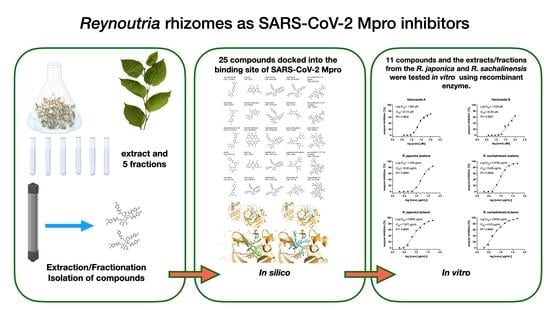
Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study
Abstract
:1. Introduction
2. Results
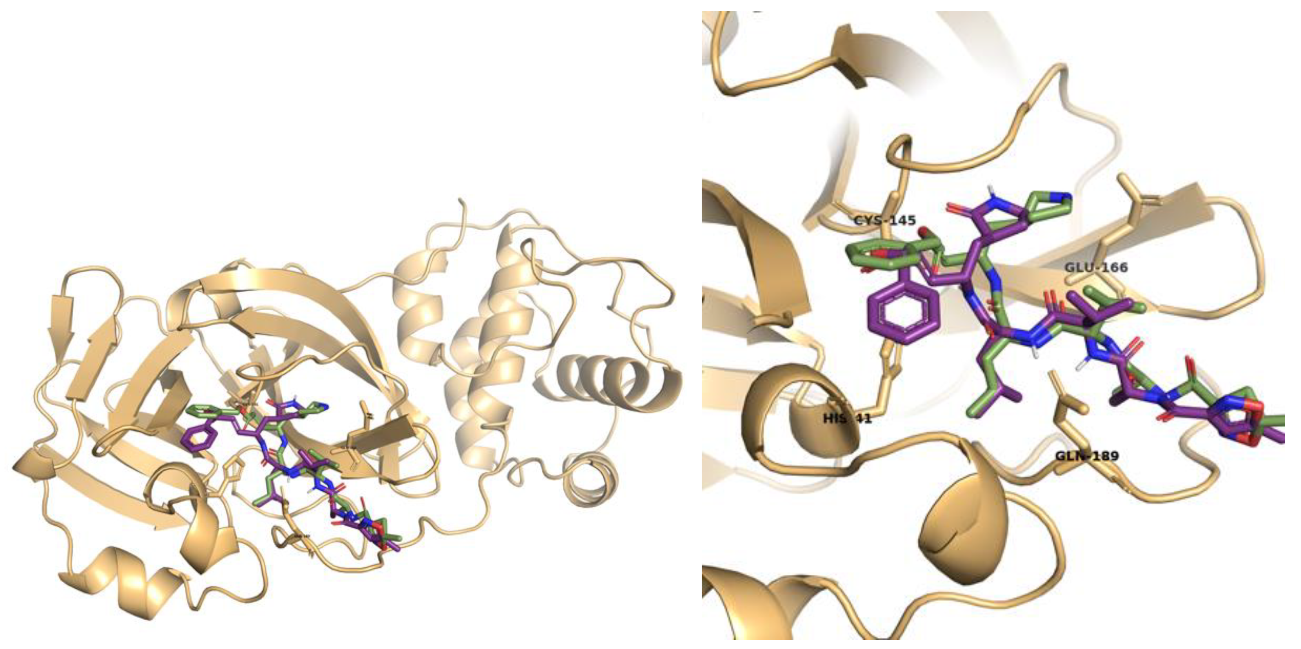
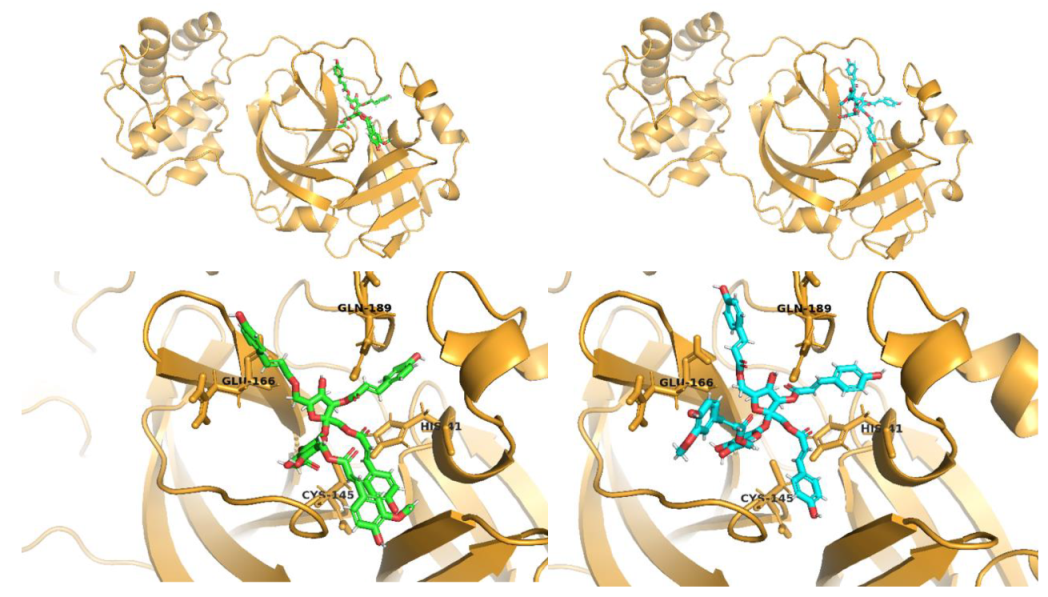
2.1. Molecular Docking Studies
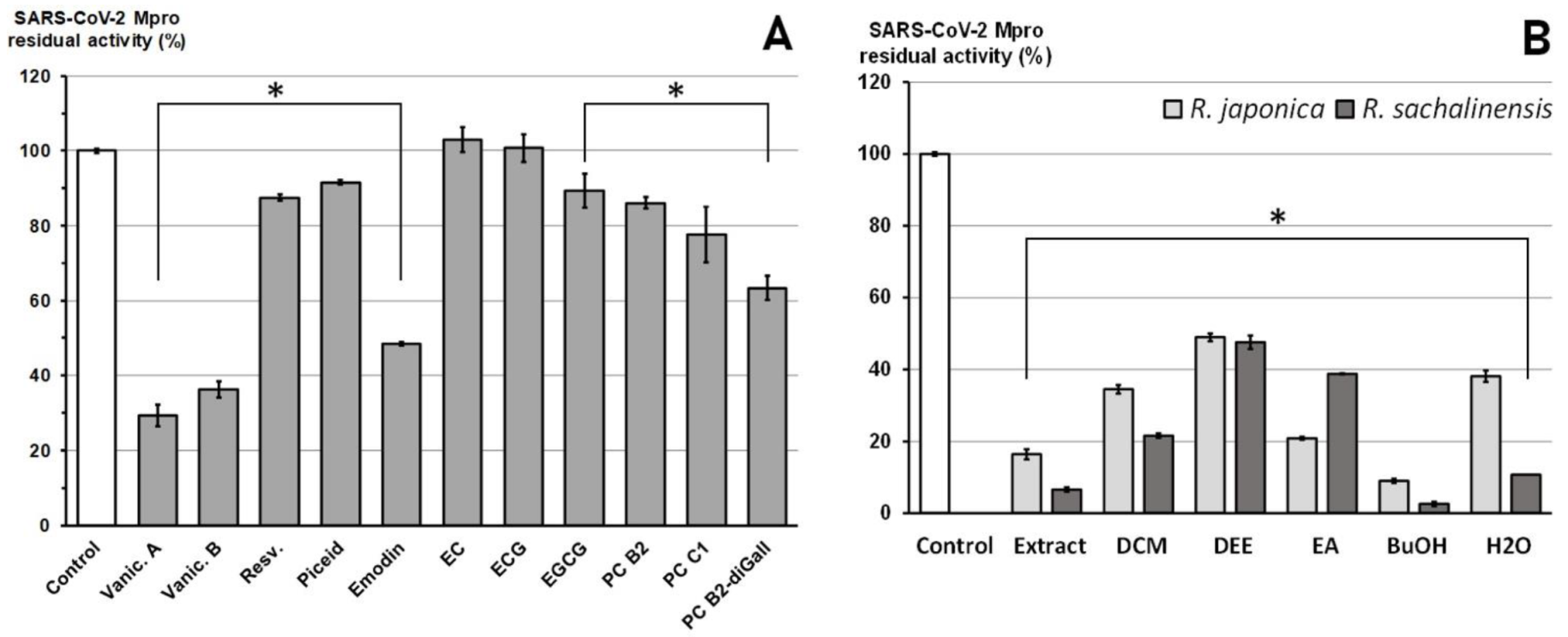
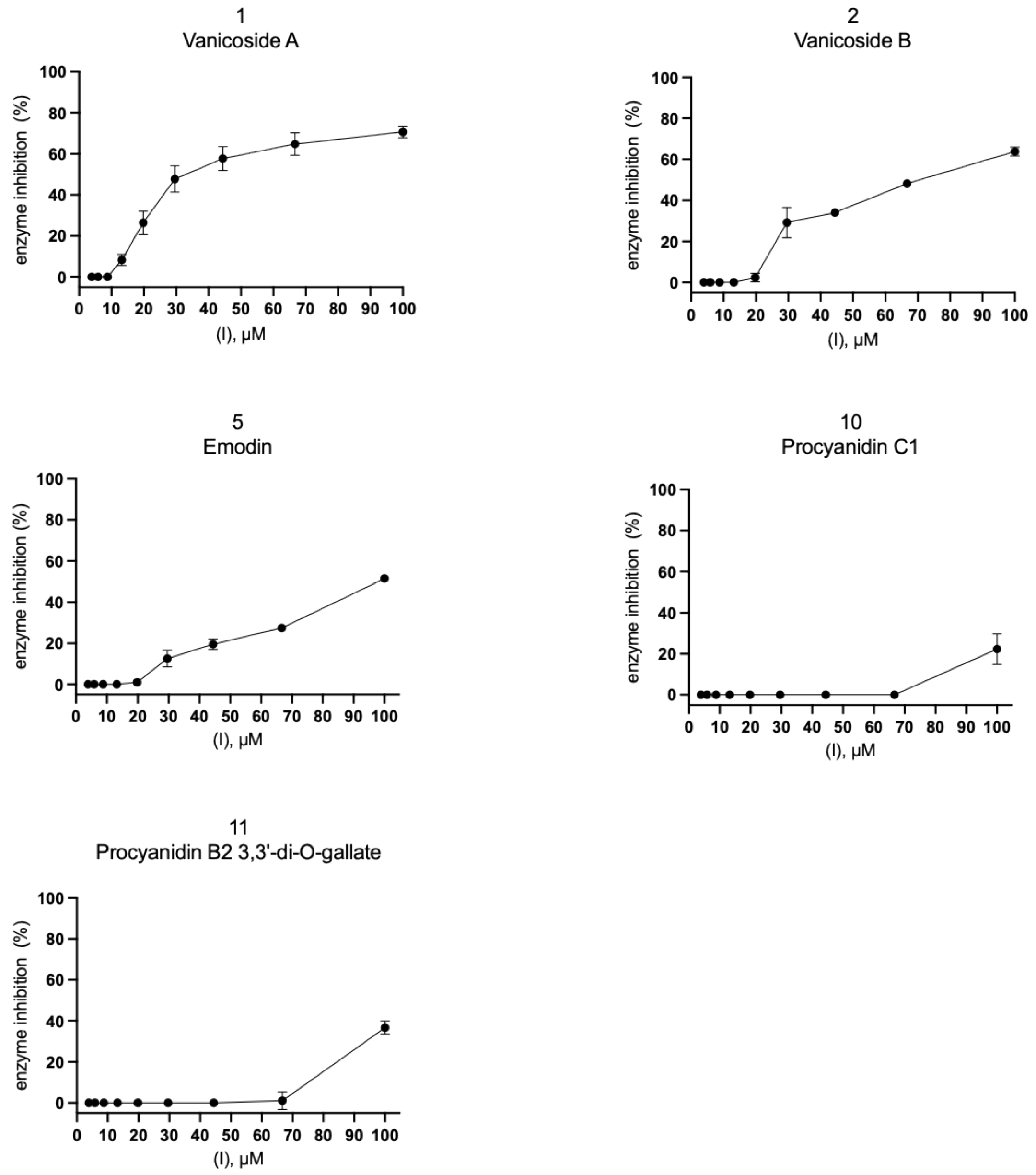
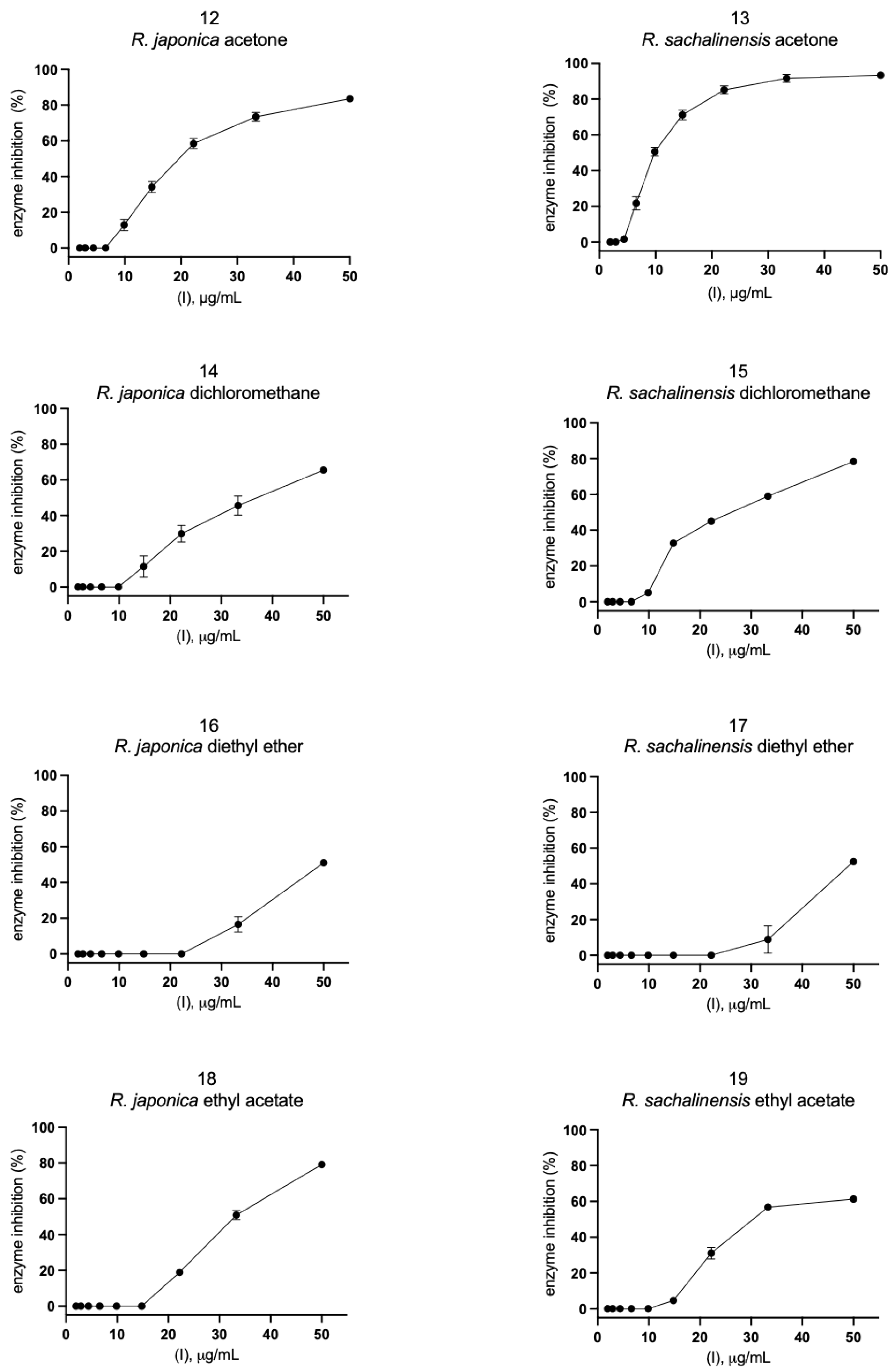
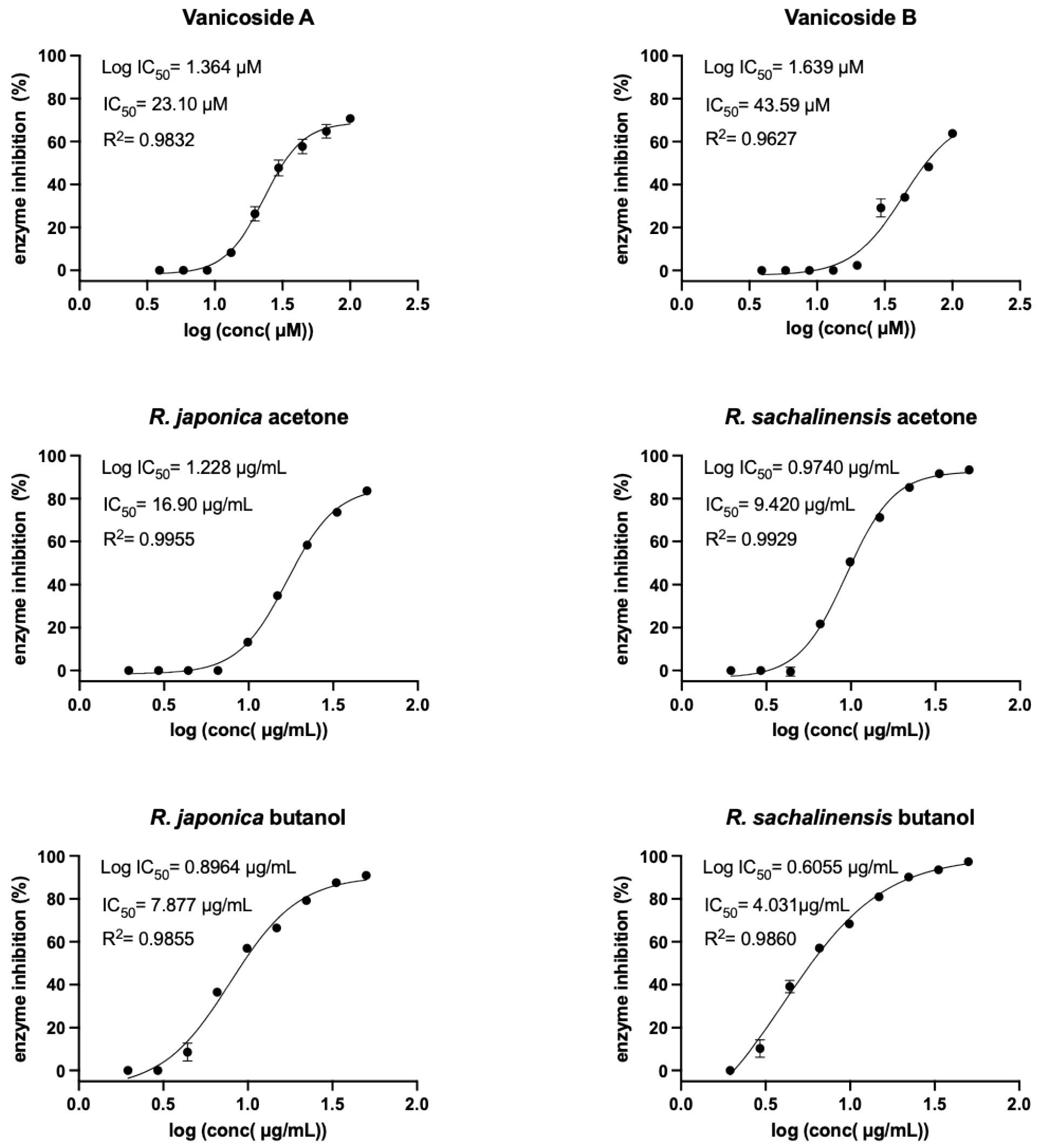
2.2. Inhibition of SARS-CoV-2 Mpro Enzyme-In Vitro Study
3. Discussion
4. Materials and Methods
4.1. Extracts and Fractions of Reynoutria Species
4.2. Compounds
4.3. Molecular Docking
4.4. Inhibition of SARS-CoV-2 Mpro Enzyme-In Vitro Study
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Valdes, A.M.; Freidin, M.B.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Ganesh, S.; Varsavsky, T.; Cardoso, M.J.; El-Sayed Moustafa, J.S.; et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020, 26, 1037–1040. [Google Scholar] [CrossRef]
- Pierron, D.; Pereda-Loth, V.; Mantel, M.; Moranges, M.; Bignon, E.; Alva, O.; Kabous, J.; Heiske, M.; Pacalon, J.; David, R.; et al. Smell and taste changes are early indicators of the COVID-19 pandemic and political decision effectiveness. Nat. Commun. 2020, 11, 5152. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, F.; Tang, J.; Nussinov, R.; Cheng, F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health 2020, 2, e667–e676. [Google Scholar] [CrossRef]
- Adeoye, A.O.; Oso, B.J.; Olaoye, I.F.; Tijjani, H.; Adebayo, A.I. Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. J. Biomol. Struct. Dyn. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Song, X.; Shi, J.; Wang, W.; He, K. Transcriptome-based drug repositioning for coronavirus disease 2019 (COVID-19). Pathog. Dis. 2020, 78. [Google Scholar] [CrossRef]
- Khan, A.; Ali, S.S.; Khan, M.T.; Saleem, S.; Ali, A.; Suleman, M.; Babar, Z.; Shafiq, A.; Khan, M.; Wei, D.-Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro). J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef]
- Available online: https://go.drugbank.com/covid-19 (accessed on 10 June 2021).
- Ton, A.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 2020, 39, 2000028. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Abel, R.; Paredes Ramos, M.; Chen, Q.; Pérez-Sánchez, H.; Coluzzi, F.; Rocco, M.; Marchetti, P.; Mura, C.; Simmaco, M.; Bourne, P.E.; et al. Computational Prediction of Potential Inhibitors of the Main Protease of SARS-CoV-2. Front. Chem. 2020, 8, 1–19. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Rut, W.; Groborz, K.; Zhang, L.; Sun, X.; Zmudzinski, M.; Pawlik, B.; Wang, X.; Jochmans, D.; Neyts, J.; Młynarski, W.; et al. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2020, 17, 222–228. [Google Scholar] [CrossRef]
- Vicidomini, C.; Roviello, V.; Roviello, G.N. Molecular Basis of the Therapeutical Potential of Clove (Syzygium aromaticum L.) and Clues to Its Anti-COVID-19 Utility. Molecules 2021, 26, 1880. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Chen, C.; Zhang, H.-Q.; Guo, H.-Y.; Wang, H.; Wang, L.; Zhang, X.; Hua, S.-N.; Yu, J.; Xiao, P.-G.; et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005, 67, 18–23. [Google Scholar] [CrossRef]
- Vicidomini, C.; Roviello, V.; Roviello, G.N. In Silico Investigation on the Interaction of Chiral Phytochemicals from Opuntia ficus-indica with SARS-CoV-2 Mpro. Symmetry 2021, 13, 1041. [Google Scholar] [CrossRef]
- Dwarka, D.; Agoni, C.; Mellem, J.J.; Soliman, M.E.; Baijnath, H. Identification of potential SARS-CoV-2 inhibitors from South African medicinal plant extracts using molecular modelling approaches. S. Afr. J. Bot. 2020, 133, 273–284. [Google Scholar] [CrossRef]
- Verma, S.; Twilley, D.; Esmear, T.; Oosthuizen, C.B.; Reid, A.-M.; Nel, M.; Lall, N. Anti-SARS-CoV Natural Products With the Potential to Inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 2020, 11, 1514. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.A.; Nagarajan, S.K.; Ramesh, V.; Palaniyandi, V.; Selvam, S.P.; Madhavan, T. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J. Mol. Struct. 2020, 1221, 128823. [Google Scholar] [CrossRef]
- Shree, P.; Mishra, P.; Selvaraj, C.; Singh, S.K.; Chaube, R.; Garg, N.; Tripathi, Y.B. Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants—Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)—A molecular docking study. J. Biomol. Struct. Dyn. 2020, 1–14. [Google Scholar] [CrossRef]
- Peng, W.; Qin, R.; Li, X.; Zhou, H. Botany, phytochemistry, pharmacology, and potential application of Polygonum cuspidatum Sieb.et Zucc.: A review. J. Ethnopharmacol. 2013, 148, 729–745. [Google Scholar] [CrossRef]
- Tao, Z.; Gao, J.; Zhang, G.; Xue, M.; Yang, W.; Tong, C.; Yuan, Y. Shufeng Jiedu Capsule protect against acute lung injury by suppressing the MAPK/NF-κB pathway. Biosci. Trends 2014, 8, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Tao, Z.; Meng, X.; Han, Y.Q.; Xue, M.M.; Wu, S.; Wu, P.; Yuan, Y.; Zhu, Q.; Zhang, T.J.; Wong, C.C.L. Therapeutic Mechanistic Studies of ShuFengJieDu Capsule in an Acute Lung Injury Animal Model Using Quantitative Proteomics Technology. J. Proteome Res. 2017, 16, 4009–4019. [Google Scholar] [CrossRef]
- Luo, L.; Jiang, J.; Wang, C.; Fitzgerald, M.; Hu, W.; Zhou, Y.; Zhang, H.; Chen, S. Analysis on herbal medicines utilized for treatment of COVID-19. Acta Pharm. Sin. B 2020, 10, 1192–1204. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, X.; Lu, Y.; Chen, F.; Zhang, W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci. Trends 2020, 14, 64–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahedi, H.M.; Ahmad, S.; Abbasi, S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 3225–3234. [Google Scholar] [CrossRef]
- Ho, T.-Y.; Wu, S.-L.; Chen, J.-C.; Li, C.-C.; Hsiang, C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007, 74, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Muchtaridi, M.; Fauzi, M.; Ikram, N.K.K.; Gazzali, A.M.; Wahab, H.A. Natural Flavonoids as Potential Angiotensin-Converting Enzyme 2 Inhibitors for Anti-SARS-CoV-2. Molecules 2020, 25, 3980. [Google Scholar] [CrossRef] [PubMed]
- Maroli, N.; Bhasuran, B.; Natarajan, J.; Kolandaivel, P. The potential role of procyanidin as a therapeutic agent against SARS-CoV-2: A text mining, molecular docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef]
- Nawrot-Hadzik, I.; Granica, S.; Domaradzki, K.; Pecio, Ł.; Matkowski, A. Isolation and Determination of Phenolic Glycosides and Anthraquinones from Rhizomes of Various Reynoutria Species. Planta Med. 2018, 84, 1118–1126. [Google Scholar] [CrossRef] [Green Version]
- Nawrot-Hadzik, I.; Slusarczyk, S.; Granica, S.; Hadzik, J.; Matkowski, A. Phytochemical diversity in rhizomes of three Reynoutria species and their antioxidant activity correlations elucidated by LC-ESI-MS/MS analysis. Molecules 2019, 24, 1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot-Hadzik, I.; Choromańska, A.; Abel, R.; Preissner, R.; Saczko, J.; Matkowski, A.; Hadzik, J. Cytotoxic effect of vanicosides a and b from reynoutria sachalinensis against melanotic and amelanotic melanoma cell lines and in silico evaluation for inhibition of brafv600e and mek1. Int. J. Mol. Sci. 2020, 21, 4611. [Google Scholar] [CrossRef]
- Chen, X.Y.; Wang, R.F.; Liu, B. An update on oligosaccharides and their esters from traditional Chinese medicines: Chemical structures and biological activities. Evid. Based Complement. Altern. Med. 2015, 2015, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, J.I.; Actis-Goretta, L.; Villordo, J.J.; Fraga, C.G. Procyanidin structure defines the extent and specificity of angiotensin I converting enzyme inhibition. Biochimie 2006, 88, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Kilmister, R.L.; Faulkner, P.; Downey, M.O.; Darby, S.J.; Falconer, R.J. The complexity of condensed tannin binding to bovine serum albumin—An isothermal titration calorimetry study. Food Chem. 2016, 190, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Iahtisham-Ul-Haq; Patel, S.; Pan, X.; Naz, S.; Sanches Silva, A.; Saeed, F.; Rasul Suleria, H.A. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Zhuang, M.; Jiang, H.; Suzuki, Y.; Li, X.; Xiao, P.; Tanaka, T.; Ling, H.; Yang, B.; Saitoh, H.; Zhang, L.; et al. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antiviral Res. 2009, 82, 73–81. [Google Scholar] [CrossRef]
- Conzelmann, C.; Weil, T.; Gross, R.; Jungke, P.; Frank, B.; Eggers, M.; Mueller, J.A.; Muench, J. Antiviral activity of plant juices and green tea against SARS-CoV-2 and influenza virus in vitro. bioRxiv 2020. [Google Scholar] [CrossRef]
- Sidor, A.; Gramza-Michałowska, A. Black Chokeberry Aronia Melanocarpa L.—A Qualitative Composition, Phenolic Profile and Antioxidant Potential. Molecules 2019, 24, 3710. [Google Scholar] [CrossRef] [Green Version]
- Derksen, A.; Kühn, J.; Hafezi, W.; Sendker, J.; Ehrhardt, C.; Ludwig, S.; Hensel, A. Antiviral activity of hydroalcoholic extract from Eupatorium perfoliatum L. Against the attachment of influenza A virus. J. Ethnopharmacol. 2016, 188, 144–152. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, J.W.; Eun, Y.G.; Lee, K.H.; Yeo, S.G.; Kim, S.W. Pretreatment with a grape seed proanthocyanidin extract downregulates proinflammatory cytokine expression in airway epithelial cells infected with respiratory syncytial virus. Mol. Med. Rep. 2019, 19, 3330–3336. [Google Scholar] [CrossRef] [Green Version]
- Derksen, A.; Hensel, A.; Hafezi, W.; Herrmann, F.; Schmidt, T.J.; Ehrhardt, C.; Ludwig, S.; Kühn, J. 3-O-galloylated procyanidins from Rumex acetosa L. inhibit the attachment of influenza A virus. PLoS ONE 2014, 9, e110089. [Google Scholar] [CrossRef]
- Gescher, K.; Hensel, A.; Hafezi, W.; Derksen, A.; Kühn, J. Oligomeric proanthocyanidins from Rumex acetosa L. inhibit the attachment of herpes simplex virus type-1. Antiviral Res. 2011, 89, 9–18. [Google Scholar] [CrossRef]
- Tsukuda, S.; Watashi, K.; Hojima, T.; Isogawa, M.; Iwamoto, M.; Omagari, K.; Suzuki, R.; Aizaki, H.; Kojima, S.; Sugiyama, M.; et al. A new class of hepatitis B and D virus entry inhibitors, proanthocyanidin and its analogs, that directly act on the viral large surface proteins. Hepatology 2017, 65, 1104–1116. [Google Scholar] [CrossRef] [Green Version]
- Hensel, A.; Bauer, R.; Heinrich, M.; Spiegler, V.; Kayser, O.; Hempel, G.; Kraft, K. Challenges at the Time of COVID-19: Opportunities and Innovations in Antivirals from Nature. Planta Med. 2020, 86, 659–664. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Meng, Z. Immunomodulation for Severe COVID-19 Pneumonia: The State of the Art. Front. Immunol. 2020, 11, 577442. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Bai, C.; He, F.; Xie, Y.; Zhou, H. Review on the potential action mechanisms of Chinese medicines in treating Coronavirus Disease 2019 (COVID-19). Pharmacol. Res. 2020, 158, 104939. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking 1 1Edited by F. E. Cohen. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2019, 47, D1388–D1395. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego- Dassault Systèmes. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 15 January 2020).
- The PyMOL Molecular Graphics System. Available online: Citeulike-article-id:240061%5Cnhttp://www.pymol.org (accessed on 31 July 2020).
- Zmudzinski, M.; Rut, W.; Olech, K.; Granda, J.; Giurg, M.; Burda-Grabowska, M.; Zhang, L.; Sun, X.; Lv, Z.; Nayak, D.; et al. Ebselen derivatives are very potent dual inhibitors of SARS-CoV-2 proteases—PLpro and Mpro in in vitro studies. bioRxiv 2020. [Google Scholar] [CrossRef]


| Phenylpropanoid Disaccharide Esters | |
|---|---|
Vanicoside B Conventional Hydrogen Bond: Cys145, Gln189, His164, Asn142, Leu141, Tyr54, Cys44; Pi-interactions: His41, Met165 | Vanicoside A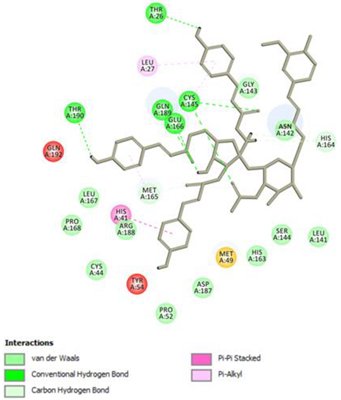 Conventional Hydrogen Bond: Cys145, Glu166, Gln189, Thr190, Thr26; Pi-interactions: His41, Leu27 |
| Procyanidins | |
Procyanidin C1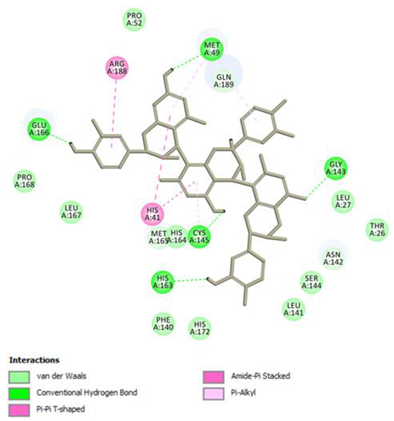 Conventional Hydrogen Bond: Cys145, Met49, Glu166, Gly143, His163; Pi-interactions: His41, Arg188, Met49 | Procyanidin B2 3,3’-di-O-gallate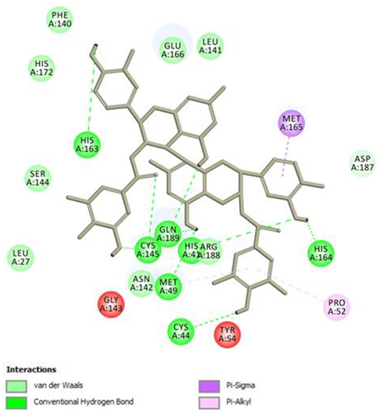 Conventional Hydrogen Bond: His41, Cys145, His163, His164, Cys44, Met49, Gln189; Pi-interactions: Pro52, Met165 |
| Anthraquinone | |
Emodin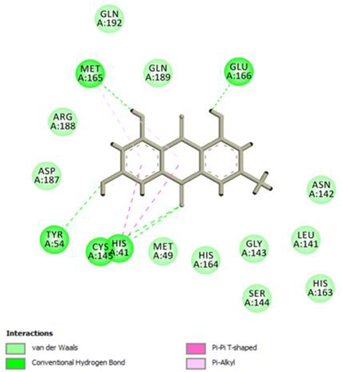 Conventional Hydrogen Bond: Cys145, Glu166, Met165, Tyr54, His41; Pi-interactions: His41; Met165 | |
| Stilbenes | |
|---|---|
Resveratrol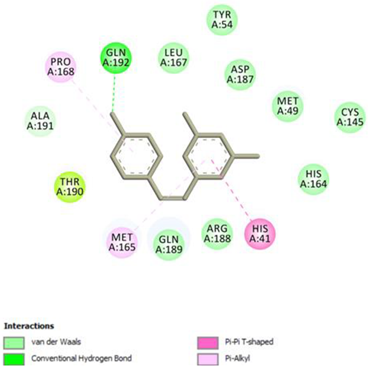 Conventional Hydrogen Bond: Gln 192; Pi-interactions: His41, Pro168, Met165 | Piceid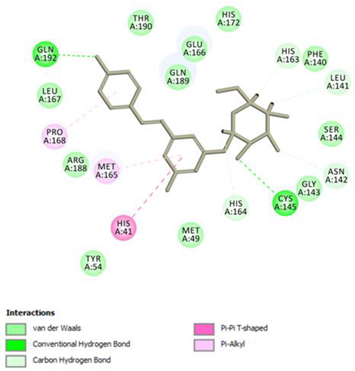 Conventional Hydrogen Bond: Cys145, Gln192; Pi-interactions: His41, Met165, Pro168 |
| Flavanols and Procyanidins | |
(−)-Epigallocatechin gallate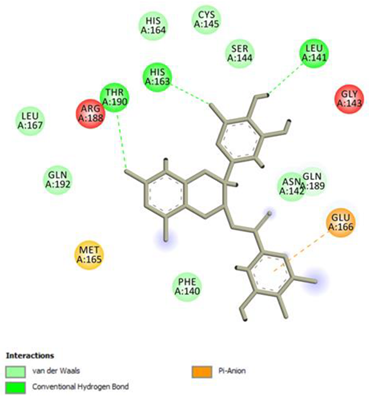 Conventional Hydrogen Bond: Thr190, Leu141, His163; Pi-interactions: Glu166 | Epicatechin Conventional Hydrogen Bond: Ser144, Glu166, Gly143, His164, Thr190; Pi-interactions: Met165, His163, Cys145 |
Epicatechin gallate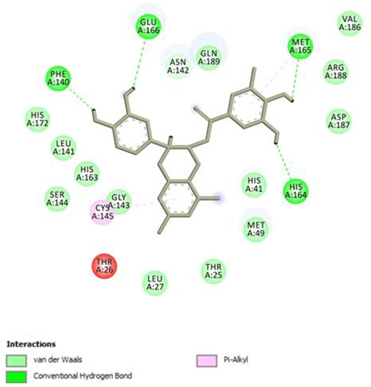 Conventional Hydrogen Bond: Met165, His164, Glu166, Phe140; Pi-interactions: Cys145, Met165 | Procyanidin B2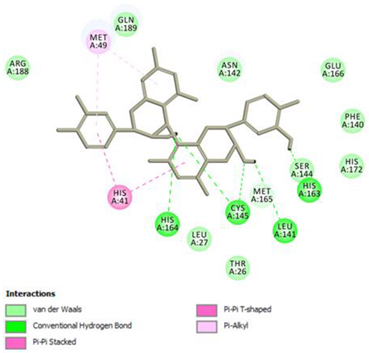 Conventional Hydrogen Bond: Cys145, Leu141, His163, His164; Pi-interactions: His41, Met49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawrot-Hadzik, I.; Zmudzinski, M.; Matkowski, A.; Preissner, R.; Kęsik-Brodacka, M.; Hadzik, J.; Drag, M.; Abel, R. Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study. Pharmaceuticals 2021, 14, 742. https://doi.org/10.3390/ph14080742
Nawrot-Hadzik I, Zmudzinski M, Matkowski A, Preissner R, Kęsik-Brodacka M, Hadzik J, Drag M, Abel R. Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study. Pharmaceuticals. 2021; 14(8):742. https://doi.org/10.3390/ph14080742
Chicago/Turabian StyleNawrot-Hadzik, Izabela, Mikolaj Zmudzinski, Adam Matkowski, Robert Preissner, Małgorzata Kęsik-Brodacka, Jakub Hadzik, Marcin Drag, and Renata Abel. 2021. "Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study" Pharmaceuticals 14, no. 8: 742. https://doi.org/10.3390/ph14080742
APA StyleNawrot-Hadzik, I., Zmudzinski, M., Matkowski, A., Preissner, R., Kęsik-Brodacka, M., Hadzik, J., Drag, M., & Abel, R. (2021). Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study. Pharmaceuticals, 14(8), 742. https://doi.org/10.3390/ph14080742