Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Virus Collection, Cells and Drugs
4.2. Antiviral Activity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 365–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.; Pandit, P.; McArthur, A.G.; Banerjee, A.; Mossman, K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021, 18, 166. [Google Scholar] [CrossRef] [PubMed]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Haider, N.; Abbasi, A.F.; Jaferi, U.; Prakash, S.; Balendra, V. The emerging SARS-CoV-2 variants of concern. Ther. Adv. Infect. Dis. 2021, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, K.L.; MacIntyre, C.R.; McIntyre, P.B.; Nelson, M.R. SARS-CoV-2 vaccines: Where are we now? J. Allergy Clin. Immunol. Pract. 2021, 9, 3535–3543. [Google Scholar] [CrossRef] [PubMed]
- Fathizadeh, H.; Afshar, S.; Masoudi, M.R.; Gholizadeh, P.; Asgharzadeh, M.; Ganbarov, S.; Yousefi, M.; Kafil, H.S. SARS-CoV-2 (COVID-19) vaccines structure, mechanisms and effectiveness: A review. Int. J. Biol. Macromol. 2021, 188, 740–750. [Google Scholar] [CrossRef]
- Gendrot, M.; Andreani, J.; Boxberger, M.; Jardot, P.; Fonta, I.; Le Bideau, M.; Duflot, I.; Mosnier, J.; Rolland, C.; Bogreau, H.; et al. Antimalarial drugs inhibit the replication of SARS-CoV-2: An in vitro evaluation. Travel Med. Infect. Dis. 2020, 37, 101873. [Google Scholar] [CrossRef]
- Gendrot, M.; Duflot, I.; Boxberger, M.; Delandre, O.; Jardot, P.; Le Bideau, M.; Andreani, J.; Fonta, I.; Mosnier, J.; Rolland, C.; et al. Antimalarial artemisinin-based combination therapies (ACT) and COVID-19 in Africa: In vitro inhibition of SARS-CoV-2 replication by mefloquine-artesunate. Int. J. Infect. Dis. 2020, 99, 437–440. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCov) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Holwerda, M.; V’kovski, P.; Wider, M.; Thiel, V.; Djikman, R. Identification of an antiviral compound from the pandemic response box that efficiently inhibits SARS-CoV-2 infection in vitro. Microorganisms 2020, 8, 1872. [Google Scholar] [CrossRef]
- Andreani, J.; Le Bideau, M.; Duflot, I.; Jardot, P.; Rolland, C.; Boxberger, M.; Wurtz, N.; Rolain, J.M.; Colson, P.; La Scola, B.; et al. In vitro testing of hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb. Pathog. 2020, 145, 104228. [Google Scholar] [CrossRef]
- Gendrot, M.; Andreani, J.; Jardot, P.; Hutter, S.; Delandre, O.; Boxberger, M.; Mosnier, J.; Le Bideau, M.; Duflot, I.; Fonta, I.; et al. In vitro antiviral activity of doxycycline against SARS-CoV-2. Molecules 2020, 25, 5064. [Google Scholar] [CrossRef] [PubMed]
- Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Sia, S.F.; Chen, D.; Hui, K.P.Y.; Chu, D.K.W.; Chan, M.C.W.; Cheung, P.P.H.; Huang, X.; et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020, 178, 104786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Cao, R.; Xu, M.; Wu, Y.; Shang, W.; Wang, X.; Zhang, H.; Jiang, X.; Sun, Y.; et al. Comparative antiviral efficacy of viral protease inhibitors against the novel SARS-CoV-2 in vitro. Virol. Sin. 2020, 35, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yin, X.; Meng, X.; Chan, J.F.W.; Ye, Z.W.; Riva, L.; Pache, L.; Chan, C.C.Y.; Lai, P.M.; Chan, C.C.S.; et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature 2021, 593, 418–423. [Google Scholar] [CrossRef]
- Weston, S.; Coleman, C.M.; Haupt, R.; Logue, J.; Matthews, K.; Li, Y.; Reyes, H.M.; Weiss, S.R.; Frieman, M.B. Broad anti-coronavirus activity of Food and Drug Administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J. Virol. 2020, 94, e01218-20. [Google Scholar] [CrossRef]
- Drayman, N.; Jones, K.A.; Azizi, S.A.; Froggatt, H.M.; Tan, K.; Maltseva, N.I.; Chen, S.; Nicolaescu, V.; Dvorkin, S.; Furlong, K.; et al. Drug repurposing screen identifies masitinib as a 3CLpro inhibitor that blocks replication of SARS-CoV-2 in vitro. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrissey, E.E.; et al. Drug repurposing screens reveal cell-type-specific entry and FDA-approved drugs active against SARS-CoV-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef]
- Kato, F.; Matsuyama, S.; Kawase, M.; Hishiki, T.; Katoh, H.; Takeda, M. Antiviral activities of mycophenolic acid and IMD-0354 against SARS-CoV-2. Microbiol. Immunol. 2020, 64, 635–639. [Google Scholar] [CrossRef]
- Ko, M.; Jeon, S.; Ryu, W.S.; Kim, S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells. J. Med. Virol. 2020, 93, 1403–1408. [Google Scholar] [CrossRef]
- Gendrot, M.; Andreani, J.; Duflot, I.; Boxberger, M.; Le Bideau, M.; Mosnier, J.; Jardot, P.; Fonta, I.; Rolland, C.; Bogreau, H.; et al. Methylene blue inhibits replication of SARS-CoV-2 in vitro. Int. J. Antimicrob. Agents 2020, 56, 106202. [Google Scholar] [CrossRef]
- Gendrot, M.; Jardot, P.; Delandre, O.; Boxberger, M.; Andreani, J.; Duflot, I.; Le Bideau, M.; Mosnier, J.; Fonta, I.; Hutter, S.; et al. In vitro evaluation of the antiviral activity of methylene blue alone or in combination against SARS-CoV-2. J. Clin. Med. 2021, 10, 3007. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chan, J.F.W.; Wang, S.; Li, H.; Zhao, J.; Ip, T.K.Y.; Zuo, Z.; Yuen, K.Y.; Yuan, S.; Sun, H. Orally administered bismuth drug together with N-acetyl cysteine as a broad-spectrum anti-coronavirus therapy. Chem. Sci. 2022, 13, 2238–2248. [Google Scholar] [CrossRef] [PubMed]
- Heidary, F.; Gharebaghi, R. Ivermectin: A systematic review from antiviral effects to COVID-19 complementary regimen. J. Antibiot. 2020, 73, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Arévalo, A.P.; Pagotto, R.; Porfido, J.L.; Daghero, H.; Segovia, M.; Yamasaki, K.; Varela, B.; Hill, M.; Verdes, J.M.; Duhalde Vega, M.; et al. Ivermectin reduced in vivo coronavirus in a mouse experimental model. Sci. Rep. 2021, 11, 7132. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.L.; Tan, K.S.W.; Chu, J.J.H.; Chow, V.T. Combination treatment with remdesivir and ivermectin exerts highly synergistic and potent antiviral activity against murine coronavirus infection. Front. Cell. Infect. Microbiol. 2021, 11, 700502. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Anviral Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.L.; Hoang, V.T.; Nguyen, N.N.; Delerce, J.; Chaudet, H.; Levasseur, A.; Lagier, J.C.; Raoult, D.; Colson, P.; Gautret, P. Clinical outcomes in COVID-19 patients infected with different SARS-CoV-2 variants in Marseille, France. Clin. Microbiol. Infect. 2021, 27, 1516.e1–1516.e6. [Google Scholar] [CrossRef]
- Gautret, P.; Houhamdi, L.; Nguyen, N.N.; Hoang, V.T.; Giraud-Gatineau, A.; Raoult, D. Does SARS-CoV-2 re-infection depend on virus variant? Clin. Microb. Infect. 2021, 27, 1374–1375. [Google Scholar] [CrossRef]
- Dao, T.L.; Hoang, V.T.; Colson, P.; Lagier, J.C.; Million, M.; Raoult, D.; Levasseur, A.; Gautret, P. SARS-CoV-2 infectivity and severity of COVID-19 according to SARS-CoV-2 variants: Current evidence. J. Clin. Med. 2021, 10, 2635. [Google Scholar] [CrossRef]
- Ikegame, S.; Siddiquey, M.N.A.; Hung, C.T.; Haas, G.; Brambilla, L.; Oguntuyo, K.Y.; Kowdle, S.; Chiu, H.P.; Stevens, C.S.; Vilardo, A.E. Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants. Nat. Commun. 2021, 12, 4598. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Al-Awaida, W.J.; Al Hourani, B.J.; Swedan, S.; Nimer, R.; Alzoughool, F.; Al-Ameer, H.J.; Al Taman, S.E.; Alashqar, R.; Al Bawareed, O.; Gushchina, Y.; et al. Correlates of SARS-CoV-2 variants on death, case incidence and case fatality ratio among the continents for the period of 1 December 2020 to 15 March 2021. Genes 2021, 12, 1061. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Zhou, H.; Samanovic, M.I.; Dcosta, B.M.; Cornelius, A.; Mulligan, M.J.; Landau, N.R. Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants. bioRxiv 2021, 6. [Google Scholar] [CrossRef]
- Jaafar, R.; Boschi, C.; Aherfi, S.; Bancod, A.; Le Bideau, M.; Edouard, S.; Colson, P.; Chahinian, H.; Raoult, D.; Yahi, N.; et al. High individual heterogeneity of neutralizing activities against the original strain and nine different variants of SARS-CoV-2. Viruses 2021, 13, 2177. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; La Scola, B.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022, 94, 2019–2025. [Google Scholar] [CrossRef]
- Pires de Souza, G.A.; Le Bideau, M.; Boschi, C.; Ferreira, L.; Wurtz, N.; Devaux, C.; Colson, P.; La Scola, B. Emerging SARS-CoV-2 genotypes show different replication patterns in human pulmonary and intestnal epithelial cells. Viruses 2022, 14, 23. [Google Scholar] [CrossRef]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2021, 602, 671–675. [Google Scholar] [CrossRef]
- Zaidi, A.K.; Dehgani-Mobaraki, P. The mechanisms of action against SARS-CoV-2: An evidence-based clinical review article. J. Antibiot. 2021, 21, 1–12. [Google Scholar]
- Caly, L.; Wagstaff, K.M.; Jans, D.A. Nuclear trafficking of proteins from RNA viruses: Potential target for antivirals. Antiviral Res. 2012, 95, 202–206. [Google Scholar] [CrossRef]
- Yang, S.N.Y.; Atkinson, S.A.; Fraser, J.E.; Wang, C.; Maher, B.; Roman, N.; Forwood, J.K.; Wagstaff, K.M.; Borg, N.A.; Jans, D.A. Novel flavivirus antiviral that targets the host nuclear tranmport importin α/β1 heterodimer. Cells 2019, 8, 281. [Google Scholar] [CrossRef] [Green Version]
- Jans, D.A.; Martin, A.J.; Wagstaff, K.M. Inhibitors of nuclear transport. Curr. Opin. Cell Biol. 2019, 58, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.N.Y.; Atkinson, S.C.; Wang, C.; Lee, A.; Bogoyevitch, M.A.; Borg, N.A.; Jans, D.A. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Res. 2020, 177, 104760. [Google Scholar] [CrossRef] [PubMed]
- Low, Z.Y.; Yip, A.J.W.; Lal, S.K. Repositioning ivermectin for COVID-19 treatment: Molecular mechanisms of action against SARS-CoV-2 replication. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166294. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Taban, I.M.; Eid, E.E.M.; Iqbal, M.; Alam, O.; Khan, S.; Mahmood, D.; Anwar, M.J.; Khalilullah, H.; Khan, M.U. An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α. J. Biomol. Struct. Dyn. 2020, 2, 1–14. [Google Scholar] [CrossRef]
- Bello, M. Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets. J. Biomol. Struct. Dyn. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Gonzalez-Paz, L.; Hurtado-Leon, M.L.; Lossada, C.; Fernandez-Materan, F.V.; Vera-Villalobas, J.; Lorono, M.; Paz, J.L.; Jeffreys, L.; Alvarado, Y.J. Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: A computational and biophysical approach. Biophys. Chem. 2021, 278, 106677. [Google Scholar] [CrossRef]
- Choudhury, A.; Das, N.C.; Patra, R.; Bhattacharya, M.; Ghosh, P.; Patra, B.C.; Mukherjee, S. Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: An in silico approach. Future Virol. 2021, 16, 277–291. [Google Scholar] [CrossRef]
- Udofia, I.A.; Gbayo, K.O.; Oloba-Whenu, O.A.; Ogunbayo, T.B.; Isanbor, C. In silico studies of selected multi-drug targeting against 3CLpro and nsp12 RNA-dependent RNA-polymerase proteins of SARS-CoV-2 and SARS-CoV. Netw. Model. Anal. Health Inform. Bioinform. 2021, 10, 22. [Google Scholar] [CrossRef]
- Parvez, M.S.A.; Karim, M.A.; Hasan, M.; Jaman, J.; Karim, Z.; Tahsin, T.; Hasan, M.N.; Hosen, M.J. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int. J. Biol. Macromol. 2020, 163, 1787–1797. [Google Scholar] [CrossRef]
- Eweas, A.F.; Alhossary, A.A.; Abdel-Moneim, A.S. Molecular docking reveals ivermectin and remdesivir as potential repurposed drugs against SARS-CoV-2. Front. Microbiol. 2021, 11, 592908. [Google Scholar] [CrossRef] [PubMed]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sencanski, M.; Perovic, V.; Pajovic, S.B.; Adzic, M.; Paessler, S.; Glisic, S. Drug repurposing for candidate SARS-CoV-2 main protease inhibitors by a novel in silico methods. Molecules 2020, 25, 3830. [Google Scholar] [CrossRef] [PubMed]
- Mody, V.; Ho, J.; Wills, S.; Mawri, A.; Lawson, L.; Ebert, M.C.C.J.C.; Fortin, G.M.; Rayalam, S.; Taval, S. Identification of 3-chymothrypsin like protease (3CLpro) inhibitors as potentail anti-SARS-CoV-2 agents. Commun. Biol. 2021, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, R.K.; Perekhoda, L.; Azam, M.; Suleiman, M.; Sarangi, A.K.; Semenets, A.; Pintilie, L.; Al-Resayes, S.I. Computational investigations of three main drugs and their comparison with synthesized compounds as potent inhibitors of SARS-CoV-2 main protease (Mpro): DFT, QSAR, molecular docking, and in silico toxicity analysis. J. King Saud Univ. Sci. 2021, 33, 101315. [Google Scholar] [CrossRef]
- Lehrer, S.; Rheinstein, P.H. Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE. Vivo 2020, 34, 3023–3026. [Google Scholar] [CrossRef]
- Saha, J.K.; Raihan, M.J. The binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2. Struct. Chem. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 55, 105960. [Google Scholar] [CrossRef]
- Baildya, N.; Ghosh, N.N.; Chattopadhyay, A.P. Inhibitory capacity of chloroquine against SARS-CoV-2 by effective binding with angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies. J. Mol. Struct. 2021, 1230, 129891. [Google Scholar] [CrossRef]
- Ribaudo, G.; Coghi, P.; Yang, L.J.; Ng, J.P.L.; Mastinu, A.; Memo, M.; Wong, V.K.W.; Gianoncelli, A. Computational and experimental insights on the interaction of artemisinin, dihydroartemisinin and chloroquine with SARS-CoV-2 spike prtein receptor-binding domain (RBD). Nat. Prod. Res. 2021, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Isaac-Lam, M.F. Molecular modeling of the interaction of ligands with ACE2-SARS-CoV-2 spike protein complex. Silico Pharmacol. 2021, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Haribabu, J.; Garisetti, V.; Malekshah, R.E.; Srividya, S.; Gayathri, D.; Bhuvanesh, N.; Mangalaraja, R.V.; Echeverria, C.; Karvembu, R. Design and synthesis of heteroclic azole based bioactive compounds: Molecular structures, quantum simulation, and mechanistic studies through docking as multi-target inhibitors of QARS-CoV-2 and cytotoxicity. J. Mol. Struct. 2022, 1250, 131782. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.; Thakur, A.; Ghadge, J.; Rath, S.L. Computational studies reveal Fluorine based quinilines to be potent inhibitors for proteins involved in SARS-CoV-2 assembly. J. Fluor. Chem. 2021, 250, 109865. [Google Scholar] [CrossRef]
- Marciniec, K.; Beberok, A.; Boryczka, S.; Wrzesniok, D. The application of in silico experimental model in the assessement of ciprofloxacin and levofloxacin interaction with man SARS-CoV-2 targets: S-, E- and TMPRSS2 proteins, RNA-dependent RNA polymerase and papain-like protease (PLpro)—Preliminary molecular docking analysis. Pharmacol. Rep. 2021, 73, 1765–1780. [Google Scholar]
- Tripathi, P.V.; Upadhyay, S.; Singh, M.; Raghavendhar, S.; Bhardwaj, M.; Sharma, P.; Patel, A.K. Sreening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int. J. Biol. Macromol. 2020, 164, 2622–2631. [Google Scholar] [CrossRef]
- Braz, H.L.B.; Silveira, A.M.; Marinho, A.D.; de Moraes, M.E.A.; Filho, M.O.M.; Monteiro, H.S.A.; Jorgr, R.J.B. In silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 56, 106119. [Google Scholar] [CrossRef]
- Willet, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; Logan, N.; de Lorenzo, G.; Furnon, W.; Scott, S.; Manali, M.; Szemiel, A.; et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibite significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv 2022. [Google Scholar] [CrossRef]
- Colson, P.; Rolain, J.M.; Lagier, J.C.; Brouqui, P.; Raoult, D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105932. [Google Scholar] [CrossRef]
- Devaux, C.; Rolain, J.M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef]
- Zhan, X.; Dowell, S.; Shen, Y.; Lee, D.L. Chloroquine to fight COVID-19: A consideration of mechanisms and adverse effects? Heliyon 2020, 6, e04900. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Won, J.J.; Graham, R.L.; Dinnon, K.H.; Sims, A.C.; Feng, J.Y.; Cihlar, T.; Denison, M.R.; Baric, R.S.; Sheahen, T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronavirus with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019, 169, 104541. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Crich, D.; Pegan, S.D.; Lou, L.; Hansen, M.C.; Booth, C.; Desrochers, E.; Mullininx, L.N.; Starling, E.B.; Chang, K.Y.; et al. Polyphenols as potential inhibitors of SARS-CoV-2 RNA dependent RNA polymerase (RdRp). Molecules 2021, 26, 7438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, P.; Rajpoot, S.; Saqib, U.; Yu, P.; Li, Y.; Li, Y.; Ma, Z.; Baig, M.S.; Pan, Q. Comparative assessement of favipiravir and remdesivir againt human coronavirus NL63 in molecular docking and cell culture lodels. Sci. Rep. 2021, 11, 23465. [Google Scholar] [CrossRef]
- Elfiky, A.A.; Azzam, E.B.; Shafaa, M.W. The anti-HCV, Sofosbuvir, versus the anti-EBOV Remdesivir against SARS-CoV-2 RNA dependent RNA polymerase in silico. Mol. Divers. 2021, 3, 1–11. [Google Scholar] [CrossRef]
- Deshpande, R.R.; Tiwari, A.P.; Nyayanit, N.; Modak, M. In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2. Eur. J. Pharmacol. 2020, 886, 173430. [Google Scholar] [CrossRef]
- Adhikari, N.; Banerjee, S.; Baidya, S.K.; Ghosh, B.; Jha, T. Ligand-based quantitative structural assessements of SARS-CoV-2 3CLpro inhibitors: An analysis in light of structure-based multi-molecular modeling evidences. J. Mol. Struct. 2022, 1251, 132041. [Google Scholar] [CrossRef]
- Audus, K.L.; Knaub, S.R.; Guillot, F.L.; Schaeffer, J.M. The effect of protein binding on ivermectin uptake by bovine brain microvessel endothelial cell. Vet. Res. Commun. 1992, 16, 365–377. [Google Scholar] [CrossRef]
- Pena-Silva, R.; Duffull, S.B.; Steer, A.C.; Jaramillo-Rincon, S.X.; Gwee, A.; Zhu, X. Pharmacokinetic considerations on the repurposing of ivermectin for treatment of COVID-19. Br. J. Clin. Pharmacol. 2020, 87, 1589–1590. [Google Scholar] [CrossRef]
- Jermain, B.; Hanafin, P.O.; Cao, Y.; Lifschitz, A.; Lanusse, C.; Rao, G.G. Development of a minimal physiologically-based pharmacokinetic model to simulate lung exposure in humans following oral administration of ivermectin for COVID-19 drug repurposing. J. Pharm. Sci. 2020, 109, 3574–3578. [Google Scholar] [CrossRef]
- Schmith, V.D.; Zhou, J.J.; Lohmer, L.R.L. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther. 2020, 108, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Kern, C.; Schöning, V.; Chaccour, C.; Hammann, F. Modeling of SARS-CoV-2 treatment effects for informed drug repurposing. Front. Pharmacol. 2021, 12, 625678. [Google Scholar] [CrossRef] [PubMed]
- Arshed, U.; Pertinez, H.; Box, H.; Tatham, L.; Rajoli, R.K.R.; Curley, P.; Neary, M.; Sharp, J.; Liptrott, N.J.; Valentijn, A.; et al. priorisation of anti-SARS-CoV-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 2020, 108, 775–790. [Google Scholar] [CrossRef]
- Segatory, V.I.; Garona, J.; Caligiuri, L.G.; Bizzotto, J.; Lavignolle, R.; Toro, A.; Sanchis, P.; Spitzer, E.; Krolewiecki, A.; Gueron, G.; et al. Effect of ivermectin and atorvastatin on nuclear localization of importin Alpha and drug target expression profiling in host cells from nasopharyngeal swabs of SARS-CoV-2-positive patients. Viruses 2021, 13, 2084. [Google Scholar] [CrossRef]
- Mahmud, R.; Rahman, M.M.; Alam, I.; Ahmed, K.G.U.; Kabir, A.K.M.H.; Sayeed, S.K.J.B.; Rassel, M.A.; Monayem, F.B.; Islam, M.S.; Islam, M.M.; et al. Ivermectin in combination with doxycycline for treating COVID-19 symptoms: A randomized trial. J. Int. Med. Res. 2021, 49, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gorial, F.I.; Mashhadani, S.; Sayaly, H.M.; Dakhil, B.D.; AlMashhadani, M.M.; Aljabory, A.M.; Abbas, H.M.; Ghanim, M.; Rasheed, J.I. Effectiveness of Ivermectin as add-on therapy in COVID-19 management. medRxiv 2020. [Google Scholar] [CrossRef]
- Chaccour, C.; Abizanda, G.; Irigoyen-Barrio, A.; Casellas, A.; Aldaz, A.; Martinez-Galan, F.; Hammann, F.; Gil, A.G. Nebulized ivermectin for COVID-19 and other respiratory diseases, a proof of concept, does-ranging study in rats. Sci. Rep. 2020, 10, 170073. [Google Scholar] [CrossRef]
- Errecalde, J.; Lifschitz, A.; Veccioli, G.; Ceballos, L.; Errecalde, F.; Bellent, M.; Marin, G.; Daniele, M.; Turic, E.; Spitzer, E.; et al. Safety and phramacokinetic assessments of a novel ivermectin nasal spray formulation in a pig model. J. Pharm. Sci. 2021, 110, 2501–2507. [Google Scholar] [CrossRef]
- Camprubi, D.; Almuedo-Riera, A.; Marti-Soler, H.; Soriano, A.; Hurtado, J.C.; Subira, C.; Grau-Pujol, B.; Krolewiecki, A.; Munoz, J. Lack of efficacy of stanadrd doses of ivermectin in severe COVID-19 patients. PLoS ONE 2020, 15, e0242184. [Google Scholar] [CrossRef]
- Galan, L.E.B.; Santos, N.M.D.; Asato, M.S.; Araujo, J.V.; de Lima Moreira, A.; Araujo, A.M.M.; Paiva, A.D.P.; Portella, D.G.S.; Marques, F.S.S.; Silva, G.M.A.; et al. Phase 2 randomized study on chloroquine, hydroxychloroquine or ivermectin in hospitalized patients with severe manifestations of SARS-CoV-2 infection. Pathog. Glob. Health 2021, 115, 235–242. [Google Scholar] [CrossRef]
- Kirti, R.; Roy, R.; Pattadar, C.; Ray, R.; Agarwal, N.; Biswas, B.; Majhi, P.K.; Rai, D.K.; Shyama; Kumar, A.; et al. Ivermectin as a potential treatment for mild to moderate COVID-19—A double blind randomized placebo-controlled trial. medRxiv 2021. [Google Scholar] [CrossRef]
- Vallejos, J.; Zoni, R.; Bangher, M.; Villamandos, S.; Bobadilla, A.; Plano, F.; Campias, C.; Campias, E.C.; Medina, M.F.; Achinalli, F.; et al. Ivermectin to prevent hospitalizations in patients with COVD-19 (IVERCOR-COVID19) a randomized, double-blind, placebo-controlled trial. BMC Infect. Dis. 2021, 21, 635. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Medina, E.; Lopez, P.; Hurtado, I.C.; Davalos, D.M.; Ramirez, O.; Martinez, E.; Diazgranados, J.A.; Onate, J.M.; Chavarriaga, H.; Herrera, S.; et al. Effect if ivermectin on time to resolution of symptoms among adults with mild COVID-19. JAMA 2021, 325, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Tiwari, P.; Suri, T.M.; Mittal, S.; Patel, A.; Jain, A.; Velpandiam, T.; Das, U.S.; Boppana, T.K.; Pandey, R.M.; et al. Single-dose oral ivermectin in mild and moderate COVID-19 (RIVET-COV): A single-centre randomized, placebo-controlled trial. J. Infect. Chemother. 2021, 27, 1743–1749. [Google Scholar] [CrossRef]
- Lim, S.C.L.; Hor, C.P.; Tay, K.H.; Jelani, A.M.; Tan, W.H.; Ker, H.B.; Chow, T.S.; Zaid, M.; Cheah, W.K.; Lim, H.H.; et al. Efficacy of ivermectin treatment on disease progression among adults with mild to moderate COVID-19 and comorbidities: The I-TECH randomized clinical trial. JAMA Intern. Med. 2022. [Google Scholar] [CrossRef]
- Lima-Morales, R.; Mendez-Hernandez, P.; Flores, Y.N.; Osorno-Romero, P.; Sancho-Hernandez, C.R.; Cuecuecha-Rugerio, E.; Nava-Zamora, A.; Hernandez-Galdamez, D.R.; Romo-Duenas, D.K.; Salmeron, J. Effectiveness of multidrug therapy consisting of Ivermectin, Azithromycin, Montelukast, and Acethylsalicylic acid to prevent hospitalization and death among ambulatory COVID-19 cases in Tlaxcala, Mexico. Int. J. Infect. Dis. 2021, 105, 598–605. [Google Scholar] [CrossRef]
- Chaccour, C.; Casellas, A.; Matteo, A.B.D.; Pineda, I.; Fernandez-Montero, A.; Ruiz-Castillo, P.; Richardson, M.A.; Rodriguez-Mateos, M.; Jordan-Iborra, C.; Brew, J.; et al. The effect of eraly treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: A pilot, doule-blind, placebo-controlled, randomized clinical trial. EClinicalMedicine 2021, 32, 100720. [Google Scholar] [CrossRef]
- Ozer, M.; Goksu, S.Y.; Conception, R.; Ulker, E.; Balderas, R.M.; Mahdi, M.; Manning, Z.; To, K.; Effendi, M.; Anandakrishnan, R.; et al. Effectiveness and safety of ivermectin in COVID-19 patients: A prospecive study at a safety-net hospital. J. Med. Virol. 2021, 94, 1473–1480. [Google Scholar] [CrossRef]
- Rajter, J.C.; Sherman, M.S.; Fatteh, N.; Vogel, F.; Sacks, J.; Rajter, J.J. Use of ivermectin is associated with lower mortality in hospitalized patients with coronavirus disease 2019. Chest 2021, 159, 85–92. [Google Scholar] [CrossRef]
- Hill, A.; Mirchandani, M.; Pilkington, V. Ivermectin for COVID-19: Adressing potential bias and medical fraud. Open Forum Infect. Dis. 2022, 9, ofab645. [Google Scholar] [CrossRef]
- Izcovich, A.; Peiris, S.; Ragusa, M.; Tortosa, F.; Rda, G.; Aldighieri, S.; Reveiz, L. Bias as a source of inconsistency in ivermectin trials for COVID-19: A systematic review. Ivermectin’s suggested benefits are mainly based on potentially biased results. J. Clin. Epidemiol. 2022, 144, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.; Garratt, A.; Levi, J.; Falconner, J.; Ellis, L.; McCann, K.; Pilkington, V.; Qavi, A.; Wang, J.; Wentzel, H. Meta-analysis of randomized trials of ivermectin to treat SARS-CoV-2 infection. Open Forum Infect. Dis. 2021, 8, ofab358. [Google Scholar] [CrossRef] [PubMed]
- Hoang, V.T.; Colson, P.; Levasseur, A.; Delerce, J.; Lagier, J.C.; Parola, P.; Million, M.; Fournier, P.E.; Raoult, D.; Gautret, P. Clinical outcomes in patients infected with different SARS-CoV-2 variants at one hospital during three phases of the COVID-19 epidemic in Marseille. France. Infect. Genet. Evol. 2021, 95, 105092. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Levasseur, A.; Gautret, P.; Fenollar, F.; Hoang, V.T.; Delerce, J.; Bitam, I.; Saile, R.; Maaloum, M.; Pdane, A.; et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: An investigational study. Travel Med. Infect. Dis. 2021, 40, 101980. [Google Scholar] [CrossRef]
- Fournier, P.E.; Colson, P.; Levasseur, A.; Devaux, C.A.; Gautret, P.; Bedotto, M.; Delerce, J.; Brechard, L.; Pinault, L.; Lagier, J.C.; et al. Emergence and outcomes of the SARS-CoV-2 ‘Marseille-4’ variant. Int. J. Infect. Dis. 2021, 106, 228–236. [Google Scholar] [CrossRef]
- Colson, P.; Levasseur, A.; Delerce, J.; Pinault, L.; Dudouet, P.; Devaux, C.; Fournier, P.E.; La Scola, B.; Lagier, J.C.; Raoult, D. Spreading of new SARS-CoV-2 N501Y spike variant in a new lineage. Clin. Microb. Infect. 2021, 27, 1352.e1–1352.e5. [Google Scholar] [CrossRef]
- La Scola, B.; Lavrad, P.; Fournier, P.E.; Colson, P.; Lacoste, A.; Raoult, D. SARS-CoV-2 variant from India to Marseille: The still active role of ports in the introduction of epidemics. Travel Med. Infect. Dis. 2021, 42, 102085. [Google Scholar] [CrossRef]
- Wurtz, N.; Penant, G.; Jardot, P.; Duclos, N.; La Scola, B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 477–484. [Google Scholar] [CrossRef]
- Kumar, S.; Sarma, P.; Kaur, H.; Prajapat, M.; Bhattacharyya, A.; Avti, P.; Sehkhar, N.; Kaur, H.; Bansal, S.; Mahendiratta, S.; et al. Clinically relevant cell culture models and their significance in isolation, pathogenesis, vaccine development, repurposing and screening of nex drugs for SARS-CoV-2: A systematic review. Tissue Cell 2021, 70, 101497. [Google Scholar] [CrossRef]
- Amrane, S.; Tissot-Dupont, H.; Doudier, B.; Eldin, C.; Hocquart, M.; Mailhe, M.; Dudouet, P.; Ormières, E.; Ailhaud, L.; Parila, P.; et al. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious disease referral hospital in Marseille, France, -January 31st to March 1st, 2020: A respiratory virus snapshot. Travel Med. Infect. Dis. 2020, 36, 101632. [Google Scholar] [CrossRef]
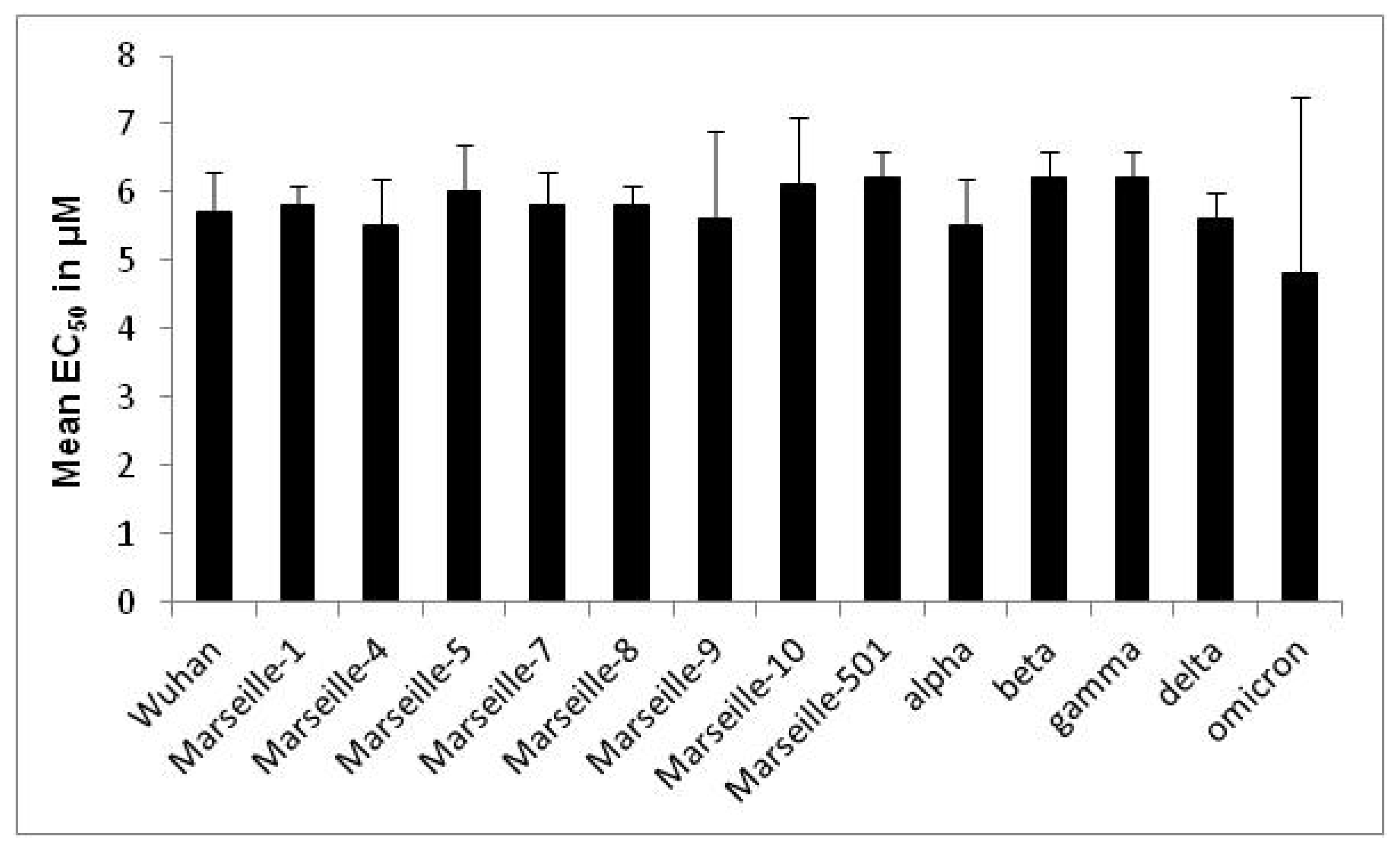
| Variant | Origin | Strain Name | EC50 in µM (Mean ± SD a) | Variant EC50 in µM (Mean ± SD a) | p-Value |
|---|---|---|---|---|---|
| Wuhan | IHU-MI-003 | 5.8 ± 0.5 | 5.7 ± 0.6 | 0.09 | |
| IHU-MI-006 | 5.9 ± 0.7 | ||||
| IHU-MI-717 | 6.1 ± 0.4 | ||||
| IHU-MI-845 | 5.1 ± 0.5 | ||||
| IHU-MI-847 | 5.4 ± 0.5 | ||||
| Marseille-1 | Algeria | IHU-MI-2122 | 5.3 ± 1.7 | 5.8 ± 0.3 | 0.10 |
| IHU-MI-2123 | 5.9 ± 0.4 | ||||
| IHU-MI-2177 | 5.5 ± 0.1 | ||||
| IHU-MI-2178 | 5.9 ± 0.4 | ||||
| Marseille-4 | France | IHU-MI-2096 | 5.7 ± 0.4 | 5.5 ± 0.7 | 0.29 |
| IHU-MI-2129 | 5.5 ± 0.2 | ||||
| IHU-MI-2179 | 5.1 ± 1.1 | ||||
| Marseille-5 | IHU-MI-2137 | 6.0 ± 0.7 | 6.0 ± 0.7 | ||
| Marseille-7 | IHU-MI-2519 | 5.8 ± 0.5 | 5.8 ± 0.5 | ||
| Marseille-8 | IHU-MI-2555 | 5.8 ± 0.3 | 5.8 ± 0.3 | ||
| Marseille-9 | IHU-MI-2615 | 5.6 ± 1.3 | 5.6 ± 1.3 | ||
| Marseille-10 | IHU-MI-2403 | 6.1 ± 1.0 | 6.1 ± 1.0 | ||
| Marseille-501 | Comoros | IHU-MI-3217 | 6.2 ± 0.4 | 6.2 ± 0.4 | |
| alpha | UK | IHU-MI-3076 | 5.8 ± 0.8 | 5.5 ± 0.7 | 0.10 |
| IHU-MI-3100 | 5.8 ± 0.7 | ||||
| IHU-MI-3127 | 5.2 ± 0.4 | ||||
| IHU-MI-3128 | 5.1 ± 0.6 | ||||
| beta | South Africa | IHU-MI-3147 | 6.2 ± 0.4 | 6.2 ± 0.4 | |
| gamma | Brazil | IHU-MI-3191 | 6.2 ± 0.4 | 6.2 ± 0.4 | |
| delta | India | IHU-MI-3396 | 5.6 ± 0.2 | 5.6 ± 0.4 | 0.70 |
| IHU-MI-3630 | 5.7 ± 0.6 | ||||
| IHU-MI-4654 | 5.6 ± 0.4 | ||||
| omicron | South Africa | IHU-MI-5227 | 1.3 ± 0.5 | 4.8 ± 2.6 | 0.003 |
| IHU-MI-5245 | 6.7 ± 0.4 | ||||
| IHU-MI-5253 | 6.3 ± 0.5 |
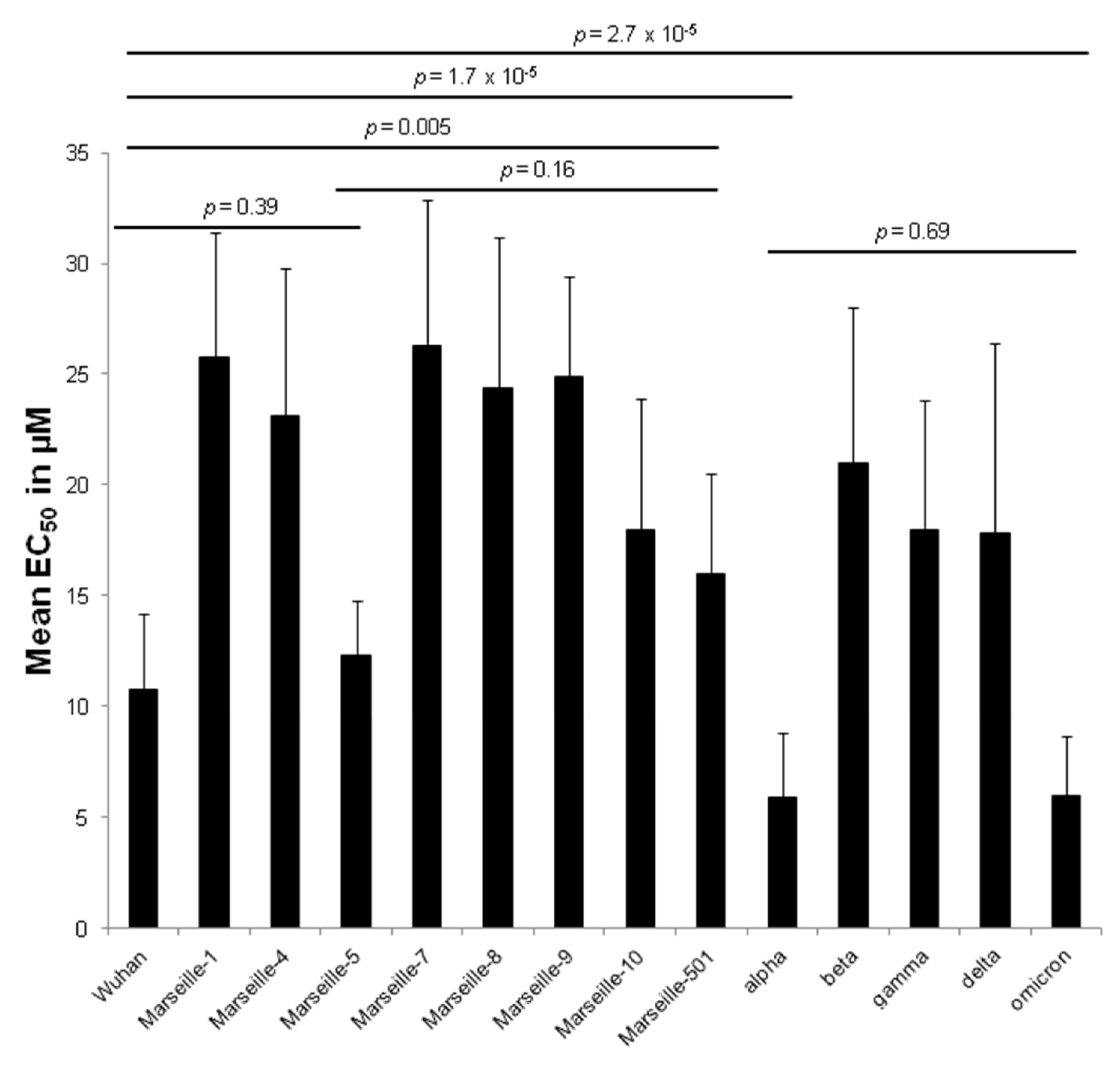
| Variant | Origin | Strain Name | EC50 in µM (Mean ± SD a) | Variant EC50 in µM (Mean ± SD a) | p-Value |
|---|---|---|---|---|---|
| Wuhan | IHU-MI-003 | 8.9 ± 4.4 | 10.8 ± 3.4 | 0.21 | |
| IHU-MI-006 | 11.4 ± 2.4 | ||||
| IHU-MI-717 | 12.4 ± 4.0 | ||||
| IHU-MI-845 | 7.7 ± 1.5 | ||||
| IHU-MI-847 | 11.7 ± 1.9 | ||||
| Marseille-1 | Algeria | IHU-MI-2122 | 25.5 ± 5.2 | 25.8 ± 5.6 | 0.59 |
| IHU-MI-2123 | 24.6 ± 6.1 | ||||
| IHU-MI-2177 | 29.3 ± 5.2 | ||||
| IHU-MI-2178 | 23.8 ± 7.1 | ||||
| Marseille-4 | France | IHU-MI-2096 | 21.9 ± 5.7 | 23.1 ± 6.7 | 0.82 |
| IHU-MI-2129 | 24.1 ± 8.3 | ||||
| IHU-MI-2179 | 24.6 ± 4.5 | ||||
| Marseille-5 | IHU-MI-2137 | 12.3 ± 2.5 | 12.3 ± 2.5 | ||
| Marseille-7 | IHU-MI-2519 | 26.3 ± 6.6 | 26.3 ± 6.6 | ||
| Marseille-8 | IHU-MI-2555 | 24.4 ± 6.8 | 24.4 ± 6.8 | ||
| Marseille-9 | IHU-MI-2615 | 24.9 ± 4.5 | 24.9 ± 4.5 | ||
| Marseille-10 | IHU-MI-2403 | 18.0 ± 5.9 | 18.0 ± 5.9 | ||
| Marseille-501 | Comoros | IHU-MI-3217 | 16.0 ± 4.5 | 16.0 ± 4.5 | |
| alpha | UK | IHU-MI-3076 | 8.4 ± 2.8 | 5.9 ± 2.9 | 0.06 |
| IHU-MI-3100 | 6.5 ± 1.8 | ||||
| IHU-MI-3127 | 4.4 ± 2.7 | ||||
| IHU-MI-3128 | 4.3 ± 2.5 | ||||
| beta | South Africa | IHU-MI-3147 | 21.0 ± 7.0 | 21.0 ± 7.0 | |
| gamma | Brazil | IHU-MI-3191 | 18.0 ± 5.8 | 18.0 ± 5.8 | |
| delta | India | IHU-MI-3396 | 22.0 ± 11.3 | 17.8 ± 8.6 | 0.48 |
| IHU-MI-3630 | 14.3 ± 2.8 | ||||
| IHU-MI-4654 | 15.0 ± 5.6 | ||||
| omicron | South Africa | IHU-MI-5227 | 5.2 ± 2.0 | 6.0 ± 2.7 | 0.06 |
| IHU-MI-5245 | 8.2 ± 2.1 | ||||
| IHU-MI-5253 | 4.5 ± 2.4 |
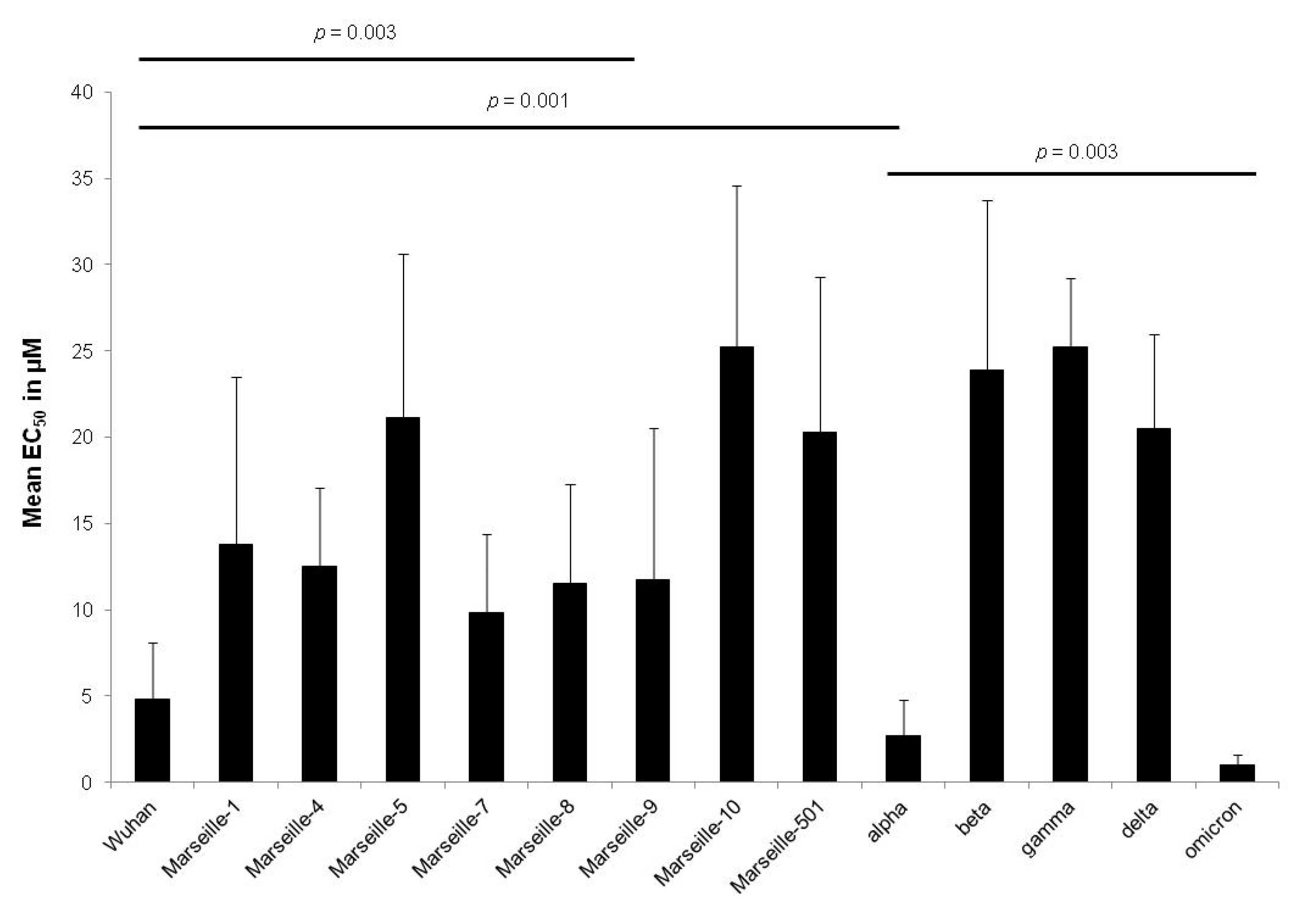
| Variant | Origin | Strain Name | EC50 in µM (Mean ± SD a) | Variant EC50 in µM (Mean ± SD a) | p-Value |
|---|---|---|---|---|---|
| Wuhan | IHU-MI-003 | 2.7 ± 1.6 | 4.8 ± 3.3 | 0.61 | |
| IHU-MI-006 | 4.3 ± 1.6 | ||||
| IHU-MI-717 | 7.3 ± 4.9 | ||||
| IHU-MI-845 | 4.7 ± 2.7 | ||||
| IHU-MI-847 | 3.7 ± 2.8 | ||||
| Marseille-1 | Algeria | IHU-MI-2122 | 12.9 ± 7.6 | 13.8 ± 9.7 | 0.08 |
| IHU-MI-2123 | 5.1± 1.2 | ||||
| IHU-MI-2177 | 21.8 ± 8.0 | ||||
| IHU-MI-2178 | 16.4 ± 8.6 | ||||
| Marseille-4 | France | IHU-MI-2096 | 12.2 ± 2.7 | 12.5 ± 4.6 | 0.94 |
| IHU-MI-2129 | 12.5 ± 7.3 | ||||
| IHU-MI-2179 | 13.0 ± 7.4 | ||||
| Marseille-5 | IHU-MI-2137 | 21.1 ± 9.5 | 21.1 ± 9.5 | ||
| Marseille-7 | IHU-MI-2519 | 9.8 ± 4.6 | 9.8 ± 4.6 | ||
| Marseille-8 | IHU-MI-2555 | 11.5 ± 5.8 | 11.5 ± 5.8 | ||
| Marseille-9 | IHU-MI-2615 | 11.7 ± 8.8 | 11.7 ± 8.8 | ||
| Marseille-10 | IHU-MI-2403 | 25.2 ± 9.4 | 25.2 ± 9.4 | ||
| Marseille-501 | Comoros | IHU-MI-3217 | 20.3 ± 9.0 | 20.3 ± 9.0 | |
| alpha | UK | IHU-MI-3076 | 3.5 ± 2.2 | 2.7 ± 2.1 | 0.77 |
| IHU-MI-3100 | 2.0 ± 1.0 | ||||
| IHU-MI-3127 | 3.4 ± 2.8 | ||||
| IHU-MI-3128 | 2.6 ± 2.0 | ||||
| beta | South Africa | IHU-MI-3147 | 23.9 ± 9.8 | 23.9 ± 9.8 | |
| gamma | Brazil | IHU-MI-3191 | 25.2 ± 4.0 | 25.2 ± 4.0 | |
| delta | India | IHU-MI-3396 | 21.4 ± 5.5 | 20.5 ± 5.5 | 0.71 |
| IHU-MI-3630 | 21.9 ± 2.7 | ||||
| IHU-MI-4654 | 17.6 ± 6.9 | ||||
| omicron | South Africa | IHU-MI-5227 | 0.4 ± 0.3 | 1.0 ± 0.6 | 0.006 |
| IHU-MI-5245 | 1.2 ± 0.4 | ||||
| IHU-MI-5253 | 1.3 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delandre, O.; Gendrot, M.; Jardot, P.; Le Bideau, M.; Boxberger, M.; Boschi, C.; Fonta, I.; Mosnier, J.; Hutter, S.; Levasseur, A.; et al. Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants. Pharmaceuticals 2022, 15, 445. https://doi.org/10.3390/ph15040445
Delandre O, Gendrot M, Jardot P, Le Bideau M, Boxberger M, Boschi C, Fonta I, Mosnier J, Hutter S, Levasseur A, et al. Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants. Pharmaceuticals. 2022; 15(4):445. https://doi.org/10.3390/ph15040445
Chicago/Turabian StyleDelandre, Océane, Mathieu Gendrot, Priscilla Jardot, Marion Le Bideau, Manon Boxberger, Céline Boschi, Isabelle Fonta, Joel Mosnier, Sébastien Hutter, Anthony Levasseur, and et al. 2022. "Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants" Pharmaceuticals 15, no. 4: 445. https://doi.org/10.3390/ph15040445
APA StyleDelandre, O., Gendrot, M., Jardot, P., Le Bideau, M., Boxberger, M., Boschi, C., Fonta, I., Mosnier, J., Hutter, S., Levasseur, A., La Scola, B., & Pradines, B. (2022). Antiviral Activity of Repurposing Ivermectin against a Panel of 30 Clinical SARS-CoV-2 Strains Belonging to 14 Variants. Pharmaceuticals, 15(4), 445. https://doi.org/10.3390/ph15040445






