Current Insights into miRNA and lncRNA Dysregulation in Diabetes: Signal Transduction, Clinical Trials and Biomarker Discovery
Abstract
1. Introduction
2. Dysregulated Human ncRNAs in Diabetes
2.1. Dysregulated ncRNAs in T1D
2.1.1. Upregulated miRNAs in T1D
2.1.2. Upregulated lncRNAs in T1D
2.1.3. Downregulated miRNAs in T1D
2.2. Dysregulated ncRNAs in T2D
2.2.1. Upregulated miRNAs in T2D
2.2.2. Upregulated lncRNAs in T2D
2.2.3. Downregulated miRNAs in T2D
2.2.4. Downregulated lncRNAs in T2D
2.3. Dysregulated ncRNAs in Diabetic Retinopathy
2.3.1. Upregulated miRNAs in Diabetic Retinopathy
2.3.2. Upregulated lncRNAs in Diabetic Retinopathy
2.3.3. Downregulated miRNAs in Diabetic Retinopathy
2.3.4. Downregulated lncRNAs in Diabetic Retinopathy
2.4. Dysregulated ncRNAs in Diabetic Nephropathy
2.4.1. Upregulated miRNAs in Diabetic Nephropathy
2.4.2. Upregulated lncRNAs in Diabetic Nephropathy
2.4.3. Downregulated miRNAs in Diabetic Nephropathy
2.4.4. Downregulated lncRNAs in Diabetic Nephropathy
2.5. Dysregulated ncRNAs in Diabetic Neuropathy
2.5.1. Upregulated miRNAs in Diabetic Neuropathy
2.5.2. Upregulated lncRNAs in Diabetic Neuropathy
2.5.3. Downregulated miRNAs in Diabetic Neuropathy
2.6. Dysregulated ncRNAs in Gestational Diabetes
2.6.1. Upregulated miRNAs in Gestational Diabetes
2.6.2. Upregulated lncRNAs in Gestational Diabetes
2.6.3. Downregulated miRNAs in Gestational Diabetes
2.6.4. Downregulated lncRNAs in Gestational Diabetes
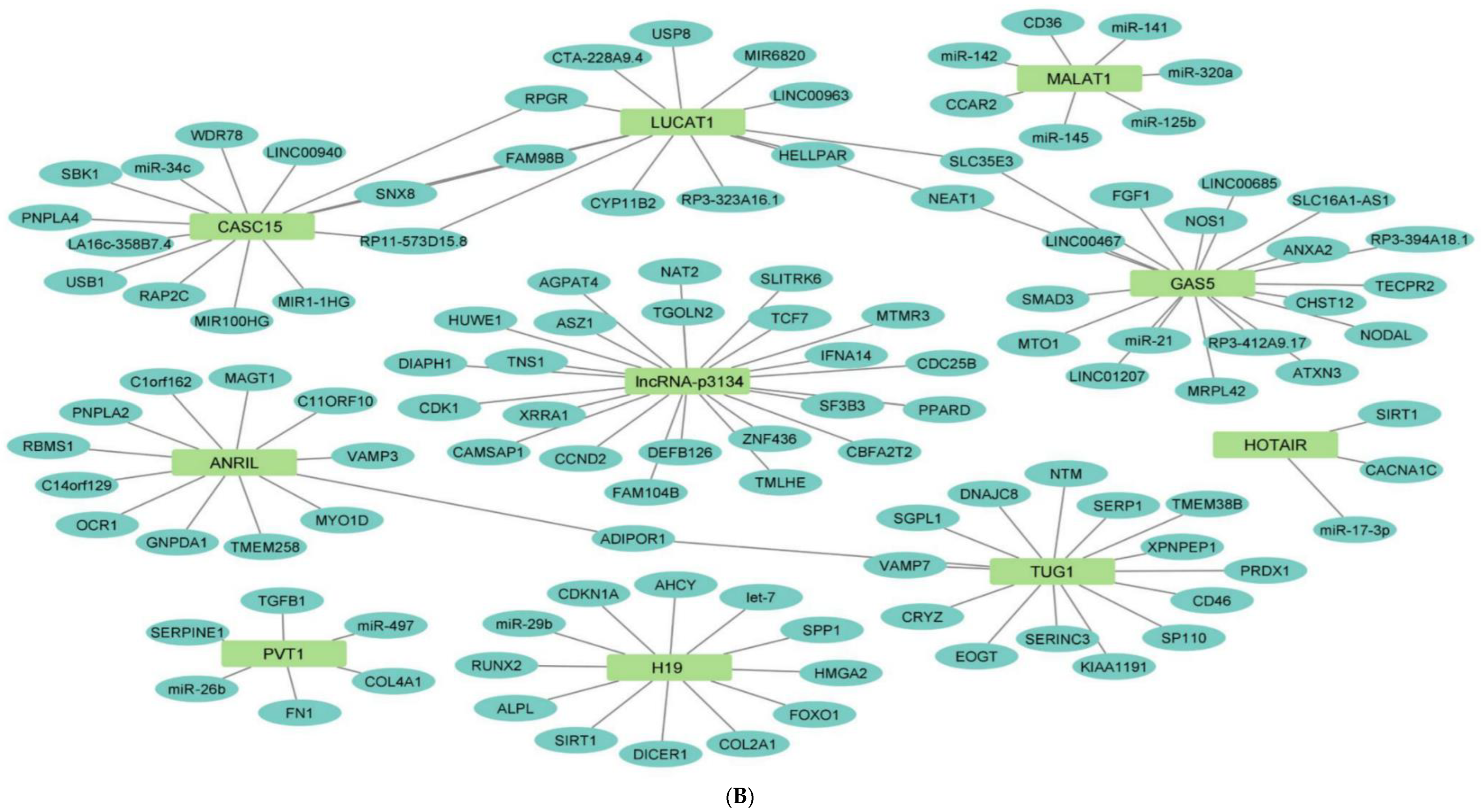
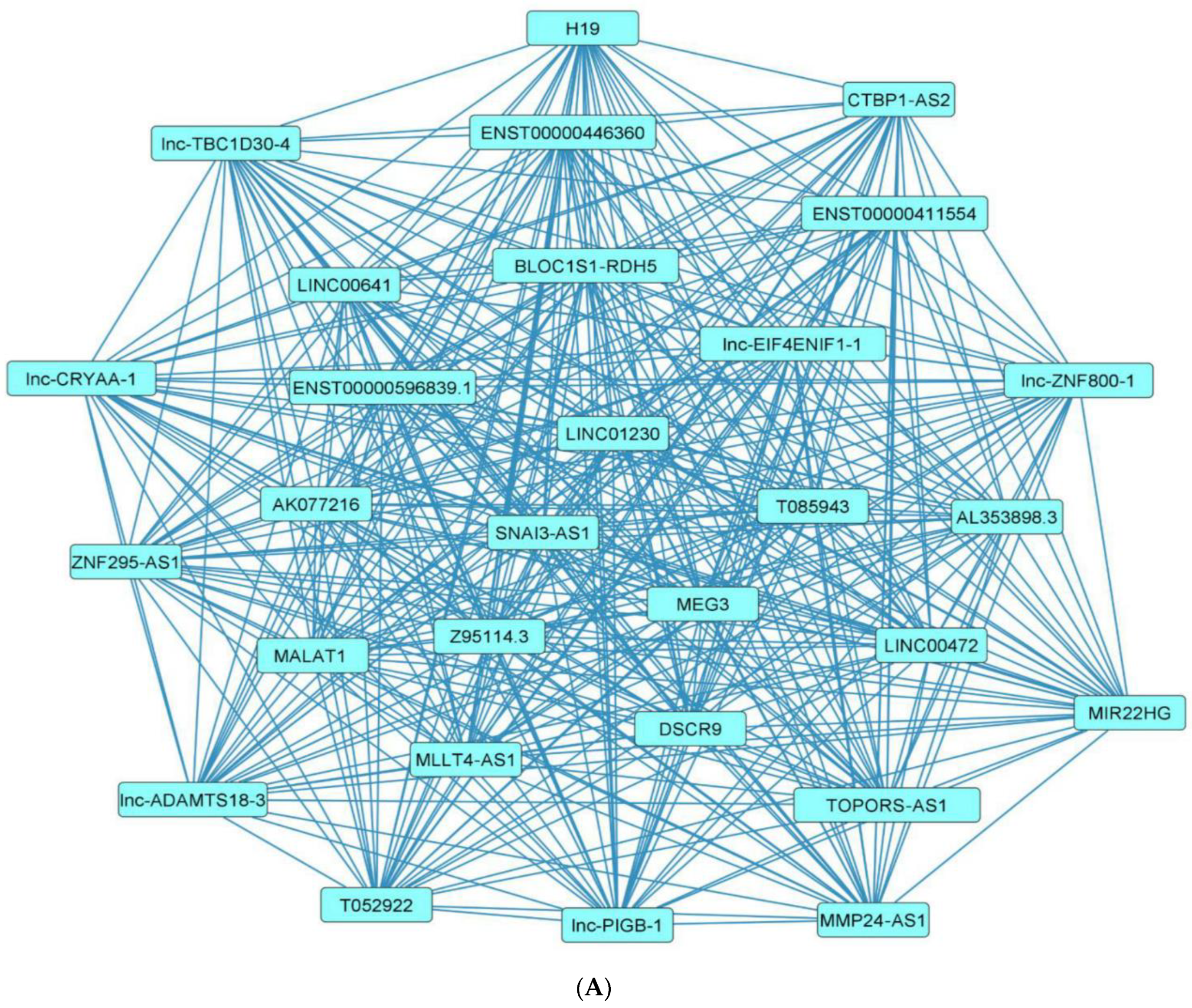
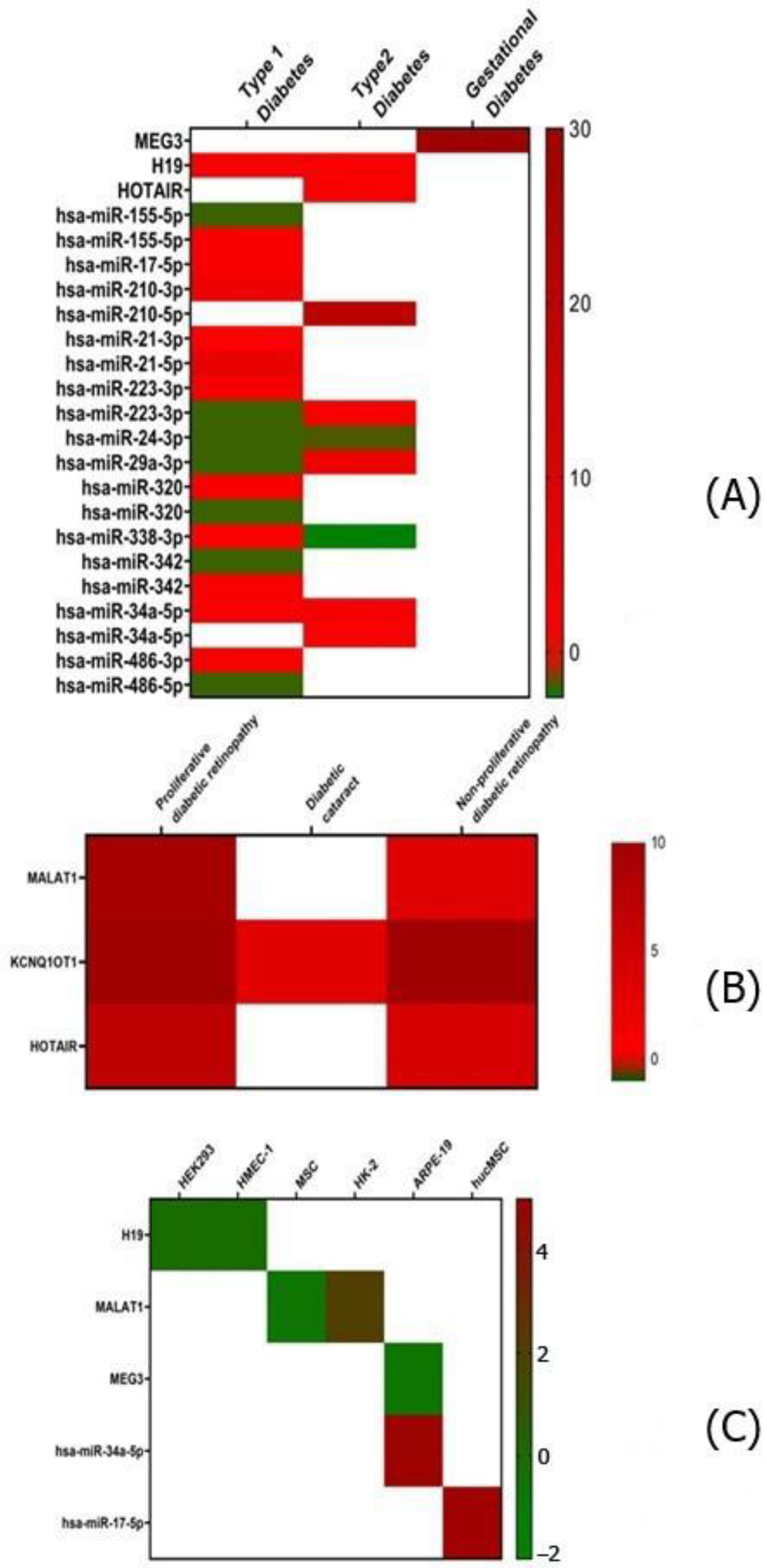
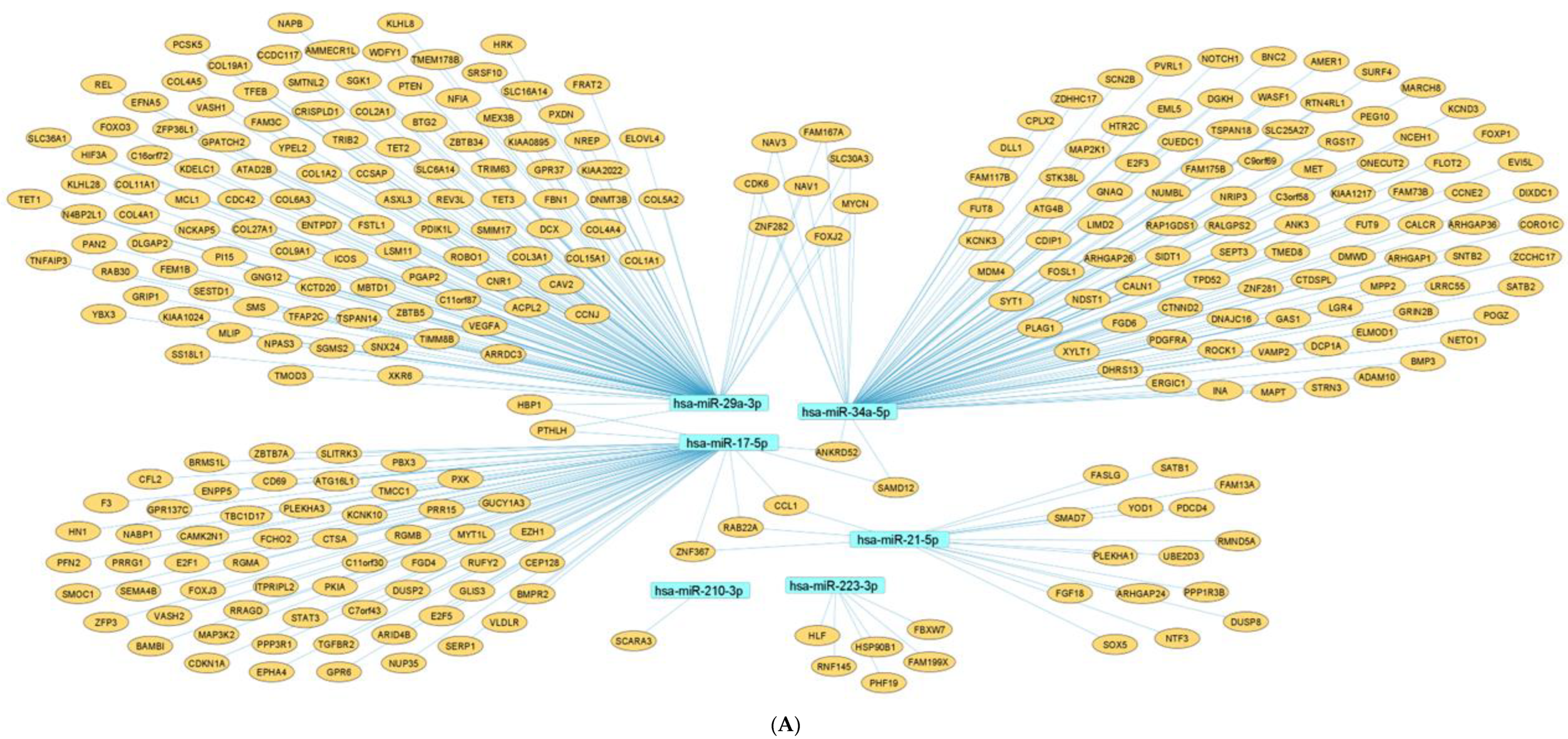
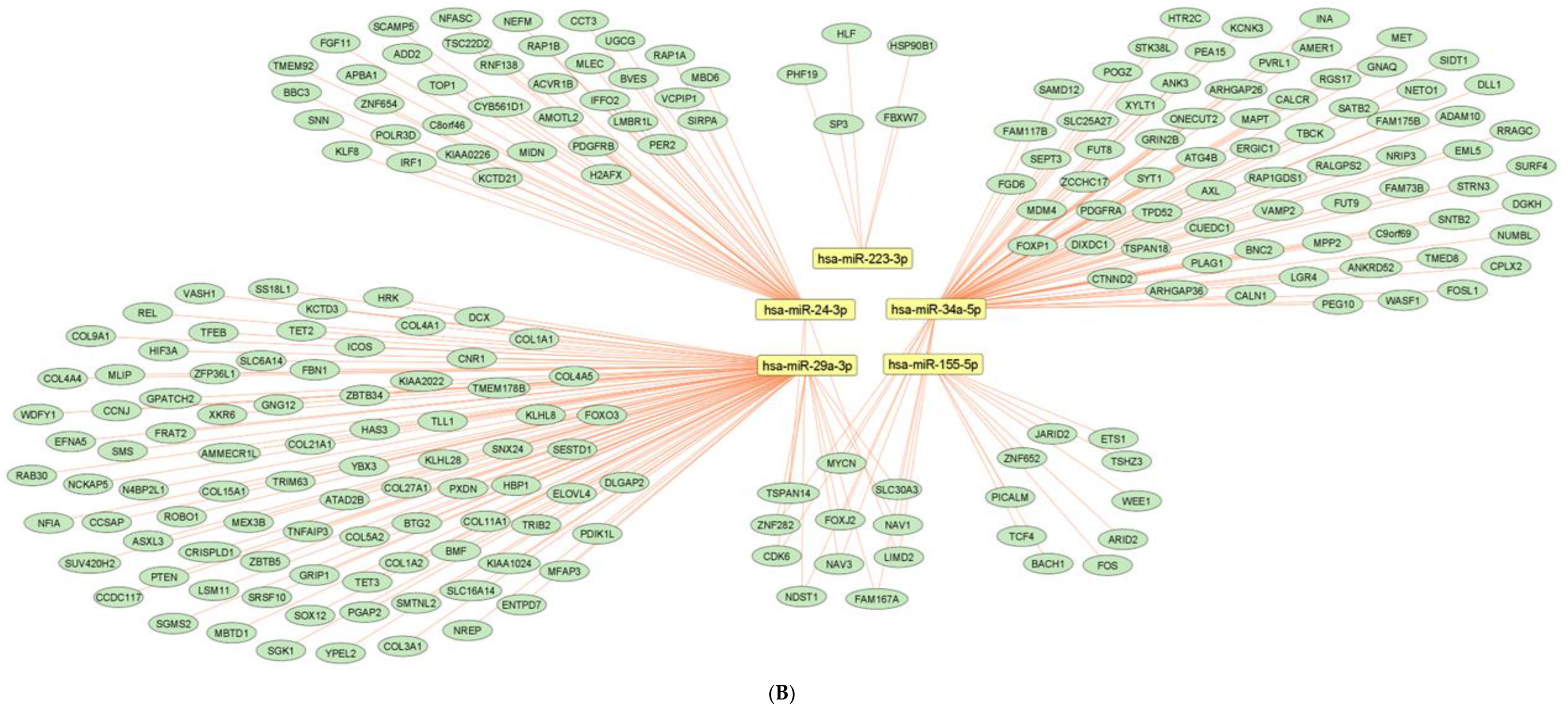
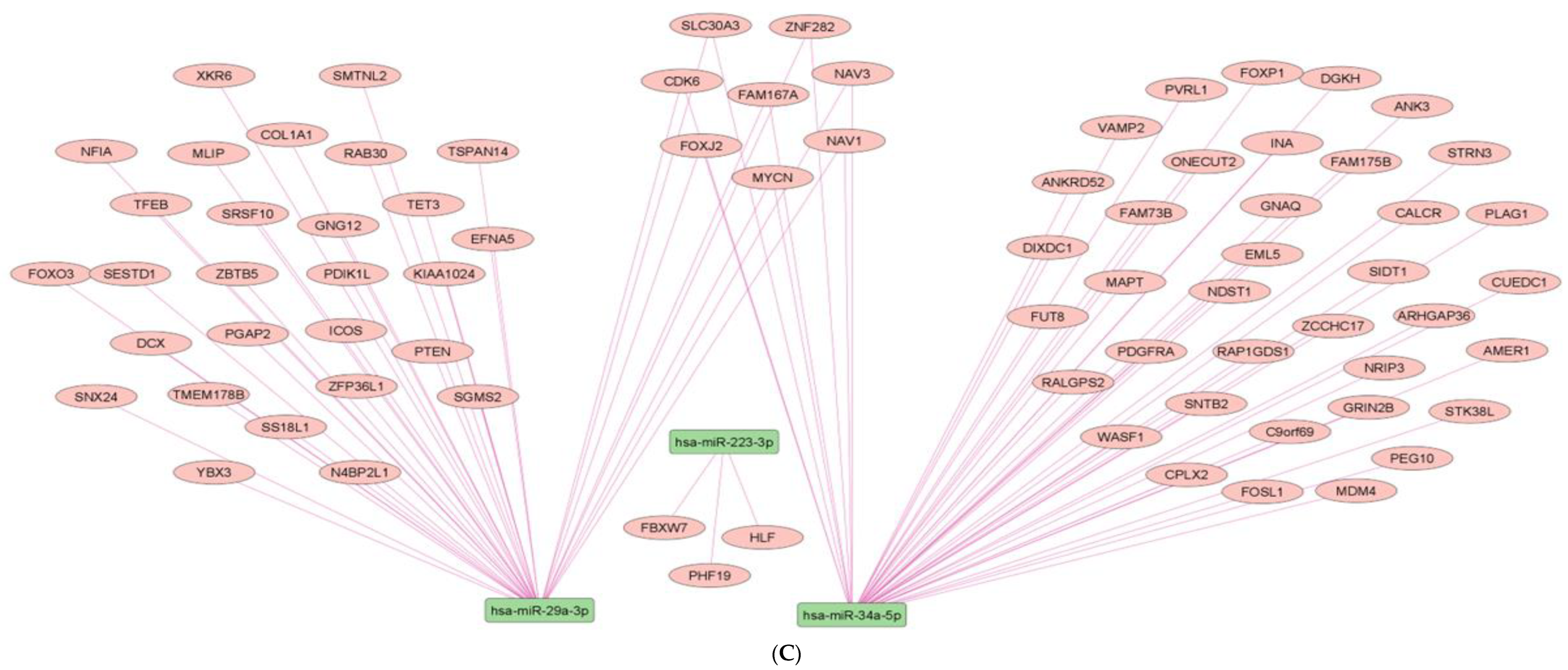
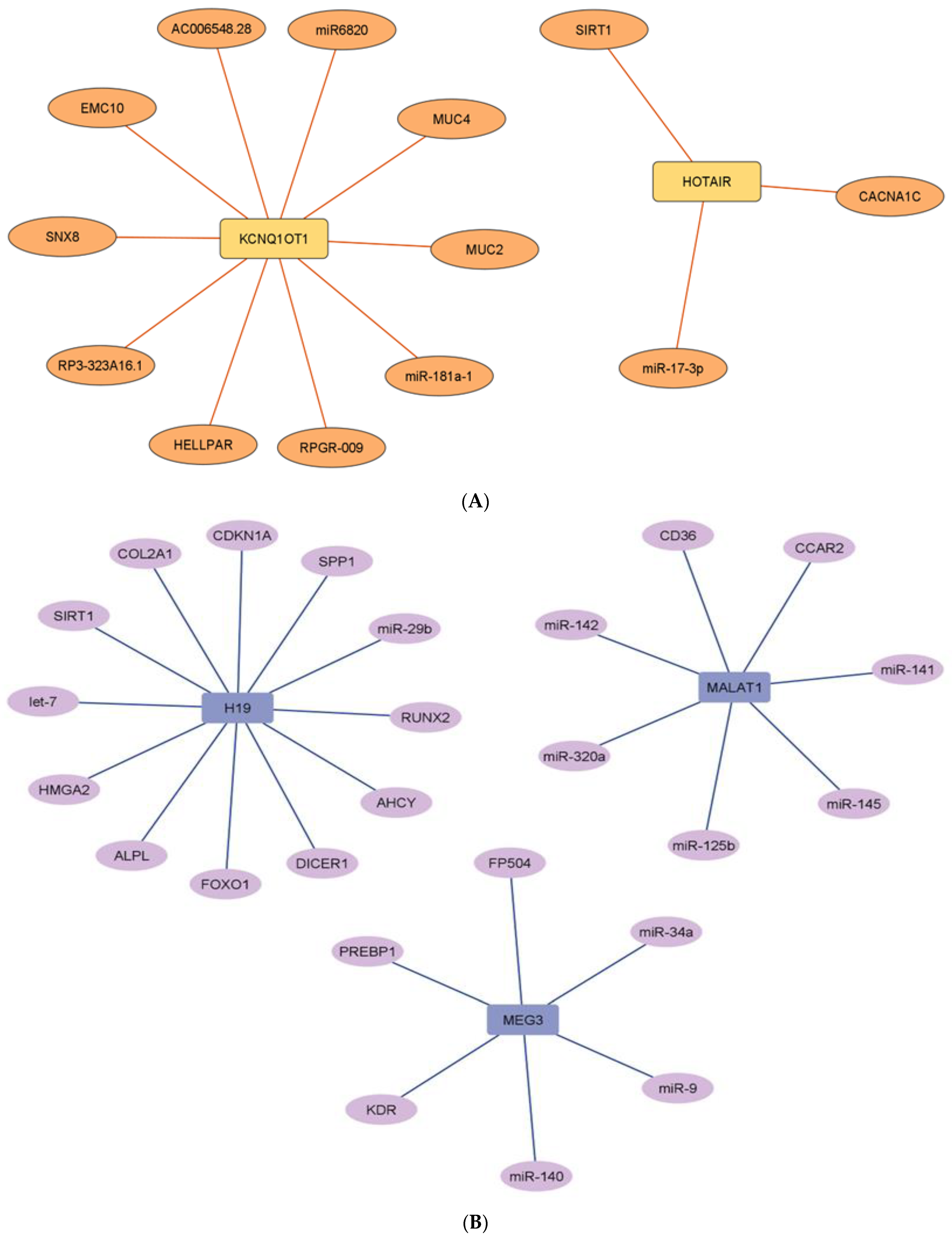
2.7. Network Architecture of Dysregulated ncRNAs in Diabetes
- (a)
- (b)
- (c)
- (d)
| Sr. No. | miRNA | Biological Matrix (Cell Line/Patient) | Targets | References |
|---|---|---|---|---|
| 1 | hsa-miR-29a | Serum samples of 18 subjects with newly diagnosed with T2D. | - | [45] |
| 2 | hsa-miR-34a | Plasma samples of prediabetic subjects (n = 12), and T2D (n = 31) and T1D patients (n = 16). | - | [46] |
| 3 | hsa-miR-1 | Serum samples of 78 males with uncomplicated T2D | - | [48] |
| 4 | hsa-miR-148b-3p | Plasma samples of 33 T1D mellitus patients | TP53, PTPN1, FADD, IRS2, CXCL8, IL8, PIK3R1, MAPK8/JNK, AKT3, IKBKB/IKKB | [33] |
| 5 | hsa-miR-1275 | |||
| 6 | hsa-miR-103a-3p | |||
| 7 | hsa-miR-200a-3p | |||
| 8 | hsa-miR-342 | |||
| 9 | hsa-miR-148a-3p | |||
| 10 | hsa-miR-21-5p | |||
| 11 | hsa-miR-210-3p | |||
| 12 | hsa-miR-338-3p | |||
| 13 | hsa-miR-200c-3p | |||
| 14 | hsa-miR-155-5p | |||
| 15 | hsa-miR-320 | |||
| 16 | hsa-miR-21-3p | |||
| 17 | hsa-miR-340-5p | |||
| 18 | hsa-miR-133a-5p | |||
| 19 | hsa-miR-2116-5p | Serum samples of 45 patients with diabetic retinopathy | NOTCH2 | [57] |
| 20 | hsa-miR-142-3p | Isolated human CD4+ T cells from serum samples of children with and without islet autoimmunity | TET2 | [40] |
| 21 | hsa-miR-23a | Serum samples of 22 children at onset of T1D | - | [38] |
| 22 | hsa-miR-98 | |||
| 23 | hsa-miR-197-3p | Plasma samples of 40 children diagnosed with new-onset T1D | TUSC2, NSUN5, CD82, BMF, PMAIP1, MTHFD1 | [39] |
| 24 | hsa-miR-199a-3p | Plasma samples of 60 patients with T2D; lower limb skin samples from 30 patients with diabetic neuropathy | SERPINE2 | [76] |
| 25 | hsa-miR-20a-5p | Serum samples of T2D patients with diabetic retinopathy | - | [58] |
| 26 | hsa-miR-223-3p | |||
| 27 | hsa-let-7a-5p | |||
| 28 | hsa-miR-885-5p | Serum samples of 455 type 1 diabetic patients | - | [35] |
| 29 | hsa-miR-486-3p | |||
| 30 | hsa-miR-574-3p | |||
| 31 | hsa-miR-140-3p | |||
| 32 | hsa-miR-17 | |||
| 33 | hsa-miR-222 | |||
| 34 | hsa-miR-16 | |||
| 35 | hsa-miR-106a | |||
| 36 | hsa-miR-139-5p | |||
| 37 | hsa-miR-210 | Plasma samples of 54 type 2 diabetic patients without coronary artery disease and 46 type 2 diabetic patients with coronary artery disease | - | [49] |
| 38 | hsa-miR-190a-5p | Serum samples of 21 T2D patients | - | [56] |
| 39 | miR-27a-3p | Human HK-2 cells | Prohibitin, TMBIM6 | [71] |
| 40 | miR-340 | Maternal and fetal WBCs from 30 women with gestational diabetes | PAIP1 | [80] |
| Sr. No. | miRNA | Biological Matrix (Cell Line/Patient) | Targets | References |
|---|---|---|---|---|
| 1 | hsa-miR-130b | Serum samples obtained from 327 T2D patients | - | [52] |
| 2 | let-7b | Whole peripheral blood from 40 T2D patients | IGF2BP2 | [53] |
| 3 | hsa-miR-320a | Plasma samples of 48 diabetes patients without diabetic retinopathy and 62 diabetes patients with diabetic retinopathy | FOXM1, YWHAZ, GTPBP2, YOD1, YWHAE, TSC1, CPD, PBX3, MAPK8IP3, GXYLT1, ARPP19, SYNGR2, CDK6, IGF2BP3, PTRN, KITLG | [64] |
| 4 | hsa-miR-214-3p | Whole-blood samples from 40 T2D patients | MAP2K5, CDK6, RASA1, MAPK14, FLOT1, MPL, IL13, ACVR1B, TGFB1, NOS3, PTPRF, ATF4, TRIB3, FGFR3 | [54] |
| 5 | hsa-miR-27b-3p | |||
| 6 | miR-23b-3p | |||
| 7 | miR-24-3p | |||
| 8 | miR-29b-3p | |||
| 9 | miR-451a | |||
| 10 | let-7f-5p | |||
| 11 | miR-34a | Serum samples of 18 subjects newly diagnosed with T2D and 19 pre-diabetic individuals | - | [45] |
| 12 | miR-4448 | Serum samples of 21 T2D patients | - | [56] |
| 13 | miR-9-5p | |||
| 14 | miR-338-3p | |||
| 15 | hsa-miR-485-5p | |||
| 16 | miR-423 | Serum samples of 10 T2D patients without retinopathy, 22 with non-proliferative diabetic retinopathy and 15 with proliferative diabetic retinopathy | - | [69] |
| 17 | hsa-miR-155 | Serum samples of 455 type 1 diabetic patients | - | [35] |
| 18 | hsa-miR-92a | |||
| 19 | hsa-miR-483-5p | |||
| 20 | hsa-miR-29a | |||
| 21 | hsa-miR-320 | |||
| 22 | hsa-miR-145 | |||
| 23 | hsa-miR-146a | |||
| 24 | hsa-miR-191 | |||
| 25 | hsa-miR-342 | |||
| 26 | hsa-miR-223 | |||
| 27 | hsa-miR-24 | |||
| 28 | hsa-miR-150 | |||
| 29 | miR-484 | |||
| 30 | miR-486-5p | |||
| 31 | hsa-miR-126 | Plasma samples of 54 T2D patients without coronary artery disease and 46 type 2 diabetic patients with coronary artery disease | - | [49] |
| 32 | miR-31 | Serum samples of 31 diabetic patients (18 with no diabetic complications and 13 with diabetic nephropathy) | TNFα, ICAM-1, IL-6 | [73] |
| 33 | miR-146 | PBMCs of T2D patients with diabetic neuropathy | TNF-α, IL-6, NF-κB | [79] |
| 34 | miR-29b | Placental tissues from 204 gestational diabetes patients | HIF3A | [82] |

| Sr. No. | lncRNA | Biological Matrix (Cell Line/Patient) | Targets | References |
|---|---|---|---|---|
| 1 | lncRNA-p3134 | Whole-blood samples of 30 T2D patients | TNS1, ASZ1, DIAPH1, IFNA14, ZNF436, MTMR3, CDK1, PPARD, TCF7, HUWE1, CCND2 | [50] |
| 2 | ANRIL | Peripheral-blood mononuclear cells (PBMCs) from 32 patients with T2D | - | [51] |
| 3 | ENST00000550337 | |||
| 4 | GAS5 | |||
| 5 | HOTAIR | |||
| 6 | lincRNA-p21 | |||
| 7 | PLUTO | |||
| 8 | HOTAIR | Serum samples of 30 diabetic patients, 30 with non-proliferative diabetic retinopathy (NPDR) and 20 with retinopathy (PDR) | - | [59] |
| 9 | MALAT1 | |||
| 10 | lncRNA-MIAT | Plasma samples of 52 patients with NPDR and ARPE-19 cells | TGFβ1 | [62] |
| 11 | KCNQ1OT1 | Serum samples of 20 patients with proliferative diabetic retinopathy and 20 with non-proliferative diabetic retinopathy, human retinal endothelial cells (hRECs) | miR-1470, EGFR | [63] |
| 12 | BANCR | Plasma samples of 64 patients with diabetic retinopathy, human retinal pigment epithelial cell line ARPE-19 | - | [88] |
| 13 | HEIH | Serum samples of 36 patients with diabetic retinopathy, human adult retinal pigment epithelial cells (ARPE-19) | miR-939 | [89] |
| 14 | CASC15 | Plasma samples of 50 diabetic patients with chronic renal failure | miR-34c | [90] |
| 15 | lnc-COX17-2:3 | Differentially expressed lncRNAs in umbilical cord-blood exosomes of 23 gestational diabetes patients | - | [83] |
| 16 | lnc-ZBTB46-3:6 | |||
| 17 | lnc-RXYLT13:2 | |||
| 18 | lnc-TFDP2-7:2 | |||
| 19 | AK023948 | Plasma samples from 25 patients with T1D | - | [41] |
| 20 | BC043430 | |||
| 21 | DGCR5 | |||
| 22 | MEG3 | |||
| 23 | PCAT-32 | |||
| 24 | SRA | |||
| 25 | ST7OT3 | |||
| 26 | TU0017629 | |||
| 27 | LincRNA-VLDLR | |||
| 28 | H19 | Plasma samples obtained from 30 patients with diabetes mellitus, human umbilical vein endothelial cells (HUVECs) | miR-29b | [91] |
| 29 | DNM3OS | CD14+ monocytes obtained from T2D patients | ILF-2 | [92] |
| 30 | SRA | Plasma samples of 25 patients with T1D | miR-146b | [41] |
| 31 | NR_038323 | Human HK-2 cells | miR-324-3p | [72] |
| 32 | MALAT1 | PBMCs of patients with diabetic neuropathy | CXCR4 | [77] |
| 33 | MALAT1 | Placental tissues from 78 gestational diabetes patients and 30 normal pregnant women | TGF, NF-κB | [81] |
| Sr. No. | lncRNA | Biological Matrix (Cell Line/Patient) | Targets | References |
|---|---|---|---|---|
| 1 | CTBP1-AS2 | Peripheral-blood mononuclear cell (PBMC) of 100 type 2 diabetic patients | TGF-β | [55] |
| 2 | AK077216 | Plasma samples from 60 diabetic retinopathy patients; human adult retinal pigment epithelial cells (ARPE-19) | miR-383 | [70] |
| 3 | ENST00000596839.1 | Differentially expressed lncRNAs in umbilical cord-blood exosomes of 23 gestational diabetes patients | miR-362-5p | [83] |
| 4 | lnc-EIF4ENIF1-1:1 | |||
| 5 | lnc-TBC1D30-4:1 | |||
| 6 | lnc-ZNF800-1:1 | |||
| 7 | LINK-A | Renal biopsies of patients with diabetic nephropathy | HIF1α | [74] |

3. Spatial Dysregulation of ncRNAs in Diabetes
3.1. Pancreatic Beta Cells
3.2. Mesenchymal Stem Cells
3.3. Human Retinal Microvascular Endothelial Cells (hRMECs)
3.4. Human Lens Epithelial Cells
3.5. Human Adult Retinal Pigment Epithelial (ARPE-19) Cells
3.6. Liver Cells and Diabetic Patient Livers
3.7. Human Umbilical Vein Endothelial Cells (HUVEC)
3.8. Human Kidney-2 (HK-2) Cells
3.9. Preretinal Fibrovascular Membranes
3.10. Human Dermal Microvascular Endothelial Cells (HMEC-1)
| Sr. No. | ncRNA | Biological Matrix (Cell Line/Patient) | Targets | Upregulated/ Downregulated | References |
|---|---|---|---|---|---|
| 1 | hsa-miR-452-5p | Human umbilical vein endothelial cell (HUVEC) samples isolated from pregnant women with gestational diabetes mellitus and normal glucose tolerance; placenta samples from women with gestational diabetes mellitus; BeWo cells initiated from malignant gestational choriocarcinoma of the fetal placentae | - | Upregulated | [111] |
| 2 | hsa-miR-34a | Human adult retinal pigment epithelial cells (ARPE-19) | SIRT1 | Upregulated | [104] |
| 3 | microRNA-205-5p | Human mesenchymal stem cells (MSCs) | VEGF | Upregulated | [97] |
| 4 | hsa-miR-223 | HepG2 human hepatic carcinoma cells | FOXO1 | Upregulated | [105] |
| 5 | hsa-miR-192-5p | Human umbilical vein endothelial cells (HUVECs) | BCL2, MCL1 | Upregulated | [107] |
| 6 | hsa-miR-221-3p | ||||
| 7 | hsa-miR-29c-3p | ||||
| 8 | hsa-miR-26b | Articular cartilage obtained from primary osteoarthritis patients, including 20 patients with diabetes | CTGF | Upregulated | [112] |
| 9 | hsa-miR-216a-5p | Kidney tissues from kidney biopsies of 20 patients with diabetic nephropathy; human kidney 2 (HK-2) cells | BMP7 | Upregulated | [108] |
| 10 | hsa-miR-181c-5p | Human induced pluripotent stem cells (hiPSCs) | TGIF2, Smad7 | Upregulated | [101] |
| 11 | miR-26a | Human hepatoma-derived HuH-7 cells, human liver samples from patients with T2D | - | Downregulated | [106] |
| 12 | ENST00000446360 | Preretinal fibrovascular membranes harvested from 20 patients diagnosed with proliferative diabetic retinopathy who underwent pars plana vitrectomy | - | Upregulated | [109] |
| 13 | ENST00000532530 | ||||
| 14 | ENST00000553754 | ||||
| 15 | ENST00000569661 | ||||
| 16 | NR_026567 | ||||
| 17 | NR_038970 | ||||
| 18 | T048173 | ||||
| 19 | T052922 | ||||
| 20 | T053053 | ||||
| 21 | T085943 | ||||
| 22 | T093217 | ||||
| 23 | T103904 | ||||
| 24 | T275433 | ||||
| 25 | T284419 | ||||
| 26 | TCONS_00029045 | ||||
| 27 | TCONS_00020046 | ||||
| 29 | MALAT1 | Human retinal microvascular endothelial cells (hRMECs) | miR-125b, VE-cadherin | Upregulated | [102] |
| 30 | MALAT1 | HK-2 (human kidney 2) cells | miR-145 | Upregulated | [113] |
| 31 | NR_047507 | Human umbilical vein endothelial cells (HUVECs) | - | Upregulated | [114] |
| 32 | NR_125792 | ||||
| 33 | T261470 | ||||
| 34 | BM807096 | HK-2 (human kidney 2) cells | miR-324-3p | Upregulated | [72] |
| 35 | DB268790 | ||||
| 36 | ENST00000418149 | ||||
| 37 | ENST00000429530 | ||||
| 38 | ENST00000430530 | ||||
| 39 | ENST00000442326 | ||||
| 40 | ENST00000452283 | ||||
| 41 | ENST00000490856 | ||||
| 42 | ENST00000511712 | ||||
| 43 | ENST00000511962 | ||||
| 44 | ENST00000527083 | ||||
| 45 | ENST00000556637 | ||||
| 46 | ENST00000567374 | ||||
| 47 | HMlincRNA215- | ||||
| 48 | NR_026830 | ||||
| 49 | NR_033950 | ||||
| 50 | NR_038323 | ||||
| 51 | NR_040017 | ||||
| 52 | TCONS_00006916 | ||||
| 53 | TCONS_00011702 | ||||
| 54 | TCONS_00020354 | ||||
| 55 | XIST | Renal tissues of 53 patients with diabetic nephropathy, HK-2 (human kidney 2) cells | miR-93-5p | Upregulated | [115] |
| 56 | MSTRG.15047.3 | Human retinal endothelial cells (hRECs), serum samples and aqueous humor from 30 diabetic retinopathy patients | PBRM1, FGF13 | Upregulated | [116] |
| 57 | AC008403.3 | ||||
| 58 | PVT1 | Human chondrocytes isolated from knee cartilage of 20 osteoarthritis patients with and without diabetes | miR-26b | Upregulated | [112] |
| 59 | LUCAT1 | AC16 cardiomyocyte cells | CYP11B2 | Upregulated | [117] |
| 60 | ARAP1-AS2 | HK-2 (human kidney 2) cells | CDC42 | Upregulated | [118] |
| 61 | H19 | Human dermal microvascular endothelial cells (HMEC-1) and human embryonic kidney cells 293 (HEK293) | - | Downregulated | [110] |
| 62 | MEG3 | Human adult retinal pigment epithelial cells (ARPE-19) | miR-34a | Downregulated | [104] |
| 63 | ENST00000446360 | Preretinal fibrovascular membranes harvested from 20 patients diagnosed with proliferative diabetic retinopathy who underwent pars plana vitrectomy | - | Downregulated | [109] |
| 64 | ENST00000569661 | ||||
| 65 | NR_038970 | ||||
| 66 | T052922 | ||||
| 67 | T085943 | ||||
| 68 | TCONS_00029045 | ||||
| 69 | MALAT1 | Human umbilical vein endothelial cells (HUVEC); human bone marrow-derived mesenchymal stem cells (MSCs) | miR-205-5p | Downregulated | [98] |
| 70 | hsa-miR-101-3p | Serum samples of 50 patients with recent-onset diabetes | HGF, MET proto-oncogene, GAB1, RAC1, COX2, c-FOS | Upregulated | [95] |
| 71 | miR-375 | Human induced pluripotent stem cells (hiPSCs) | - | Upregulated | [96] |
| 72 | miR-17-5p | Human umbilical cord mesenchymal stem cells (hucMSCs) | PTEN | Upregulated | [99] |
| 73 | miR-375 | Adipose-derived stem cells (ADSCs) | - | Upregulated | [100] |
| 74 | KCNQ1OT1 | Diabetic cataract posterior lens-capsule tissues and high-glucose (HG)-treated lens epithelial cells SRA01/04 | miR-26a-5p | Upregulated | [103] |
| 75 | miR-26a-5p | Diabetic cataract posterior lens-capsule tissues and high-glucose (HG)-treated lens epithelial cells SRA01/04 | ITGAV | Downregulated | [103] |
4. ncRNAs as Prognostic Markers in Diabetes
5. Signal Transduction with Diabetes-Associated Noncoding RNAs
5.1. miR-144
5.2. miR-26b
5.3. miR-96
5.4. miR-145
5.5. miR-375
5.6. miR-222
5.7. miR-148a
5.8. miR-18a-5p
5.9. miR-27a-3p
5.10. miR-23a
5.11. lncRNA MALAT1
5.12. lncRNA MEG3
6. ncRNAs and Epigenetic Mechanisms of Diabetes
7. Clinical Trials on Diabetes-Associated Noncoding RNAs
8. Functional Relevance of Dysregulated Noncoding RNAs
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| T1D | Type 1 diabetes |
| T2D | Type 2 diabetes |
| GDM | Gestational diabetes mellitus |
| IGF | Insulin-like growth factors |
| GAS6 | Growth arrest-specific gene 6 protein |
| VKDP | Vitamin K-dependent protein |
| ncRNA | Noncoding RNA |
| miRNA | microRNA |
| lncRNA | Long noncoding RNA |
| ceRNA | Competing endogenous RNA |
| MSC | Mesenchymal stem cells |
| HK-2 | Human Kidney-2 cells |
| HUVEC | Human umbilical vein endothelial cells |
| HepG2 | Human hepatocellular carcinoma |
| hRECs | Human retinal endothelial cells |
| hRMECs | Human retina microvascular endothelial cells |
| ARPE-19 | Adult retinal pigment epithelial cells |
| PBMCs | Peripheral-blood mononuclear cells |
| NPDR | Non-proliferative diabetic retinopathy |
| HMEC-1 | Human dermal microvascular endothelial cells |
| IRS1 | Insulin receptor substrate 1 |
| HPA-V | Human preadipocytes-visceral |
| INSR | Insulin receptor |
| PDPK1 | Phosphoinositide-dependent kinase-1 |
| ER-α | Estrogen receptor-α |
| PTC | Papillary thyroid cancer |
| PDE4 | Phosphodiesterase 4 |
| PRC2 | Polycomb-repressive complex 2 |
| 5-aza-dC | 5-aza-2′-deoxycytidine |
| DNMT | DNA methyltransferase |
| ARPE-19 | Human adult retinal pigment epithelial cells |
| PDR | Proliferative diabetic retinopathy |
| DKD | Diabetic kidney disease |
| DR | Diabetic retinopathy |
| DN | Diabetic nephropathy |
References
- Behl, T.; Kaur, I.; Sehgal, A.; Sharma, E.; Kumar, A.; Grover, M.; Bungau, S. Unfolding Nrf2 in Diabetes Mellitus. Mol. Biol. Rep. 2021, 48, 927–939. [Google Scholar] [CrossRef]
- Thomas, C.C.; Philipson, L.H. Update on Diabetes Classification. Med. Clin. N. Am. 2015, 99, 1–16. [Google Scholar] [CrossRef]
- CDC What Is Diabetes? Available online: https://www.cdc.gov/diabetes/basics/diabetes.html (accessed on 13 September 2021).
- National Diabetes Statistics Report 2020. Estimates of Diabetes and Its Burden in the United States. 2020; Volume 32. Available online: https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf (accessed on 16 June 2021).
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th Edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Shah, D.; Gandhi, M.; Kumar, A.; Cruz-Martins, N.; Sharma, R.; Nair, S. Current Insights into Epigenetics, Noncoding RNA Interactome and Clinical Pharmacokinetics of Dietary Polyphenols in Cancer Chemoprevention. Crit. Rev. Food Sci. Nutr. 2021, 1–37. [Google Scholar] [CrossRef]
- Gandhi, M.; Nair, S. New Vistas in Malignant Mesothelioma: MicroRNA Architecture and NRF2/MAPK Signal Transduction. Life Sci. 2020, 257, 118–123. [Google Scholar] [CrossRef]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of Translation and MRNA Degradation by MiRNAs and SiRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef]
- Kalis, M.; Bolmeson, C.; Esguerra, J.L.S.; Gupta, S.; Edlund, A.; Tormo-Badia, N.; Speidel, D.; Holmberg, D.; Mayans, S.; Khoo, N.K.S.; et al. Beta-Cell Specific Deletion of Dicer1 Leads to Defective Insulin Secretion and Diabetes Mellitus. PLoS ONE 2011, 6, e29166. [Google Scholar] [CrossRef]
- Eliasson, L. Micro(RNA) Management and Mismanagement of the Islet. J. Mol. Biol. 2019, 432, 1419–1428. [Google Scholar] [CrossRef]
- Nair, S.; Liew, C.; Khor, T.-O.; Cai, L.; Kong, A.-N.T. Differential Signaling Regulatory Networks Governing Hormone Refractory Prostate Cancers. J. Chin. Pharm. Sci. 2014, 23, 511–524. [Google Scholar] [CrossRef]
- Zhang, P.; Itan, Y. Biological Network Approaches and Applications in Rare Disease Studies. Genes 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Neelakandan, K.; Babu, P.; Nair, S. Emerging Roles for Modulation of MicroRNA Signatures in Cancer Chemoprevention. CCDT 2012, 12, 716–740. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Kong, A.-N.T. Architecture of Signature MiRNA Regulatory Networks in Cancer Chemoprevention. Curr. Pharm. Rep. 2015, 1, 89–101. [Google Scholar] [CrossRef]
- Nair, S.; Liew, C.; Khor, T.-O.; Cai, L.; Tony Kong, A.-N. Elucidation of Regulatory Interaction Networks Underlying Human Prostate Adenocarcinoma. J. Chin. Pharm. Sci. 2015, 24, 12–27. [Google Scholar] [CrossRef]
- Gada, Y.; Pandey, A.; Jadhav, N.; Ajgaonkar, S.; Mehta, D.; Nair, S. New Vistas in MicroRNA Regulatory Interactome in Neuropathic Pain. Front. Pharmacol. 2022, 12, 778014. [Google Scholar] [CrossRef] [PubMed]
- Nair, S. Current Insights into the Molecular Systems Pharmacology of LncRNA-MiRNA Regulatory Interactions and Implications in Cancer Translational Medicine. AIMS Mol. Sci. 2016, 3, 104–124. [Google Scholar] [CrossRef]
- Drag, M.H.; Kilpeläinen, T.O. Cell-Free DNA and RNA—Measurement and Applications in Clinical Diagnostics with Focus on Metabolic Disorders. Physiol. Genom. 2021, 53, 33–46. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and Future Perspectives of Liquid Biopsies in Genomics-Driven Oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Roskams-Hieter, B.; Kim, H.J.; Anur, P.; Wagner, J.T.; Callahan, R.; Spiliotopoulos, E.; Kirschbaum, C.W.; Civitci, F.; Spellman, P.T.; Thompson, R.F.; et al. Plasma Cell-Free RNA Profiling Distinguishes Cancers from Pre-Malignant Conditions in Solid and Hematologic Malignancies. NPJ Precis. Oncol. 2022, 6, 1–11. [Google Scholar] [CrossRef]
- Alfaifi, M.; Verma, A.K.; Alshahrani, M.Y.; Joshi, P.C.; Alkhathami, A.G.; Ahmad, I.; Hakami, A.R.; Beg, M.M.A. Assessment of Cell-Free Long Non-Coding RNA-H19 and MiRNA-29a, MiRNA-29b Expression and Severity of Diabetes. DMSO 2020, 13, 3727–3737. [Google Scholar] [CrossRef]
- Xiong, Y.; Chen, L.; Yan, C.; Zhou, W.; Endo, Y.; Liu, J.; Hu, L.; Hu, Y.; Mi, B.; Liu, G. Circulating Exosomal MiR-20b-5p Inhibition Restores Wnt9b Signaling and Reverses Diabetes-Associated Impaired Wound Healing. Small 2020, 16, 1904044. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jia, Z.; Zhao, X.; Xu, M.; Chen, M. Expression of MiR-210 in the Peripheral Blood of Patients with Newly Diagnosed Type 2 Diabetes Mellitus and Its Effect on the Number and Function of Endothelial Progenitor Cells. Microvasc. Res. 2020, 131, 104032. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Ames, R.M.; Moore, K.; Lee, B.P.; Jeffery, N.; Harries, L.W. Islet-Expressed Circular RNAs Are Associated with Type 2 Diabetes Status in Human Primary Islets and in Peripheral Blood. BMC Med. Genom. 2020, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Z.; Jian, D.; Li, W.; Tang, H.; Li, M. Hsa-CircRNA11783-2 in Peripheral Blood Is Correlated with Coronary Artery Disease and Type 2 Diabetes Mellitus. Diabetes Vasc. Dis. Res. 2017, 14, 510–515. [Google Scholar] [CrossRef]
- Jadhav, N.; Ajgaonkar, S.; Saha, P.; Gurav, P.; Pandey, A.; Basudkar, V.; Gada, Y.; Panda, S.; Jadhav, S.; Mehta, D.; et al. Molecular Pathways and Roles for Vitamin K2-7 as a Health-Beneficial Nutraceutical: Challenges and Opportunities. Front. Pharmacol. 2022, 13, 896920. [Google Scholar] [CrossRef]
- Vaidya, R.; Godse, C.; Jadhav, S.; Saha, P.; Ajgaonkar, S.; Pandey, A.; Gurav, P.; Jadhav, N.; Mehta, D.; Nair, S. An Intrinsic Need for K2-7 Supplementation: A Narrative Review of K2-7 and Peripheral Neuropathy. Biomed. J. Sci. Tech. Res. 2022, 42, 33679–33687. [Google Scholar]
- Tan, S.Y.; Mei Wong, J.L.; Sim, Y.J.; Wong, S.S.; Mohamed Elhassan, S.A.; Tan, S.H.; Ling Lim, G.P.; Rong Tay, N.W.; Annan, N.C.; Bhattamisra, S.K.; et al. Type 1 and 2 Diabetes Mellitus: A Review on Current Treatment Approach and Gene Therapy as Potential Intervention. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 364–372. [Google Scholar] [CrossRef]
- Nair, S.; Kong, A.-N.T. Emerging Roles for Clinical Pharmacometrics in Cancer Precision Medicine. Curr. Pharm. Rep. 2018, 4, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; LLerena, A. New Perspectives in Personalised Medicine for Ethnicity in Cancer: Population Pharmacogenomics and Pharmacometrics. Drug Metab. Pers. Ther. 2018, 33, 61–64. [Google Scholar] [CrossRef]
- Vasistha, A.; Kothari, R.; Mishra, A.; De Andrés, F.; LLerena, A.; Nair, S. Current Insights into Interethnic Variability in Testicular Cancers: Population Pharmacogenetics, Clinical Trials, Genetic Basis of Chemotherapy- Induced Toxicities and Molecular Signal Transduction. CTMC 2020, 20, 1824–1838. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; Puñales, M.; Tschiedel, B.; Canani, L.H.; Crispim, D. MicroRNA Expression Profile in Plasma from Type 1 Diabetic Patients: Case-Control Study and Bioinformatic Analysis. Diabetes Res. Clin. Pract. 2018, 141, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Roggli, E.; Britan, A.; Gattesco, S.; Lin-Marq, N.; Abderrahmani, A.; Meda, P.; Regazzi, R. Involvement of MicroRNAs in the Cytotoxic Effects Exerted by Proinflammatory Cytokines on Pancreatic β-Cells. Diabetes 2010, 59, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Barutta, F.; Bruno, G.; Matullo, G.; Chaturvedi, N.; Grimaldi, S.; Schalkwijk, C.; Stehouwer, C.D.; Fuller, J.H.; Gruden, G. MicroRNA-126 and Micro-/Macrovascular Complications of Type 1 Diabetes in the EURODIAB Prospective Complications Study. Acta Diabetol. 2017, 54, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhao, S. MiRNA-16-5p Inhibits the Apoptosis of High Glucose-Induced Pancreatic β Cells via Targeting of CXCL10: Potential Biomarkers in Type 1 Diabetes Mellitus. Endokrynol. Pol. 2020, 71, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.W.; Bakhashab, S.; Bastaman, I.T.; Crossland, R.E.; Glanville, M.; Weaver, J.U. Anti-Angiogenic MiR-222, MiR-195, and MiR-21a Plasma Levels in T1DM Are Improved by Metformin Therapy, Thus Elucidating Its Cardioprotective Effect: The MERIT Study. Int. J. Mol. Sci. 2018, 19, 3242. [Google Scholar] [CrossRef]
- Marchand, L.; Jalabert, A.; Meugnier, E.; van den Hende, K.; Fabien, N.; Nicolino, M.; Madec, A.-M.; Thivolet, C.; Rome, S. MiRNA-375 a Sensor of Glucotoxicity Is Altered in the Serum of Children with Newly Diagnosed Type 1 Diabetes. J. Diabetes Res. 2016, 2016, 1869082. [Google Scholar] [CrossRef] [PubMed]
- Samandari, N.; Mirza, A.H.; Nielsen, L.B.; Kaur, S.; Hougaard, P.; Fredheim, S.; Mortensen, H.B.; Pociot, F. Circulating MicroRNA Levels Predict Residual Beta Cell Function and Glycaemic Control in Children with Type 1 Diabetes Mellitus. Diabetologia 2017, 60, 354–363. [Google Scholar] [CrossRef]
- Scherm, M.G.; Serr, I.; Zahm, A.M.; Schug, J.; Bellusci, S.; Manfredini, R.; Salb, V.K.; Gerlach, K.; Weigmann, B.; Ziegler, A.-G.; et al. MiRNA142-3p Targets Tet2 and Impairs Treg Differentiation and Stability in Models of Type 1 Diabetes. Nat. Commun. 2019, 10, 5697. [Google Scholar] [CrossRef]
- Huang, Y.-N.; Chiang, S.-L.; Lin, Y.-J.; Liu, S.-C.; Li, Y.-H.; Liao, Y.-C.; Lee, M.-R.; Su, P.-H.; Tsai, F.-J.; Hung, H.-C.; et al. Long, Noncoding RNA SRA Induces Apoptosis of β-Cells by Promoting the IRAK1/LDHA/Lactate Pathway. IJMS 2021, 22, 1720. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Zheng, Y.S.; Li, Z.G.; Cui, Y.M.; Jiang, J.C. MiR-92a Contributes to the Cardiovascular Disease Development in Diabetes Mellitus through NF-ΚB and Downstream Inflammatory Pathways. Eur. Rev. 2019, 23, 3070–3079. [Google Scholar]
- Van Solingen, C.; Bijkerk, R.; de Boer, H.C.; Rabelink, T.J.; van Zonneveld, A.J. The Role of MicroRNA-126 in Vascular Homeostasis. Curr. Vasc. Pharm. 2015, 13, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Asgeirsdóttir, S.A.; van Solingen, C.; Kurniati, N.F.; Zwiers, P.J.; Heeringa, P.; van Meurs, M.; Satchell, S.C.; Saleem, M.A.; Mathieson, P.W.; Banas, B.; et al. MicroRNA-126 Contributes to Renal Microvascular Heterogeneity of VCAM-1 Protein Expression in Acute Inflammation. Am. J. Physiol. Ren. Physiol. 2012, 302, F1630–F1639. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of Serum MicroRNAs in Pre-Diabetes and Newly Diagnosed Type 2 Diabetes: A Clinical Study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-Enriched MiRNAs Are Altered in the Circulation of Subjects with Diabetes: A Pilot Cross-Sectional Study. Sci. Rep. 2016, 6, 31479. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Roy, S.; Dhas, Y.; Mishra, N. Senescence-Associated MiR-34a and MiR-126 in Middle-Aged Indians with Type 2 Diabetes. Clin. Exp. Med. 2020, 20, 149–158. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo-Calvo, D.; van der Meer, R.W.; Rijzewijk, L.J.; Smit, J.W.A.; Revuelta-Lopez, E.; Nasarre, L.; Escola-Gil, J.C.; Lamb, H.J.; Llorente-Cortes, V. Serum MicroRNA-1 and MicroRNA-133a Levels Reflect Myocardial Steatosis in Uncomplicated Type 2 Diabetes. Sci. Rep. 2017, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Amr, K.; Abdelmawgoud, H.; Ali, Z.; Shehata, S.; Raslan, H. Potential Value of Circulating MicroRNA-126 and MicroRNA-210 as Biomarkers for Type 2 Diabetes with Coronary Artery Disease. Br. J. Biomed. Sci. 2018, 75, 82–87. [Google Scholar] [CrossRef]
- Ruan, Y.; Lin, N.; Ma, Q.; Chen, R.; Zhang, Z.; Wen, W.; Chen, H.; Sun, J. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function. Cell. Physiol. Biochem. 2018, 46, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Sathishkumar, C.; Prabu, P.; Mohan, V.; Balasubramanyam, M. Linking a Role of LncRNAs (Long Non-Coding RNAs) with Insulin Resistance, Accelerated Senescence, and Inflammation in Patients with Type 2 Diabetes. Hum. Genom. 2018, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Zhou, Y.; Wu, C.; Shao, Y.; Lu, C.; Wang, Q. The Changes in MiR-130b Levels in Human Serum and the Correlation with the Severity of Diabetic Nephropathy: Serum MiR-130b and Diabetic Nephropathy. Diabetes Metab. Res. Rev. 2015, 31, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.-I. Decreased Expression of MicroRNAs Targeting Type-2 Diabetes Susceptibility Genes in Peripheral Blood of Patients and Predisposed Individuals. Endocrine 2019, 66, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Scorilas, A.; Fragoulis, E.G.; Christodoulou, M.-I. Blood-Based Analysis of 84 MicroRNAs Identifies Molecules Deregulated in Individuals with Type-2 Diabetes, Risk Factors for the Disease or Metabolic Syndrome. Diabetes Res. Clin. Pract. 2020, 164, 108187. [Google Scholar] [CrossRef] [PubMed]
- Erfanian Omidvar, M.; Ghaedi, H.; Kazerouni, F.; Kalbasi, S.; Shanaki, M.; Miraalamy, G.; Zare, A.; Rahimipour, A. Clinical Significance of Long Noncoding RNA VIM-AS1 and CTBP1-AS2 Expression in Type 2 Diabetes. J. Cell. Biochem. 2019, 120, 9315–9323. [Google Scholar] [CrossRef]
- Li, Z.; Dong, Y.; He, C.; Pan, X.; Liu, D.; Yang, J.; Sun, L.; Chen, P.; Wang, Q. RNA-Seq Revealed Novel Non-Proliferative Retinopathy Specific Circulating MiRNAs in T2DM Patients. Front. Genet. 2019, 10, 531. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Yi, Q.; Chen, L.; Wong, L.; Liu, Y.; Xu, G.; Zhao, J.; Huang, T.; Li, B.; Yang, Y.; et al. Circulating MiR-3197 and MiR-2116-5p as Novel Biomarkers for Diabetic Retinopathy. Clin. Chim. Acta 2020, 501, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Gao, K.P.; Wang, Y.X.; Liu, Z.C.; Tian, L.; Yang, X.Z.; Ding, J.Y.; Wu, W.T.; Yang, W.H.; Li, Y.L.; et al. RNA Sequencing Identified Specific Circulating MiRNA Biomarkers for Early Detection of Diabetes Retinopathy. Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E374–E385. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Abdelaleem, O.O.; Mahmoud, R.H.; Abdelghaffar, N.K.; Ahmed, T.I.; Said, O.M.; Zaki, O.M. Diagnostic and Prognostic Role of Serum MiR-20b, MiR-17-3p, HOTAIR, and MALAT1 in Diabetic Retinopathy. IUBMB Life 2019, 71, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long Non-coding RNA MALAT 1 Regulates Hyperglycaemia Induced Inflammatory Process in the Endothelial Cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef]
- Wang, H.; Xia, Y.; Zhang, Y. Diagnostic Significance of Serum LncRNA HOTAIR and Its Predictive Value for the Development of Chronic Complications in Patients with Type 2 Diabetes Mellitus. Diabetol. Metab. Syndr. 2021, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pang, L.; Yang, W.; Liu, X.; Su, G.; Dong, Y. Long Non-Coding RNA of Myocardial Infarction Associated Transcript (LncRNA-MIAT) Promotes Diabetic Retinopathy by Upregulating Transforming Growth Factor-B1 (TGF-B1) Signaling. Med. Sci. Monit. 2018, 24, 9497–9503. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Pan, X.; Yin, X.; Fan, G.; Tan, C.; Yao, Y.; Xin, Y.; Sun, C. KCNQ1OT1 Affects the Progression of Diabetic Retinopathy by Regulating MiR-1470 and Epidermal Growth Factor Receptor. J. Cell. Physiol. 2019, 234, 17269–17279. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.S.G.; de Jesus, M.L.; de Goes, T.C.; Mendonça, L.S.O.; Kaneto, C.M. Downregulation of Circulating MiR-320a and Target Gene Prediction in Patients with Diabetic Retinopathy. BMC Res. Notes 2020, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Thameem, F.; Wolford, J.K.; Bogardus, C.; Prochazka, M. Analysis of PBX1 as a Candidate Gene for Type 2 Diabetes Mellitus in Pima Indians. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2001, 1518, 215–220. [Google Scholar] [CrossRef][Green Version]
- Monica, K.; Galili, N.; Nourse, J.; Saltman, D.; Cleary, M.L. PBX2 and PBX3, New Homeobox Genes with Extensive Homology to the Human Proto-Oncogene PBX1. Mol. Cell. Biol. 1991, 11, 6149–6157. [Google Scholar] [PubMed]
- Liu, J.; Lang, G.; Shi, J. Epigenetic Regulation of PDX-1 in Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2021, 14, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Dawed, A.Y.; Yee, S.W.; Zhou, K.; van Leeuwen, N.; Zhang, Y.; Siddiqui, M.K.; Etheridge, A.; Innocenti, F.; Xu, F.; Li, J.H.; et al. Genome-Wide Meta-Analysis Identifies Genetic Variants Associated with Glycemic Response to Sulfonylureas. Diabetes Care 2021, 44, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Blum, A.; Meerson, A.; Rohana, H.; Jabaly, H.; Nahul, N.; Celesh, D.; Romanenko, O.; Tamir, S. MicroRNA-423 May Regulate Diabetic Vasculopathy. Clin. Exp. Med. 2019, 19, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, E.; Yang, L.; Fu, W.; Hu, F.; Zhou, X. LncRNA AK077216 Is Downregulated in Diabetic Retinopathy and Inhibited the Apoptosis of Retinal Pigment Epithelial Cells by Downregulating MiR-383. Endocr. J. 2019, 66, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Q.; Guo, F.; Ma, X.; Wang, J.; Zhao, Y.; Yan, Y.; Qin, G. Involvement of MiR-27a-3p in Diabetic Nephropathy via Affecting Renal Fibrosis, Mitochondrial Dysfunction, and Endoplasmic Reticulum Stress. J. Cell. Physiol. 2021, 236, 1454–1468. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Wang, J.; Wu, D.; Zhou, Y.; Qiu, S.; Chen, J.; Zhu, X.; Xiang, X.; Li, H.; Zhang, D. LncRNA NR_038323 Suppresses Renal Fibrosis in Diabetic Nephropathy by Targeting the MiR-324-3p/DUSP1 Axis. Mol. Ther. Nucleic Acids 2019, 17, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Rovira-Llopis, S.; Escribano-Lopez, I.; Diaz-Morales, N.; Iannantuoni, F.; Lopez-Domenech, S.; Andújar, I.; Jover, A.; Pantoja, J.; Pallardo, L.M.; Bañuls, C.; et al. Downregulation of MiR-31 in Diabetic Nephropathy and Its Relationship with Inflammation. Cell. Physiol. Biochem. 2018, 50, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, L.; Hong, S.; Zhou, Z.; Fan, W. LINK-A LncRNA Activates HIF1α Signaling and Inhibits Podocyte Cell Apoptosis in Diabetic Nephropathy. Exp. Ther. Med. 2019, 18, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Nordquist, L.; Friederich-Persson, M.; Fasching, A.; Liss, P.; Shoji, K.; Nangaku, M.; Hansell, P.; Palm, F. Activation of Hypoxia-Inducible Factors Prevents Diabetic Nephropathy. JASN 2015, 26, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-B.; Wu, Q.; Liu, J.; Fan, Y.-Z.; Yu, K.-F.; Cai, Y. MiR-199a-3p Is Involved in the Pathogenesis and Progression of Diabetic Neuropathy through Downregulation of SerpinE2. Mol. Med. Rep. 2017, 16, 2417–2424. [Google Scholar] [CrossRef] [PubMed]
- Ashjari, D.; Karamali, N.; Rajabinejad, M.; Hassani, S.S.; Afshar Hezarkhani, L.; Afshari, D.; Gorgin Karaji, A.; Salari, F.; Rezaiemanesh, A. The Axis of Long Non-Coding RNA MALAT1/MiR-1-3p/CXCR4 Is Dysregulated in Patients with Diabetic Neuropathy. Heliyon 2022, 8, e09178. [Google Scholar] [CrossRef]
- Menichella, D.M.; Abdelhak, B.; Ren, D.; Shum, A.; Frietag, C.; Miller, R.J. CXCR4 Chemokine Receptor Signaling Mediates Pain in Diabetic Neuropathy. Mol. Pain 2014, 10, 1744–8069. [Google Scholar] [CrossRef]
- Wang, G.-F.; Xu, N. Expression and Clinical Significance of MicroRNA 146a in Peripheral Blood Mononuclear Cells from Type 2 Diabetic Neuropathy Patients. Int. J. Clin. Exp. Med. 2018, 11, 7165–7173. [Google Scholar]
- Stirm, L.; Huypens, P.; Sass, S.; Batra, R.; Fritsche, L.; Brucker, S.; Abele, H.; Hennige, A.M.; Theis, F.; Beckers, J.; et al. Maternal Whole Blood Cell MiRNA-340 Is Elevated in Gestational Diabetes and Inversely Regulated by Glucose and Insulin. Sci. Rep. 2018, 8, 1366. [Google Scholar] [CrossRef]
- Zhang, Y.; Qu, L.; Ni, H.; Wang, Y.; Li, L.; Yang, X.; Wang, X.; Hou, Y. Expression and Function of LncRNA MALAT1 in Gestational Diabetes Mellitus. Adv. Clin. Exp. Med. 2020, 29, 903–910. [Google Scholar] [CrossRef]
- Sun, D.-G.; Tian, S.; Zhang, L.; Hu, Y.; Guan, C.-Y.; Ma, X.; Xia, H.-F. The MiRNA-29b Is Downregulated in Placenta during Gestational Diabetes Mellitus and May Alter Placenta Development by Regulating Trophoblast Migration and Invasion through a HIF3A-Dependent Mechanism. Front. Endocrinol. 2020, 11, 169. [Google Scholar] [CrossRef]
- Cao, M.; Zhang, L.; Lin, Y.; Li, Z.; Xu, J.; Shi, Z.; Chen, Z.; Ma, J.; Wen, J. Differential MRNA and Long Noncoding RNA Expression Profiles in Umbilical Cord Blood Exosomes from Gestational Diabetes Mellitus Patients. DNA Cell Biol. 2020, 39, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. Mienturnet: An Interactive Web Tool for MicroRNA-Target Enrichment and Network-Based Analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Sun, Z.; Ren, Q.; Su, X.; Zhang, D. Long Non-Coding RNA BANCR Is Overexpressed in Patients with Diabetic Retinopathy and Promotes Apoptosis of Retinal Pigment Epithelial Cells. Med. Sci. Monit. 2019, 25, 2845–2851. [Google Scholar] [CrossRef]
- Zhao, C.; Fei, X.; Xu, B.; Lu, Y.; Zhang, Q. Long Non-Coding RNA HEIH Contributes to Diabetic Retinopathy by Regulating MiR-939/VEGF Axis. Int. J. Clin. Exp. Pathol. 2019, 12, 2022–2033. [Google Scholar] [PubMed]
- Qin, X.; Zhu, S.; Chen, Y.; Chen, D.; Tu, W.; Zou, H. Long Non-Coding RNA (LncRNA) CASC15 Is Upregulated in Diabetes-Induced Chronic Renal Failure and Regulates Podocyte Apoptosis. Med. Sci. Monit. 2020, 26, e919415. [Google Scholar] [CrossRef]
- Cheng, X.-W.; Chen, Z.-F.; Wan, Y.-F.; Zhou, Q.; Wang, H.; Zhu, H.-Q. Long Non-Coding RNA H19 Suppression Protects the Endothelium against Hyperglycemic-Induced Inflammation via Inhibiting Expression of MiR-29b Target Gene Vascular Endothelial Growth Factor a through Activation of the Protein Kinase B/Endothelial Nitric Oxide Synthase Pathway. Front. Cell Dev. Biol. 2019, 7, 263. [Google Scholar] [CrossRef]
- Das, S.; Reddy, M.A.; Senapati, P.; Stapleton, K.; Lanting, L.; Wang, M.; Amaram, V.; Ganguly, R.; Zhang, L.; Devaraj, S.; et al. Diabetes Mellitus-Induced Long Noncoding RNA Dnm3os Regulates Macrophage Functions and Inflammation via Nuclear Mechanisms. Arter. Thromb. Vasc. Biol. 2018, 38, 1806–1820. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A Comprehensive Database for Target Genes of LncRNAs in Human and Mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, T.; Iwakiri, J.; Ono, Y.; Hamada, M. LncRRIsearch: A Web Server for LncRNA-RNA Interaction Prediction Integrated with Tissue-Specific Expression and Subcellular Localization Data. Front. Genet. 2019, 10, 462. [Google Scholar] [CrossRef]
- Santos, A.S.; Cunha Neto, E.; Fukui, R.T.; Ferreira, L.R.P.; Silva, M.E.R. Increased Expression of Circulating MicroRNA 101-3p in Type 1 Diabetes Patients: New Insights Into MiRNA-Regulated Pathophysiological Pathways for Type 1 Diabetes. Front. Immunol. 2019, 10, 1637. [Google Scholar] [CrossRef] [PubMed]
- Lahmy, R.; Soleimani, M.; Sanati, M.H.; Behmanesh, M.; Kouhkan, F.; Mobarra, N. MiRNA-375 Promotes Beta Pancreatic Differentiation in Human Induced Pluripotent Stem (HiPS) Cells. Mol. Biol. Rep. 2014, 41, 2055–2066. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, G.; Fischbach, S.; Xiao, X. Suppression of MicroRNA-205-5p in Human Mesenchymal Stem Cells Improves Their Therapeutic Potential in Treating Diabetic Foot Disease. Oncotarget 2017, 8, 52294–52303. [Google Scholar] [CrossRef]
- Zhu, L.; Zhong, Q.; Yang, T.; Xiao, X. Improved Therapeutic Effects on Diabetic Foot by Human Mesenchymal Stem Cells Expressing MALAT1 as a Sponge for MicroRNA-205-5p. Aging 2019, 11, 12236–12245. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, Y.; Ma, K.; Li, Q.; Li, B.; Hu, W.; Fu, X.; Zhang, C. Extracellular Vesicles from Human Umbilical Cord Mesenchymal Stem Cells Facilitate Diabetic Wound Healing through MiR-17-5p-Mediated Enhancement of Angiogenesis. Stem Cell Rev. Rep. 2022, 18, 1025–1040. [Google Scholar] [CrossRef] [PubMed]
- Piran, M.; Enderami, S.E.; Piran, M.; Sedeh, H.S.; Seyedjafari, E.; Ardeshirylajimi, A. Insulin Producing Cells Generation by Overexpression of MiR-375 in Adipose-Derived Mesenchymal Stem Cells from Diabetic Patients. Biologicals 2017, 46, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Jiang, D.; He, Q.; He, F.; Li, Y.; Deng, C.; Li, F. MicroRNA-181c-5p Promotes the Formation of Insulin-Producing Cells from Human Induced Pluripotent Stem Cells by Targeting Smad7 and TGIF2. Cell Death Dis. 2020, 11, 462. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.-B.; Shi, J.-M.; Li, W.-J.; Tang, L.-S.; Zhu, X.-H.; Tong, P. LncRNA-MALAT1 Promotes Neovascularization in Diabetic Retinopathy through Regulating MiR-125b/VE-Cadherin Axis. BioSci. Rep. 2019, 39, BSR20181469. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dong, Y.; Wen, Y.; Shi, L.; Zhu, Z.; Ke, G.; Gu, Y. LncRNA KCNQ1OT1 Knockdown Inhibits Viability, Migration and Epithelial-Mesenchymal Transition in Human Lens Epithelial Cells via MiR-26a-5p/ITGAV/TGF-Beta/Smad3 Axis. Exp. Eye Res. 2020, 200, 108251. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Peng, Q.-H.; Gu, L.-M.; Xie, W.-W.; Li, W.-J. LncRNA-MEG3 Alleviates High Glucose Induced Inflammation and Apoptosis of Retina Epithelial Cells via Regulating MiR-34a/SIRT1 Axis. Exp. Mol. Pathol. 2019, 107, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, J.; Yao, H.; Lin, Y.; Wei, J.; Hu, G.; Guo, J.; Li, J. Ultraconserved Element Uc.333 Increases Insulin Sensitivity by Binding to MiR-223. Aging 2020, 12, 6667–6679. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Wang, X.; Lou, F.; et al. MicroRNA-26a Regulates Insulin Sensitivity and Metabolism of Glucose and Lipids. J. Clin. Investig. 2015, 125, 2497–2509. [Google Scholar] [CrossRef] [PubMed]
- Silambarasan, M.; Tan, J.; Karolina, D.; Armugam, A.; Kaur, C.; Jeyaseelan, K. MicroRNAs in Hyperglycemia Induced Endothelial Cell Dysfunction. IJMS 2016, 17, 518. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Zhai, X.; Yuan, Y.; Ji, Q.; Zhang, P. LncRNA ZEB1-AS1 Inhibits High Glucose-Induced EMT and Fibrogenesis by Regulating the MiR-216a-5p/BMP7 Axis in Diabetic Nephropathy. Braz. J. Med. Biol. Res. 2020, 53, e9288. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, X.; Liu, J.; Wang, J.; Zhang, Y.; Zhang, T.; Zhang, H. Effect of Intravitreal Conbercept Treatment on the Expression of Long Noncoding RNAs and MRNAs in Proliferative Diabetic Retinopathy Patients. Acta Ophthalmol. 2019, 97, e902–e912. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.-C.; Rui, B.-Y.; Wang, Q.-Y.; Zhou, D.; Zhang, Y.; Guo, S.-C. Extracellular Vesicle-Mimetic Nanovesicles Transport LncRNA-H19 as Competing Endogenous RNA for the Treatment of Diabetic Wounds. Drug Deliv. 2018, 25, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Tryggestad, J.B.; Vishwanath, A.; Jiang, S.; Mallappa, A.; Teague, A.M.; Takahashi, Y.; Thompson, D.M.; Chernausek, S.D. Influence of Gestational Diabetes Mellitus on Human Umbilical Vein Endothelial Cell MiRNA. Clin. Sci. 2016, 130, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.-B.; Li, Y.; Liu, G.-Y.; Li, T.-H.; Li, F.; Guan, J.; Wang, H.-J. Long Non-Coding RNA PVT1, a Molecular Sponge of MiR-26b, Is Involved in the Progression of Hyperglycemia-Induced Collagen Degradation in Human Chondrocytes by Targeting CTGF/TGF- β Signal Ways. Innate Immun. 2020, 26, 204–214. [Google Scholar] [CrossRef]
- Liu, B.; Qiang, L.; Wang, G.-D.; Duan, Q.; Liu, J. LncRNA MALAT1 Facilities High Glucose Induced Endothelial to Mesenchymal Transition and Fibrosis via Targeting MiR-145/ZEB2 Axis. Eur. Rev. Med. Pharm. Sci. 2019, 23, 3478–3486. [Google Scholar] [CrossRef]
- Luo, Y.; Fang, Z.; Ling, Y.; Luo, W. LncRNA-H19 Acts as a CeRNA to Regulate HE4 Expression by Sponging MiR-140 in Human Umbilical Vein Endothelial Cells under Hyperglycemia with or without α-Mangostin. Biomed. Pharmacother. 2019, 118, 109256. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shen, Y.; Yang, X.; Long, Y.; Chen, S.; Lin, X.; Dong, R.; Yuan, J. Silencing of Long Noncoding RNA XIST Protects against Renal Interstitial Fibrosis in Diabetic Nephropathy via MicroRNA-93-5p-Mediated Inhibition of CDKN1A. Am. J. Physiol. Ren. Physiol. 2019, 317, F1350–F1358. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Zhang, Y.; Fan, G.; Xin, Y.; Yao, Y. Transcriptome Analysis Identified a Novel 3-LncRNA Regulatory Network of Transthyretin Attenuating Glucose Induced HRECs Dysfunction in Diabetic Retinopathy. BMC Med. Genom. 2019, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yang, Z.-F.; Li, X.-H.; Zhou, L.-Q.; Zhang, Y.-J.; Yang, B. Knockdown of Long Non-Coding RNA LUCAT1 Reverses High Glucose-Induced Cardiomyocyte Injury via Targeting CYP11B2. Eur. Rev. Med. Pharm. Sci. 2019, 23, 8560–8565. [Google Scholar] [CrossRef]
- Li, L.; Xu, L.; Wen, S.; Yang, Y.; Li, X.; Fan, Q. The Effect of LncRNA-ARAP1-AS2/ARAP1 on High Glucose-Induced Cytoskeleton Rearrangement and Epithelial-Mesenchymal Transition in Human Renal Tubular Epithelial Cells. J. Cell. Physiol. 2020, 235, 5787–5795. [Google Scholar] [CrossRef] [PubMed]
- Conserva, F.; Barozzino, M.; Pesce, F.; Divella, C.; Oranger, A.; Papale, M.; Sallustio, F.; Simone, S.; Laviola, L.; Giorgino, F.; et al. Urinary MiRNA-27b-3p and MiRNA-1228-3p Correlate with the Progression of Kidney Fibrosis in Diabetic Nephropathy. Sci. Rep. 2019, 9, 11357. [Google Scholar] [CrossRef]
- Tao, L.; Huang, X.; Xu, M.; Qin, Z.; Zhang, F.; Hua, F.; Jiang, X.; Wang, Y. Value of Circulating MiRNA-21 in the Diagnosis of Subclinical Diabetic Cardiomyopathy. Mol. Cell. Endocrinol. 2020, 518, 110944. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Lucena, R.; Camargo, A.; Alcalá-Diaz, J.F.; Romero-Baldonado, C.; Luque, R.M.; van Ommen, B.; Delgado-Lista, J.; Ordovás, J.M.; Pérez-Martínez, P.; Rangel-Zúñiga, O.A.; et al. A Plasma Circulating MiRNAs Profile Predicts Type 2 Diabetes Mellitus and Prediabetes: From the CORDIOPREV Study. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- Kim, H.; Bae, Y.-U.; Jeon, J.S.; Noh, H.; Park, H.K.; Byun, D.W.; Han, D.C.; Ryu, S.; Kwon, S.H. The Circulating Exosomal MicroRNAs Related to Albuminuria in Patients with Diabetic Nephropathy. J. Transl. Med. 2019, 17, 236. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, M.; Yu, J.; Ping, F.; Zhang, H.; Li, W.; Xu, L.; Li, Y. Decreased Serum MicroRNA-21, MicroRNA-25, MicroRNA-146a, and MicroRNA-181a in Autoimmune Diabetes: Potential Biomarkers for Diagnosis and Possible Involvement in Pathogenesis. Int. J. Endocrinol. 2019, 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Xu, G.; Ji, C.; Song, G.; Zhao, C.; Shi, C.; Song, L.; Chen, L.; Yang, L.; Huang, F.; Pang, L.; et al. MiR-26b Modulates Insulin Sensitivity in Adipocytes by Interrupting the PTEN/PI3K/AKT Pathway. Int. J. Obes. 2015, 39, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Min, K.-H.; Lee, W. Induction of MiR-96 by Dietary Saturated Fatty Acids Exacerbates Hepatic Insulin Resistance through the Suppression of INSR and IRS-1. PLoS ONE 2016, 11, e0169039. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Yang, Y.; Jin, D.; Sun, J.; Yu, X.; Yang, Z. MiRNA-145 Is Involved in the Development of Resistin-Induced Insulin Resistance in HepG2 Cells. Biochem. Biophys. Res. Commun. 2014, 445, 517–523. [Google Scholar] [CrossRef] [PubMed]
- El Ouaamari, A.; Baroukh, N.; Martens, G.A.; Lebrun, P.; Pipeleers, D.; van Obberghen, E. MiR-375 Targets 3’-Phosphoinositide-Dependent Protein Kinase-1 and Regulates Glucose-Induced Biological Responses in Pancreatic Beta-Cells. Diabetes 2008, 57, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhao, C.; Guo, X.; Ding, H.; Cui, Y.; Shen, R.; Liu, J. Differential Expression of MicroRNAs in Omental Adipose Tissue from Gestational Diabetes Mellitus Subjects Reveals MiR-222 as a Regulator of ERα Expression in Estrogen-Induced Insulin Resistance. Endocrinology 2014, 155, 1982–1990. [Google Scholar] [CrossRef]
- Xu, Y.; Han, Y.-F.; Zhu, S.-J.; Dong, J.-D.; Ye, B. MiRNA-148a Inhibits Cell Growth of Papillary Thyroid Cancer through STAT3 and PI3K/AKT Signaling Pathways. Oncol. Rep. 2017, 38, 3085–3093. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.-T.; Li, X.-X.; Peng, D.-W.; Zhang, W.-M.; Qu, J.; Lu, F.; D’Amato, R.J.; Chi, Z.-L. MicroRNA-18a-5p Administration Suppresses Retinal Neovascularization by Targeting FGF1 and HIF1A. Front. Pharm. 2020, 11, 276. [Google Scholar] [CrossRef]
- Yasmeen, S.; Kaur, S.; Mirza, A.H.; Brodin, B.; Pociot, F.; Kruuse, C. MiRNA-27a-3p and MiRNA-222-3p as Novel Modulators of Phosphodiesterase 3a (PDE3A) in Cerebral Microvascular Endothelial Cells. Mol. Neurobiol. 2019, 56, 5304–5314. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xia, L.; Xie, A.; Liao, O.; Ju, F.; Zhou, Y. Low Concentration of Bupivacaine Ameliorates Painful Diabetic Neuropathy by Mediating MiR-23a/PDE4B Axis in Microglia. Eur. J. Pharm. 2021, 891, 173719. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kowluru, R.A. Long Noncoding RNA MALAT1 and Regulation of the Antioxidant Defense System in Diabetic Retinopathy. Diabetes 2021, 70, 227–239. [Google Scholar] [CrossRef]
- Nair, S.; Doh, S.T.; Chan, J.Y.; Kong, A.-N.; Cai, L. Regulatory Potential for Concerted Modulation of Nrf2- and Nfkb1-Mediated Gene Expression in Inflammation and Carcinogenesis. Br. J. Cancer 2008, 99, 2070–2082. [Google Scholar] [CrossRef]
- Zhu, W.; Shen, Y.; Liu, J.; Fei, X.; Zhang, Z.; Li, M.; Chen, X.; Xu, J.; Zhu, Q.; Zhou, W.; et al. Epigenetic Alternations of MicroRNAs and DNA Methylation Contribute to Gestational Diabetes Mellitus. J. Cell. Mol. Med. 2020, 24, 13899–13912. [Google Scholar] [CrossRef]
- Belot, M.-P.; Fradin, D.; Mai, N.; Le Fur, S.; Zélénika, D.; Kerr-Conte, J.; Pattou, F.; Lucas, B.; Bougnères, P. CpG Methylation Changes within the IL2RA Promoter in Type 1 Diabetes of Childhood Onset. PLoS ONE 2013, 8, e68093. [Google Scholar] [CrossRef] [PubMed]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA Methylation Directs MicroRNA Biogenesis in Mammalian Cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef]
- Biswas, S.; Feng, B.; Chen, S.; Liu, J.; Aref-Eshghi, E.; Gonder, J.; Ngo, V.; Sadikovic, B.; Chakrabarti, S. The Long Non-Coding RNA HOTAIR Is a Critical Epigenetic Mediator of Angiogenesis in Diabetic Retinopathy. Investig. Opthalmology Vis. Sci. 2021, 62, 20. [Google Scholar] [CrossRef]
- Biswas, S.; Thomas, A.A.; Chen, S.; Aref-Eshghi, E.; Feng, B.; Gonder, J.; Sadikovic, B.; Chakrabarti, S. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci. Rep. 2018, 8, 6526. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Tabaraie, T.; Steffens, D.; Friebel, J.; Dörner, A.; Skurk, C.; Witkowski, M.; Stratmann, B.; Tschoepe, D.; Landmesser, U.; et al. MicroRNA-19a Contributes to the Epigenetic Regulation of Tissue Factor in Diabetes. Cardiovasc. Diabetol. 2018, 17, 34. [Google Scholar] [CrossRef]
- Singh, K.; Pal, D.; Sinha, M.; Ghatak, S.; Gnyawali, S.C.; Khanna, S.; Roy, S.; Sen, C.K. Epigenetic Modification of MicroRNA-200b Contributes to Diabetic Vasculopathy. Mol. Ther. 2017, 25, 2689–2704. [Google Scholar] [CrossRef] [PubMed]
- Peñas-LLedó, E.; Terán, E.; Sosa-Macías, M.; Galaviz-Hernández, C.; Gil, J.-P.; Nair, S.; Diwakar, S.; Hernández, I.; Lara-Riegos, J.; Ramírez-Roa, R.; et al. Challenges and Opportunities for Clinical Pharmacogenetic Research Studies in Resource-Limited Settings: Conclusions from the Council for International Organizations of Medical Sciences–Ibero-American Network of Pharmacogenetics and Pharmacogenomics Meeting. Clin. Ther. 2020, 42, 1595–1610.e5. [Google Scholar] [CrossRef] [PubMed]
- Guo, X. Circulating MicroRNAs as Novel Prognostic Biomarkers in Obese Preschoolers at Risk for Type 2 Diabetes in Adulthood. 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT02843139 (accessed on 27 December 2021).
- Solini, A. Association Between Urinary and Serum Levels of MiRNA 192 and MiRNA 25 and Glomerular Filtration and Albuminuria in Patients With and Without Type 2 Diabetes. Available online: https://clinicaltrials.gov/ct2/show/NCT04176276 (accessed on 27 December 2021).
- Casa Sollievo della Sofferenza IRCCS. White Blood Cells Gene Expression Profiles as a Tool for Predicting Metformin Efficacy in Patients with Type 2 Diabetes Mellitus. 2011. Available online: https://clinicaltrials.gov/ct2/show/NCT01334684 (accessed on 27 December 2021).
- AdventHealth Translational Research Institute. Effect of AT-Derived MiRNA on the Biology and Insulin Sensitivity of Skeletal Muscle in Humans. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT02459106 (accessed on 27 December 2021).
- Benincasa, G. Perturbation of Interactome through Micro-RNA and Methylome Analyses in Diabetes Endophenotypes: The PIRAMIDE Study Design. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT03792607 (accessed on 27 December 2021).
- Hadassah Medical Organization. Biological Changes in the Adipose Tissue (RNA Profile) among Pregnant Women with Diabetes. 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT02383537 (accessed on 27 December 2021).
- National Research Centre, E. Diabetes Mellitus and MicroRNA as Risk Factors for Mild Cognitive Impairment: Impact of Life Style Modification. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04891887 (accessed on 27 December 2021).
- Nantes University Hospital. Profiling of Original Cellular and Humoral Biomarkers of Type 1 Diabetes. 2015. Available online: https://clinicaltrials.gov/ct2/show/NCT01042301 (accessed on 27 December 2021).
- Muschitz, D.C. MicroRNAs Levels in Women with Postmenopausal Osteoporosis under Antiresorptive or Osteoanabolic Treatment. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT03472846 (accessed on 27 December 2021).
- Kautzky-Willer, A. Sex-Specific Relationship of Epigenetics Based Modifications in the Saliva and Blood with the Occurence of Type 2 Diabetes. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT04011228 (accessed on 27 December 2021).
- Labayen, I. Prevention of Diabetes in Overweight/Obese Preadolescent Children through a Family-Based Intervention Program Including Supervised Exercise; the PREDIKID Study. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT03027726 (accessed on 27 December 2021).
- Rizzo, M. Effect of Dapagliflozin on Cardio-Metabolic Risk Factors in Patients with Type-2 Diabetes. 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT03377335 (accessed on 27 December 2021).
- University Hospital, M. Study of New Determinants of Type 2 Diabetes in Severe Obesity. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT02861781 (accessed on 27 December 2021).
- Centre Hospitalier Universitaire de Nice. Macrophage Phenotype in Type 2 Diabetics after Myocardial Infarction and the Potential Role of MiRNAs Secreted. 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT02768935 (accessed on 27 December 2021).
- Shitrit, S.B. Correlation between Vitamin D Levels to ADAMTS13, VWF and Micro RNA Expression in Diabetic Hemodialysis Patients-Full Text View-ClinicalTrials.Gov. 2014. Available online: https://clinicaltrials.gov/ct2/show/NCT02245633 (accessed on 29 December 2021).
- Gallelli, L. Comparison of Anti-Inflammatory Status Linked to Atherosclerosis Formation/Progression among Diabetes Mellitus Type 2 Patients under Combined Pharmacological Therapy. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04392557 (accessed on 27 December 2021).
- Findikoglu, G. Metabolic, Physical Responses to Exercise in Patients with Type 2 Diabetes Mellitus. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT03682445 (accessed on 27 December 2021).
- Ukropec, J. Randomised Placebo Controlled Study of the Effect of Carnosine Diabetes and Cardiovascular Risk Factors; 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT02011100 (accessed on 27 December 2021).
- AdventHealth Translational Research Institute. CharacterIzation of Adult Onset Autoimmune Diabetes. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT03971955 (accessed on 27 December 2021).
- Weill Medical College of Cornell University. Epigenetic Contribution to the Pathogenesis of Diabetic Nephropathy in Qatari Population. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT02316522 (accessed on 27 December 2021).
- Stefanini, G. Coronary Artery Disease Progression in Patients with Acute Coronary Syndromes and Diabetes Mellitus. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT03890822 (accessed on 27 December 2021).
- Da Pereira, M.G. Contribution of Psychological Factors in the Healing of the Diabetic Foot Ulcer, in Physiological Indicators of Healing Prognosis and Quality of Life: A Randomized Longitudinal Trial with a Nested Qualitative Study of Effectiveness Assessment. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04698720 (accessed on 27 December 2021).
- Nassar, W.F. Phase 1 Study of the Effect of Cell-Free Cord Blood Derived Microvesicles on β-Cell Mass in Type 1 Diabetes Mellitus (T1DM) Patients. 2014. Available online: https://clinicaltrials.gov/ct2/show/NCT02138331 (accessed on 27 December 2021).
- Sardu, C. Epicardial Fat Evaluation to Predict Clinical Outcomes in Patients Affected by Coronary Artery Disease and Treated by Coronary Artery Bypass Grafting: Diabetic vs. Non Diabetic Patients, and Incretin Therapy Effect; The EPI.FAT.IN Study. 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT03360981 (accessed on 27 December 2021).
- University of South China. Prospective Cohort Study of Coronary Artery Calcification in Type 2 Diabetes Mellitus (USCAC Study). 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04889053 (accessed on 28 December 2021).
- Mandarino, L. PGC-1 & Muscle Mitochondrial Dysfunction in Diabetes: AIMS 1-4. 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT03323788 (accessed on 28 December 2021).
- Puder, J. Improving Cardio-Metabolic and Mental Health in Women with Gestational Diabetes Mellitus (GDM) and Their Offspring: MySweetHeart Trial. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT02890693 (accessed on 28 December 2021).
- Benincasa, G. Network-Based Epigenome-Wide AssociaTion Study in Obesity PrecisioN Medicine: NEWTON Clinical Trial. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT03903757 (accessed on 28 December 2021).
- Sardu, C. Effect of Inflammatory Axis and Sirtuins’ Expression in a Population of Overweight Pre-Diabetics Patients from Metabolic Homeostasis Towards the Adipogonesis, and to Cardiac Redomelling. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT03491241 (accessed on 28 December 2021).
- Boston University. Assessment of Dapagliflozin on Vascular Health in Patients with Type 2 Diabetes. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT05139914 (accessed on 28 December 2021).
- Liu, K. Prognostic Role of Serum Exosomal MiRNA and Its Function in Pathogenesis of Diabetic Retinopathy (DR). 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT03264976 (accessed on 28 December 2021).
- Marfella, R. Lipid Accumulation in Heart Transplant from Non-Diabetic Donors to Diabetic Recipients. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT03546062 (accessed on 28 December 2021).
- Prato, P.S.D. Precision Medicine for Preventing Type 2 Diabetes: A Step Forward (PRE-MED2). 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT05147961 (accessed on 28 December 2021).
- Assistance Publique-Hôpitaux de Paris. Effect of GLP-1 on Angiogenesis, Angiosafe Type 2 Diabetes Study 1. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT02686177 (accessed on 28 December 2021).
- Medical University of Bialystok. Polish Registry of Diabetes-PL: Polski Rejestr Diabetologiczny (PolReD). 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04657367 (accessed on 28 December 2021).
- Hofer, C.K. Optimierung der Kardioprotektion Durch Inhalative Anästhetika Eine Untersuchung Bei Patienten Mit Diabetes Mellitus Während off-Pump Herzchirurgie. 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT02407626 (accessed on 28 December 2021).
- Medical University of Bialystok. Searching for the New Mechanisms That Activate Brow Fat Tissue. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04787952 (accessed on 28 December 2021).
- Mandarino, L. PGC-1 & Muscle Mitochondrial Dysfunction in Diabetes. 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT02282423 (accessed on 28 December 2021).
- Espinosa, L.M. Efficacy of Oral Sodium Chloride vs Iv Sodium Chloride in the Prevention of Contrast Nephropathy in Outpatients. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT03476460 (accessed on 28 December 2021).
- University of Aarhus. Hormonal and Inflammatory Changes during Pregnancy in Women with Glucose Metabolic Disorders. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04617405 (accessed on 28 December 2021).
- Weill Cornell Medical College in Qatar. Kidney Disease in Type 2 Diabetes Mellitus: Biomarker Discovery and Novel Therapeutics. 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT02410005 (accessed on 28 December 2021).
- Alexandra, C. Search for Highly Specific Predictors of Response to Different Hypoglycemic Therapy for Cardiovascular Prognosis. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT03804411 (accessed on 28 December 2021).
- Irdam, G.A. The Outcomes of Intracavernosal Umbilical Cord Mesenchymal Stem Cells Implantation in Patients with Diabetic Erectile Dysfunction. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04972890 (accessed on 28 December 2021).
- Bhatwadekar, A. Biomarker of Diabetic Retinopathy. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT05079399 (accessed on 28 December 2021).
- Shi, X. The Effect of Circulating LncRNAs on Type 2 Diabetic Peripheral Neuropathy by Regulating MiR-146a. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04638556 (accessed on 28 December 2021).
- University of Aarhus. Mechanisms Behind Severe Insulin Resistance during Pregnancy in Women with Glucose Metabolic Disorders (SIR-MET). 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04924504 (accessed on 28 December 2021).
- Sen, C. Diabetic Foot Ulcer (DFU) Biofilm Infection and Recurrence. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT05172089 (accessed on 28 December 2021).
- Isidori, A.M. PRecisiOn MEdicine to Target Frailty of Endocrine-Metabolic Origin. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04856683 (accessed on 28 December 2021).
- Yale University. Bright 1 Bodies: Extending the Bright Bodies Weight Management Program to Adolescents with Type 1 Diabetes; clinicaltrials.gov, 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT02768987 (accessed on 30 May 2022).
- Mansoura University. Role of LncRNA H19 in the Regulation of IGF-1R Expression: A Possible Association between Type 2 Diabetes and Hepatocellular Carcinoma; clinicaltrials.gov, 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT04767750 (accessed on 30 May 2022).
- Mohany, K.M.; Al Rugaie, O.; Al-Wutayd, O.; Al-Nafeesah, A. Investigation of the Levels of Circulating MiR-29a, MiR-122, Sestrin 2 and Inflammatory Markers in Obese Children with/without Type 2 Diabetes: A Case Control Study. BMC Endocr. Disord. 2021, 21, 152. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-S.; Song, L.; Wang, J.; Wu, H.; Gu, G.; Sugi, Y.; Li, Z.; Wang, H. GRP94 Is an Essential Regulator of Pancreatic β-Cell Development, Mass, and Function in Male Mice. Endocrinology 2018, 159, 1062–1073. [Google Scholar] [CrossRef] [PubMed]


| Clinical Trial Number | Phase | Status | Number of Participants | Condition | Study Type | Biological Matrix | Intervention | Comments | References |
|---|---|---|---|---|---|---|---|---|---|
| NCT02843139 | - | Completed | 535 | Obesity | Observational/ Cross-sectional | Serum, whole blood | - | Identification of microRNAs (miRNAs) as potential biomarkers to distinguish obese children with diabetes risk in adulthood | [144] |
| NCT04176276 | - | Completed | 300 | Diabetic kidney disease | Observational/ Retrospective | Urine, serum | Not required | Pathogenesis of renal disorders and related miRNAs that include miR-25, miR-192, miR-194 and miR-204 | [145] |
| NCT01334684 | NA | Unknown | 100 | Type 2 diabetes (T2D) | Interventional | White blood cells (WBCs) | Metformin | Determination of efficacy of metformin by evaluating miRNA profiles in WBCs | [146] |
| NCT02459106 | - | Active, not recruiting | 100 | T2D | Observational/ Prospective | Muscle, adipose tissue | - | Determination of effect of miRNAs released from fat tissue on skeletal muscle and their contribution to insulin resistance | [147] |
| NCT03792607 | - | Unknown | 35 | T2D, Cardiovascular disease (CVD) | Observational/ Prospective | Peripheral blood, CD4+ and CD8+ T-cells | - | Prediction of development of CVD in T2D patients by studying the interactions between miRNAs and DNA methylation | [148] |
| NCT02383537 | - | Unknown | 150 | Pregnancy and Diabetes | Observational/ Prospective | Placenta, fat tissue | - | Effect of insulin resistance, fat tissue and miRNA during pregnancy on diabetes | [149] |
| NCT04891887 | NA | Completed | 163 | T2D | Interventional | - | Health education | Determination of risk factors including miRNA and diabetes for mild cognitive impairment | [150] |
| NCT01042301 | NA | Completed | 120 | Type 1 diabetes mellitus (T1D) | Interventional/ Non-randomized | Blood | Sampling of blood | Profiles of miRNAs determined as humoral and cellular biomarkers along with other biomarkers in T1D | [151] |
| NCT03472846 | IV | Active, not recruiting | 80 | Post-menopausal osteoporosis | Interventional/ Non-randomized | Serum | Teriparatide and Prolia (denosumab) | Levels of miRNA determined under osteoanabolic treatment and anti-resorptive treatment | [152] |
| NCT04011228 | - | Unknown | 224 | Prediabetes, Gestational diabetes | Observational/ Prospective | Saliva | Magnetic resonance spectroscopy | Characterizing miRNAs in pre-diabetics and evaluating miRNAs present in heart fat, liver fat and skeletal muscle | [153] |
| NCT03027726 | NA | Completed | 84 | Obese children with T2D | Interventional/ Randomized | Peripheral mononuclear cells | Psycho-educational program | Profiling miRNA in mononuclear cells and in circulating exosomes to detect diabetes risk in obese children | [154] |
| NCT03377335 | IV | Unknown | 186 | T2D | Interventional/ Randomized | Serum | Dapagliflozin and metformin | Determination of the effects of dapagliflozin treatment on miRNAs and cardiometabolic risk | [155] |
| NCT02861781 | - | Recruiting | 270 | Obesity, Insulin resistance, T2D | Observational/ Prospective | Plasma | Sampling of tissue and blood | Comparison and changes in levels of miRNA in plasma, adipose tissue, muscle and liver | [156] |
| NCT02768935 | NA | Unknown | 20 | Myocardial infarction | Interventional/ Non-randomized | Plasma, platelets | Sampling of blood | Identification of miRNAs responsible for promoting the differentiation of monocytes | [157] |
| NCT02245633 | - | Unknown | 70 | Diabetic kidney disease | Observational/ Cross-sectional | Plasma | - | Determining the contribution of miRNA expression and levels of vitamin D in DKD treatment | [158] |
| NCT04392557 | IV | Recruiting | 36 | T2D | Interventional/ Randomized | - | Metformin/alogliptin (oral), metformin/pioglitazone (pill), metformin/alogliptin/pioglitazone (triple therapy) | To compare anti-inflammatory status in atherosclerosis by evaluating inflammatory miRNA levels | [159] |
| NCT03682445 | NA | Completed | 60 | T2D | Interventional/ Randomized | Blood | Exercise | Effect of exercise on levels of miRNA-143 and other metabolic and physical parameters | [160] |
| NCT02011100 | NA | Completed | 28 | T2D, CVD, Metabolic diseases | Interventional/ Randomized | - | Carnosine | Effect of carnosine on risk factors of CVD and diabetes; miRNAs related to action of carnosine may provide more effective therapy | [161] |
| NCT03971955 | NA | Recruiting | 40 | Auto-immune diabetes | Observational/ Cross-sectional | Blood | Mixed meal | Changes in miRNA levels to characterize autoimmune diabetes | [162] |
| NCT02316522 | - | Active, not recruiting | 158 | Diabetic nephropathy | Observational/ Prospective | Urine, blood | - | Contribution of epigenetics in diabetic nephropathy | [163] |
| NCT03890822 | - | Recruiting | 100 | Coronary artery disease (CAD) | Observational/ Prospective | Blood | - | Role of inflammatory mediators and circulating miRNAs in diabetic patients along with CAD progression | [164] |
| NCT04698720 | NA | Recruiting | 48 | Diabetic foot ulcer (DFU) | Interventional/ Randomized | Blood | Hypnosis and muscle relaxation with guided imagery | Effects of hypnosis and muscle-relaxation therapy and evaluation of physiological indicators, including miRNAs (miR-21 and miR-155), for healing DFU | [165] |
| NCT02138331 | II and III | Unknown | 20 | T1D | Interventional | Cord-blood-derived exosomes | Mesenchymal stem cell (MSC) exosomes | Effect of exosomal content, including pre-miRNA derived from MSC | [166] |
| NCT03360981 | IV | Unknown | 150 | CVD and diabetes | Interventional/ Randomized | Serum, epicardial fat | Incretins | Evaluation of epicardial fat for prediction of clinical outcomes of CVD-affected patients | [167] |
| NCT04889053 | - | Recruiting | 1400 | T2D, Vascular calcification (VC), Coronary artery calcification (CAC) | Observational/ Prospective | Serum | - | Change in levels of miR-32 for early diagnosis of VC (prognostic biomarker) | [168] |
| NCT03323788 | - | Unknown | 96 | Obesity | Observational/ Prospective | Muscle | Exercise | Increased expression of miR-128, miR-378, miR-10a, miR-422a and miR-30 family; decreased expression of miR-532 | [169] |
| NCT02890693 | NA | Recruiting | 200 | Gestational diabetes | Interventional/ Randomized | Cord blood | Psychosocial therapy | To improve mental health and cardio-metabolic health by evaluating variables, including miRNA | [170] |
| NCT03903757 | - | Unknown | 100 | Obesity and T2D | Observational/ Prospective | RNA sequencing | RNA sequencing | Identifying and comparing miRNA targets in obese and diabetic patients with controls | [171] |
| NCT03491241 | NA | Completed | 60 | Pre-diabetes and obesity | Interventional/ Randomized | Tissue biopsy | Hypocaloric diet | Effect of sirtuins and inflammatory axis expression; downregulation of miR-27 and miR-195 | [172] |
| NCT05139914 | IV | Not yet recruiting | 50 | Endothelial dysfunction | Interventional/ Randomized | Plasma | Dapagliflozin and placebo | Effect of miRNA and other biomarkers on T2D patients and vascular health | [173] |
| NCT03264976 | - | Not yet recruiting | 200 | Diabetic retinopathy (DR) | Observational/ Prospective | Serum | Diagnostic test | Role of exosomal miRNAs present in serum | [174] |
| NCT03546062 | - | Completed | 30 | T2D patients with heart transplant | Observational | Heart tissue | Heart transplant | Evaluation of miRNAs and inflammatory markers associated with cardiac disease progression | [175] |
| NCT05147961 | NA | Not yet recruiting | 300 | Pre-diabetes | Interventional/ Randomized | Lymphocytes, plasma | Standard care | Prediction of disease risk and determination of prevention measures using miRNAs as biomarkers | [176] |
| NCT02686177 | IV | Completed | 50 | T2D | Interventional/ Randomized | Progenitor cells | Liraglutide, Metformin or sulfonylurea | Effect of glucagon-like peptide receptor (GLP-1) on angiogenesis and AngiomiR-126 levels compared before and after treatment | [177] |
| NCT04657367 | - | Recruiting | 10,000 | Pre-diabetes, Diabetes, Dysglycemia, Obesity | Observational/ Prospective | Plasma | - | Plasma miRNAs assessed for diabetes registry | [178] |
| NCT02407626 | NA | Terminated | 2 | Myocardial ischemia | Interventional/ Randomized | - | Propofol and sevoflurane | Myocardial protection of diabetic patients while undergoing surgery and evaluation of miR-144, miR-125b and miR-208a | [179] |
| NCT04787952 | - | Completed | 40 | Obesity | Observational/ Prospective | Brown adipose tissue (BAT) | Cold exposure | Role of miRNA in activation of BAT | [180] |
| NCT02282423 | NA | Terminated | 26 | Obesity and Diabetes | Interventional/ Non-randomized | Muscle tissue | Instructions for diet and exercise | Muscle processed to isolate miRNA and mRNA for study | [181] |
| NCT03476460 | II | Completed | 269 | Kidney disease | Interventional/ Randomized | - | Sodium chloride (oral and intravenous) | Effect of sodium chloride to prevent contrast nephropathy; miRNAs and other biomarkers evaluated | [182] |
| NCT04617405 | - | Recruiting | 300 | Pregnancy and diabetes | Observational/ Prospective | Serum, plasma | No interventions | Changes in inflammatory and hormonal factors during pregnancy | [183] |
| NCT02410005 | II and III | Terminated | 56 | Diabetic nephropathy | Interventional/ Randomized | Urine | Calcitriol and Losartan | Discovery of biomarkers (miRNAs) for kidney disease in T2D | [184] |
| NCT03804411 | IV | Recruiting | 800 | T2D | Interventional/ Randomized | - | Standard treatment | Determination of genetic markers (miR-21, miR-27, miR-125 and miR-126) for prediction of response to hypoglycemic therapy | [185] |
| NCT04972890 | II and III | Recruiting | 12 | Erectile dysfunction with DM | Interventional/ Randomized | Mesenchymal stem cells (MSC) | Stem cells | Treating erectile dysfunction using umbilical cord MSC-derived miR-16 and miR-126 | [186] |
| NCT05079399 | - | Not yet recruiting | 192 | Diabetic retinopathy | Observational/ Prospective | Blood | Blood sample | Sequencing of miRNA from angiogenic cells | [187] |
| NCT04638556 | - | Recruiting | 300 | Peripheral neuropathy (PN) | Observational/ Cross-sectional | Plasma | Diagnostic test | Effect of lncRNAs on PN by regulation of miR-146a | [188] |
| NCT04924504 | - | Recruiting | 24 | Pregnancy and diabetes | Observational/ Prospective | Serum, plasma | No interventions | Profiling of miRNA isolated from exosomes and other proteins to determine insulin resistance | [189] |
| NCT05172089 | - | Recruiting | 420 | Diabetic foot ulcer (DFU) | Observational/ Prospective | DFU tissue | - | Determination of disruption of barrier function in the repair of DFU by miRNAs | [190] |
| NCT04856683 | - | Recruiting | 1100 | Frailty | Observational | Blood | - | Profiling of circulating miRNAs from patients suffering with gonadal and adrenal disease | [191] |
| NCT02768987 | NA | Completed | 50 | T1D | Interventional | - | Bright bodies (weight-management program) | Effect of physical activities on obese T1D patients | [192] |
| NCT04767750 | - | Completed | 101 | T2D, Cancer | Observational | Blood | Collection of blood sample | To study lncRNA H19’s role in regulating the expression of insulin-like growth factor 1 receptor (IGF-1R) | [193] |
| Sr. no. | ncRNA | Disease | Upregulated /Downregulated | References |
|---|---|---|---|---|
| 1 | H19 | Diabetes mellitus | Upregulated | [91] |
| 2 | H19 | Human dermal microvascular endothelial cells (HMEC-1) and human embryonic kidney cells 293 (HEK293) | Downregulated | [110] |
| 3 | HOTAIR | Non-proliferative diabetic retinopathy, proliferative diabetic retinopathy | Upregulated | [59] |
| 4 | HOTAIR | Type 2 diabetes | Upregulated | [51] |
| 5 | hsa-miR-155-5p | Type 1 diabetes | Downregulated | [35] |
| 6 | hsa-miR-155-5p | Type 1 diabetes | Upregulated | [33] |
| 7 | hsa-miR-17-5p | Human umbilical cord mesenchymal stem cells (hucMSCs) | Upregulated | [99] |
| 8 | hsa-miR-17-5p | Type 1 diabetes | Upregulated | [35] |
| 9 | hsa-miR-210-3p | Type 1 diabetes | Upregulated | [33] |
| 10 | hsa-miR-210-5p | Type 2 diabetes | Upregulated | [49] |
| 11 | hsa-miR-21-3p | Type 1 diabetes | Upregulated | [33] |
| 12 | hsa-miR-21-5p | Type 1 diabetes | Upregulated | [33] |
| 13 | hsa-miR-223-3p | Type 1 diabetes | Downregulated | [35] |
| 14 | hsa-miR-223-3p | Type 1 diabetes | Upregulated | [105] |
| 15 | hsa-miR-223-3p | Type 2 diabetes | Upregulated | [58] |
| 16 | hsa-miR-24-3p | Type 1 diabetes | Downregulated | [35] |
| 17 | hsa-miR-24-3p | Type 2 diabetes | Downregulated | [54] |
| 18 | hsa-miR-29a-3p | Type 1 diabetes | Downregulated | [35] |
| 19 | hsa-miR-29a-3p | Type 2 diabetes | Upregulated | [45] |
| 20 | hsa-miR-320 | Type 1 diabetes | Upregulated | [33] |
| 21 | hsa-miR-320 | Type 1 diabetes | Downregulated | [35] |
| 22 | hsa-miR-338-3p | Type 1 diabetes | Upregulated | [33] |
| 23 | hsa-miR-338-3p | Type 2 diabetes | Downregulated | [56] |
| 24 | hsa-miR-342 | Type 1 diabetes | Upregulated | [33] |
| 25 | hsa-miR-342 | Type 1 diabetes | Downregulated | [35] |
| 26 | hsa-miR-34a-5p | Human adult retinal pigment epithelial cells (ARPE-19) | Upregulated | [104] |
| 27 | hsa-miR-34a-5p | Type 2 diabetes patients, type 1 diabetes | Upregulated | [46] |
| 28 | hsa-miR-34a-5p | Type 2 diabetes | Downregulated | [45] |
| 29 | hsa-miR-486-3p | Type 1 diabetes | Upregulated | [35] |
| 30 | hsa-miR-486-5p | Type 1 diabetes | Downregulated | [35] |
| 31 | KCNQ1OT1 | Diabetic cataract | Upregulated | [103] |
| 32 | KCNQ1OT1 | Proliferative diabetic retinopathy, non-proliferative diabetic retinopathy | Upregulated | [116] |
| 33 | MALAT1 | Diabetic retinopathy | Upregulated | [102] |
| 34 | MALAT1 | HK-2 (human kidney 2) cells | Upregulated | [113] |
| 35 | MALAT1 | Human bone marrow-derived mesenchymal stem cells (MSCs) | Downregulated | [98] |
| 36 | MALAT1 | Non-proliferative diabetic retinopathy, proliferative diabetic retinopathy | Upregulated | [59] |
| 37 | MEG3 | Human adult retinal pigment epithelial cells (ARPE-19) | Downregulated | [104] |
| 38 | MEG3 | Gestational diabetes | Upregulated | [41] |
| Noncoding RNAs | Biological Functions | Reference |
|---|---|---|
| hsa-miR-155-5p | Negative correlation with glycated hemoglobin (HbA1c) levels | [33] |
| Negative correlation with insulin-dose-adjusted HbA1c (IDAA1c) levels | ||
| Regulation of Fas-associated protein with death domain (FADD) | ||
| Regulation of (C-X-C motif chemokine ligand 8/interleukin 8) CXCL8/IL8 | ||
| Targets phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) gene | ||
| hsa-miR-210-3p | Negative correlation with thyroid-stimulating hormone (TSH) levels | [33] |
| Negative correlation with insulin-dose-adjusted HbA1c (IDAA1c) levels | ||
| Targets protein tyrosine phosphatase non-receptor type 1 (PTPN1) gene participating in insulin signaling pathway | ||
| has-miR-210 | Linked with hypoxia pathway and upregulated in response to hypoxia-inducible factors (HIFs) | [49] |
| hsa-miR-29a | Negative correlation with serum sestrin2 levels | [194] |
| Suppresses IRS-1, leading to decreased uptake of glucose into the cell | ||
| hsa-miR-17-5p | Downregulates PTEN and activates AKT/HIF-1α/VEGF pathway to promote proliferation, migration and tube formation in HG-treated human umbilical vascular endothelial cells (HUVECs) | [195] |
| ANRIL GAS5 HOTAIR MALAT1 MEG3 | Positive correlation with senescence-related biomarkers (p16, p21, p53 and β-galactosidase) and pro-inflammatory biomarkers (IL1- β, MCP1, IL6 and TNF-α) | [51] |
| hsa-miR-34a | Decreased SIRT1 activity resulting in senescence of pancreatic β-cells | [47] |
| hsa-miR-223-3p | Targets HSP90P1 regulating GRP94 expression, which is essential for development of pancreatic β-cells | [196] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.; Ajgaonkar, S.; Jadhav, N.; Saha, P.; Gurav, P.; Panda, S.; Mehta, D.; Nair, S. Current Insights into miRNA and lncRNA Dysregulation in Diabetes: Signal Transduction, Clinical Trials and Biomarker Discovery. Pharmaceuticals 2022, 15, 1269. https://doi.org/10.3390/ph15101269
Pandey A, Ajgaonkar S, Jadhav N, Saha P, Gurav P, Panda S, Mehta D, Nair S. Current Insights into miRNA and lncRNA Dysregulation in Diabetes: Signal Transduction, Clinical Trials and Biomarker Discovery. Pharmaceuticals. 2022; 15(10):1269. https://doi.org/10.3390/ph15101269
Chicago/Turabian StylePandey, Amitkumar, Saiprasad Ajgaonkar, Nikita Jadhav, Praful Saha, Pranay Gurav, Sangita Panda, Dilip Mehta, and Sujit Nair. 2022. "Current Insights into miRNA and lncRNA Dysregulation in Diabetes: Signal Transduction, Clinical Trials and Biomarker Discovery" Pharmaceuticals 15, no. 10: 1269. https://doi.org/10.3390/ph15101269
APA StylePandey, A., Ajgaonkar, S., Jadhav, N., Saha, P., Gurav, P., Panda, S., Mehta, D., & Nair, S. (2022). Current Insights into miRNA and lncRNA Dysregulation in Diabetes: Signal Transduction, Clinical Trials and Biomarker Discovery. Pharmaceuticals, 15(10), 1269. https://doi.org/10.3390/ph15101269






