of an N-terminal two-domain fragment of vascular cell adhesion molecule 1 (VCAM-1): A cyclic peptide based on the domain 1 C-D loop can inhibit VCAM-1-alpha 4 integrin interaction. Proc. Natl. Acad. Sci. USA 1995, 92, 5714–5718. [Google Scholar] [CrossRef] [PubMed]
- Jones, Y.; Harlos, K.; Bottomley, M.J.; Robinson, R.; Driscoll, P.; Edwards, R.M.; Clements, J.M.; Dudgeon, T.J.; Stuart, D. Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 Å resolution. Nature 1995, 373, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Vonderheide, R.H.; Tedder, T.F.; Springer, T.A.; Staunton, D.E. Residues within a conserved amino acid motif of domains 1 and 4 of VCAM-1 are required for binding to VLA-4. J. Cell Biol. 1994, 125, 215–222. [Google Scholar] [CrossRef]
- Gao, M.; Skolnick, J. A Comprehensive Survey of Small-Molecule Binding Pockets in Proteins. PLoS Comput. Biol. 2013, 9, e1003302. [Google Scholar] [CrossRef]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. Correction to ‘The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets’. Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Asens, M.L.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Casey, M.J.; Stewart, R.A. Pediatric Cancer Models in Zebrafish. Trends Cancer 2020, 6, 407–418. [Google Scholar] [CrossRef]
- Yan, C.; Brunson, D.; Tang, Q.; Do, D.; Iftimia, N.A.; Moore, J.C.; Hayes, M.N.; Welker, A.M.; Garcia, E.G.; Dubash, T.D.; et al. Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell 2019, 177, 1903–1914. [Google Scholar] [CrossRef]
- Kumar, A.; Zhang, K.Y.J. Advances in the Development of Shape Similarity Methods and Their Application in Drug Discovery. Front. Chem. 2018, 6, 315. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Daina, A.; Bovigny, C.; Michielin, O. SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening. J. Chem. Inf. Model. 2016, 56, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, A.; McGaughey, G.B.; Sheridan, R.P.; Good, A.C.; Warren, G.; Mathieu, M.; Muchmore, S.W.; Brown, S.P.; Grant, J.A.; Haigh, J.A.; et al. Molecular Shape and Medicinal Chemistry: A Perspective. J. Med. Chem. 2010, 53, 3862–3886. [Google Scholar] [CrossRef] [PubMed]
- Disner, G.; Falcão, M.; Lima, C.; Lopes-Ferreira, M. In Silico Target Prediction of Overexpressed microRNAs from LPS-Challenged Zebrafish (Danio rerio) Treated with the Novel Anti-Inflammatory Peptide TnP. Int. J. Mol. Sci. 2021, 22, 7117. [Google Scholar] [CrossRef] [PubMed]
- Bourque, J.; Hawiger, D. Current and Future Immunotherapies for Multiple Sclerosis. Mol. Med. 2021, 118, 334–339. [Google Scholar]
- Del Gatto, A.; Saviano, M.; Zaccaro, L. AnOverview of Peptide-Based Molecules as Potential Drug Candidates for Multiple Sclerosis. Molecules 2021, 26, 5227. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorganic. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Baiula, M.; Spampinato, S.; Gentilucci, L.; Tolomelli, A. Novel Ligands Targeting α4β1 Integrin: Therapeutic Applications and Perspectives. Front. Chem. 2019, 7, 489. [Google Scholar] [CrossRef]
- Gilon, C.; Klazas, M.; Lahiani, A.; Schumacher-Klinger, A.; Merzbach, S.; Naoum, J.N.; Ovadia, H.; Rubin, L.; Cornell-Kennon, S.; Schaefer, E.M.; et al. Synthesis and Pharmacological Characterization of Visabron, a Backbone Cyclic Peptide Dual Antagonist of α4β1 (VLA-4)/α9β1 Integrin for Therapy of Multiple Sclerosis. JACS Au 2021, 1, 2361–2376. [Google Scholar] [CrossRef]
- Elices, M.J.; Osborn, L.; Takada, Y.; Crouse, C.; Luhowskyj, S.; Hemler, M.E.; Lobb, R.R. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/Fibronectin binding site. Cell 1990, 60, 577–584. [Google Scholar] [CrossRef]
- Nakao, S.; Kuwano, T.; Ishibashi, T.; Kuwano, M.; Ono, M. Synergistic Effect of TNF-α in Soluble VCAM-1-Induced Angiogenesis Through α4Integrins. J. Immunol. 2003, 170, 5704–5711. [Google Scholar] [CrossRef] [PubMed]
- Calzada, M.J.; Zhou, L.; Sipes, J.M.; Zhang, J.; Krutzsch, H.C.; Iruela-Arispe, M.L.; Annis, D.S.; Mosher, D.F.; Roberts, D.D. α4 β1 Integrin Mediates Selective Endothelial Cell Responses to Thrombospondins 1 and 2 In Vitro and Modulates Angiogenesis In Vivo. Circ. Res. 2004, 94, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Massagué, J. Molecular Pathways: VCAM-1 as a Potential Therapeutic Target in Metastasis. Clin. Cancer Res. 2012, 18, 5520–5525. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, R.; Khaket, T.P.; Dutta, C.; Chakraborty, B.; Mukherjee, T.K. Breast cancer metastasis: Putative therapeutic role of vascular cell adhesion molecule-1. Cell. Oncol. 2017, 40, 199–208. [Google Scholar] [CrossRef]
- Schlesinger, M.; Bendas, G. Contribution of very late antigen-4 (VLA-4) integrin to cancer progression and metastasis. Cancer Metastasis Rev. 2015, 34, 575–591. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Hamilton, A.M.; Aidoudi-Ahmed, S.; Sharma, S.; Kotamraju, V.R.; Foster, P.J.; Sugahara, K.N.; Ruoslahti, E.; Rutt, B.K. Nanoparticles coated with the tumor-penetrating peptide iRGD reduce experimental breast cancer metastasis in the brain. J. Mol. Med. 2015, 93, 991–1001. [Google Scholar] [CrossRef]
- Ley, K.; Rivera-Nieves, J.; Sandborn, W.J.; Shattil, S. Integrin-based therapeutics: Biological basis, clinical use and new drugs. Nat. Rev. Drug Discov. 2016, 15, 173–183. [Google Scholar] [CrossRef]
- Takagi, J.; Springer, T.A. Integrin activation and structural rearrangement. Immunol. Rev. 2002, 186, 141–163. [Google Scholar] [CrossRef]
- Alon, R.; Kassner, P.D.; Carr, M.W.; Finger, E.B.; Hemler, M.E.; Springer, T.A. The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. J. Cell Biol. 1995, 128, 1243–1253. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, C.; Eto, S.F.; Lopes-Ferreira, M. Shedding Light on the Drug–Target Prediction of the Anti-Inflammatory Peptide TnP with Bioinformatics Tools. Pharmaceuticals 2022, 15, 994. https://doi.org/10.3390/ph15080994
Lima C, Eto SF, Lopes-Ferreira M. Shedding Light on the Drug–Target Prediction of the Anti-Inflammatory Peptide TnP with Bioinformatics Tools. Pharmaceuticals. 2022; 15(8):994. https://doi.org/10.3390/ph15080994
Chicago/Turabian StyleLima, Carla, Silas Fernandes Eto, and Monica Lopes-Ferreira. 2022. "Shedding Light on the Drug–Target Prediction of the Anti-Inflammatory Peptide TnP with Bioinformatics Tools" Pharmaceuticals 15, no. 8: 994. https://doi.org/10.3390/ph15080994
APA StyleLima, C., Eto, S. F., & Lopes-Ferreira, M. (2022). Shedding Light on the Drug–Target Prediction of the Anti-Inflammatory Peptide TnP with Bioinformatics Tools. Pharmaceuticals, 15(8), 994. https://doi.org/10.3390/ph15080994







