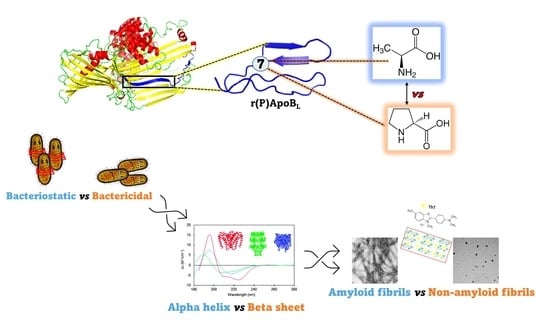
Impact of a Single Point Mutation on the Antimicrobial and Fibrillogenic Properties of Cryptides from Human Apolipoprotein B
Abstract
1. Introduction
2. Results
2.1. Design of Pro → Ala Mutation in Position 7 of r(P)ApoBL Peptide
2.2. Evaluation of the Effects of Pro → Ala Single Point Mutation on the Antimicrobial Activity of r(P)ApoBL Peptide
2.3. Evaluation of the Effects of Pro → Ala Substitution on the Anti-Biofilm Properties of r(P)ApoBL Peptide
2.4. Evaluation of the Effects of Pro → Ala Single Point Mutation on the Kinetics of Bacterial Killing
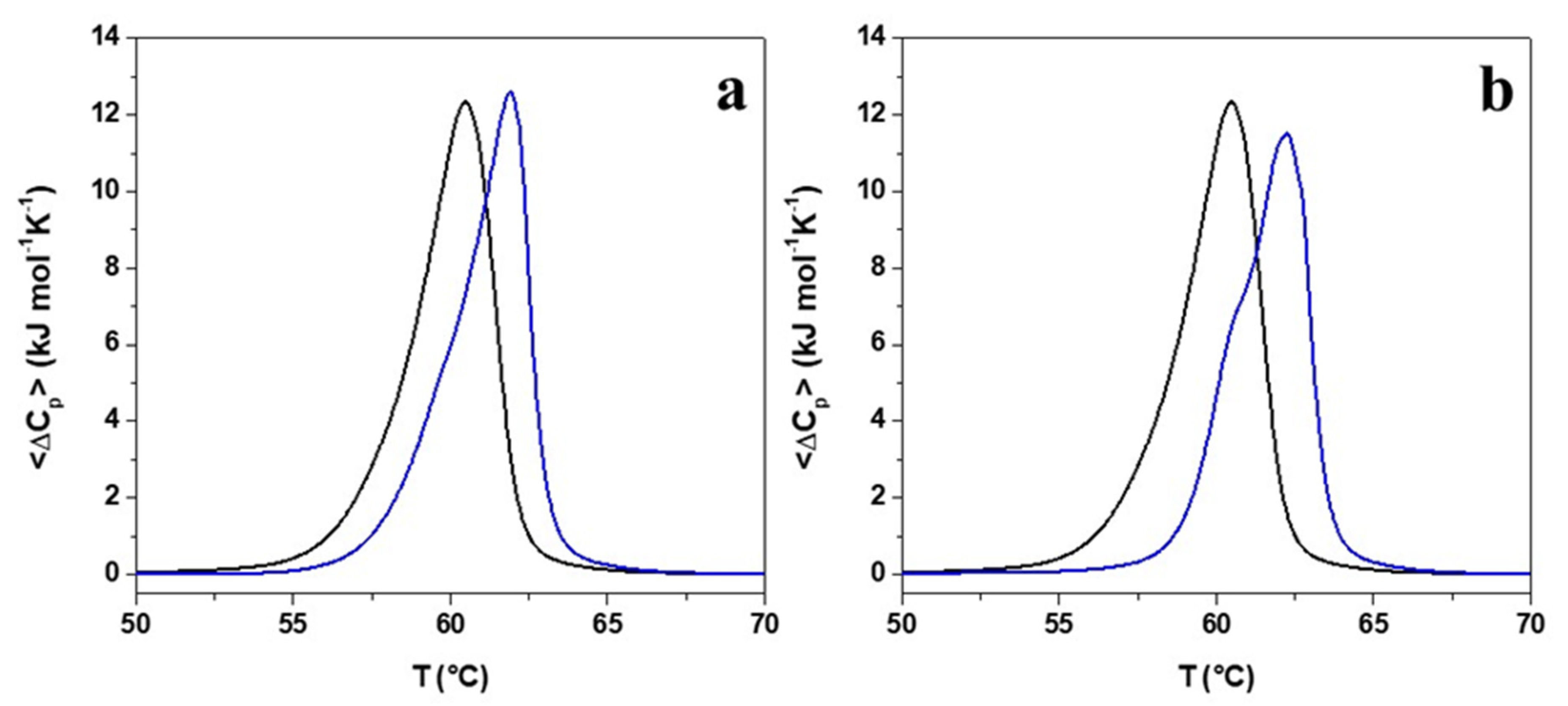
2.5. Evaluation of the Effects of Pro → Ala Single Point Mutation on Peptide Interaction with Double Lipid Layer by Differential Scanning Calorimetry (DSC) Analyses
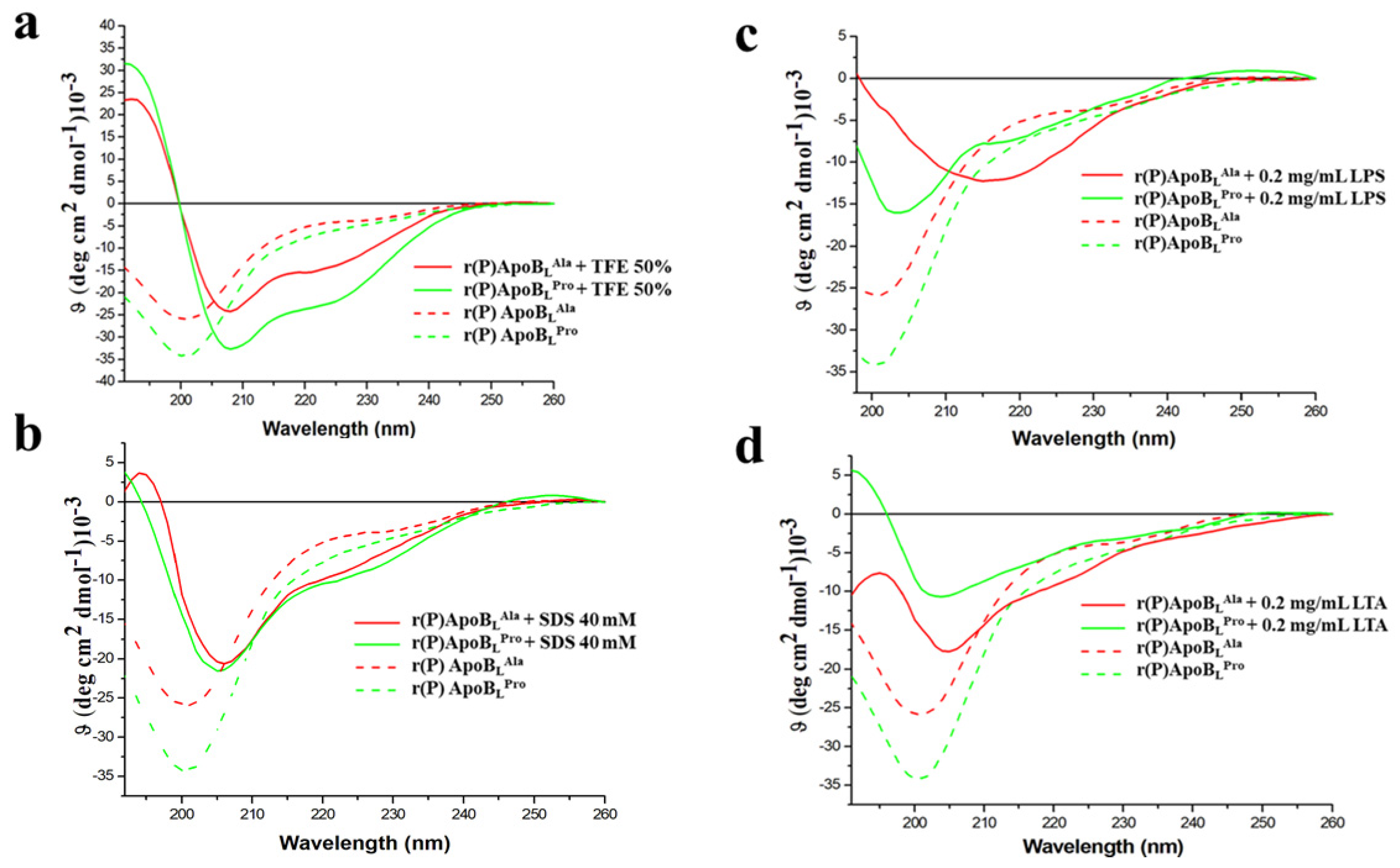
2.6. Evaluation of the Effects of Pro → Ala Single Point Mutation on r(P)ApoBL Conformation by Far-UV Circular Dichroism Analyses
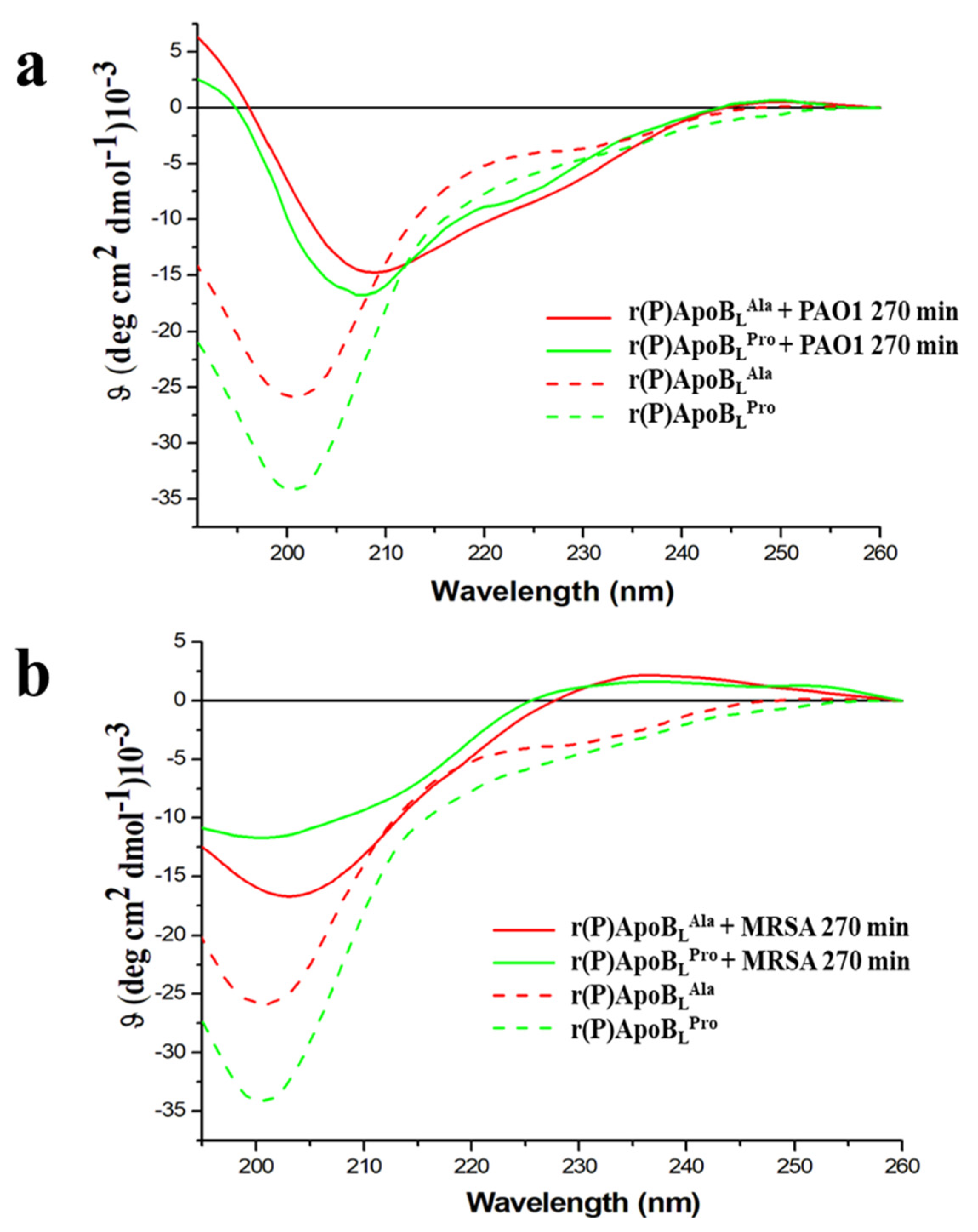
2.7. Conformational Analyses of r(P)ApoBLPro or r(P)ApoBLAla Peptides in the Presence of Susceptible Bacterial Cells
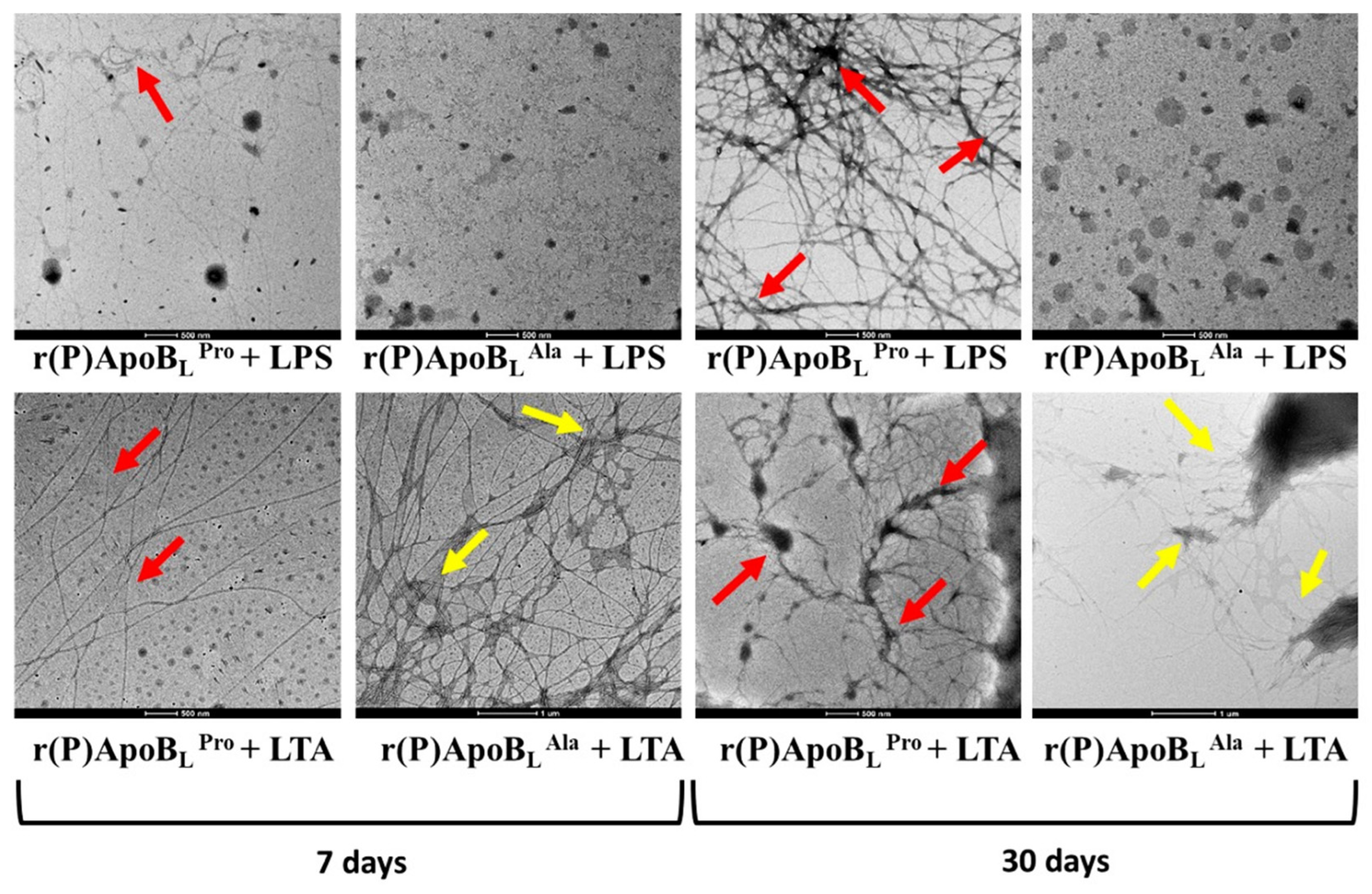
2.8. ApoB-Derived Peptides Self-Assembly
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. Production of Recombinant Peptides
5.3. Liposomes Preparation
5.4. Differential Scanning Calorimetry (DSC)
5.5. Bacterial Strains and Growth Conditions
5.6. Antimicrobial Activity Assays
5.7. Anti-Biofilm Activity
5.8. Circular Dichroism Spectroscopy
5.9. Killing Kinetic Studies
5.10. In Situ Real-Time ThT Fluorescence Assays
5.11. Statistical Analysis
5.12. Transmission Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spellberg, B.; Blaser, M.; Guidos, R.J.; Boucher, H.W.; Bradley, J.S.; Eisenstein, B.I.; Gerding, D.; Lynfield, R.; Reller, L.B.; Rex, J.; et al. Combating Antimicrobial Resistance: Policy Recommendations to Save Lives. Clin. Infect. Dis. 2011, 52, S397–S428. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Jain, A.; Goyal, M.; Singh, T.; Sood, H.; Malviya, H. Antibiotic Abuse during Endodontic Treatment: A Contributing Factor to Antibiotic Resistance. J. Fam. Med. Prim. Care 2019, 8, 3518. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic Resistance of Bacterial Biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Mishra, B.; Reiling, S.; Zarena, D.; Wang, G. Host Defense Antimicrobial Peptides as Antibiotics: Design and Application Strategies. Curr. Opin. Chem. Biol. 2017, 38, 87–96. [Google Scholar] [CrossRef]
- Phoenix, D.A.; Dennison, S.R.; Harris, F. Antimicrobial Peptides: Their History, Evolution, and Functional Promiscuity. In Antimicrobial Peptides; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; pp. 1–37. [Google Scholar]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef]
- Pletzer, D.; Hancock, R.E.W. Antibiofilm Peptides: Potential as Broad-Spectrum Agents. J. Bacteriol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef] [PubMed]
- Bosso, A.; Pirone, L.; Gaglione, R.; Pane, K.; del Gatto, A.; Zaccaro, L.; di Gaetano, S.; Diana, D.; Fattorusso, R.; Pedone, E.; et al. A New Cryptic Host Defense Peptide Identified in Human 11-Hydroxysteroid Dehydrogenase-1 β-like: From in Silico Identification to Experimental Evidence. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2342–2353. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Dell’Olmo, E.; Bosso, A.; Chino, M.; Pane, K.; Ascione, F.; Itri, F.; Caserta, S.; Amoresano, A.; Lombardi, A.; et al. Novel Human Bioactive Peptides Identified in Apolipoprotein B: Evaluation of Their Therapeutic Potential. Biochem. Pharmacol. 2017, 130, 34–50. [Google Scholar] [CrossRef]
- Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Della Ventura, B.; Casillo, A.; Di Girolamo, R.; Velotta, R.; Notomista, E.; Veldhuizen, E.J.A.; Corsaro, M.M.; et al. Effects of Human Antimicrobial Cryptides Identified in Apolipoprotein B Depend on Specific Features of Bacterial Strains. Sci. Rep. 2019, 9, 6728. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Cesaro, A.; Dell’olmo, E.; Di Girolamo, R.; Tartaglione, L.; Pizzo, E.; Arciello, A. Cryptides Identified in Human Apolipoprotein b as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. Int. J. Mol. Sci. 2020, 21, 2049. [Google Scholar] [CrossRef] [PubMed]
- Zanfardino, A.; Bosso, A.; Gallo, G.; Pistorio, V.; Di Napoli, M.; Gaglione, R.; Dell’Olmo, E.; Varcamonti, M.; Notomista, E.; Arciello, A.; et al. Human Apolipoprotein E as a Reservoir of Cryptic Bioactive Peptides: The Case of ApoE 133-167. J. Pept. Sci. 2018, 24, e3095. [Google Scholar] [CrossRef]
- Gaglione, R.; Pirone, L.; Farina, B.; Fusco, S.; Smaldone, G.; Aulitto, M.; Dell’Olmo, E.; Roscetto, E.; Del Gatto, A.; Fattorusso, R.; et al. Insights into the Anticancer Properties of the First Antimicrobial Peptide from Archaea. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2155–2164. [Google Scholar] [CrossRef]
- Matsumoto, K.; Kusaka, J.; Nishibori, A.; Hara, H. Lipid Domains in Bacterial Membranes. Mol. Microbiol. 2006, 61, 1110–1117. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Mahalka, A.K.; Kinnunen, P.K.J. Binding of Amphipathic α-Helical Antimicrobial Peptides to Lipid Membranes: Lessons from Temporins B and L. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Valle, J.; Nogués, M.V.; Boix, E.; Andreu, D. The Generation of Antimicrobial Peptide Activity: A Trade-off between Charge and Aggregation? Angew. Chem. Int. Ed. 2011, 50, 10686–10689. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.R.; Dobson, C.M. In Vitro Characterization of Lactoferrin Aggregation and Amyloid Formation †. Biochemistry 2003, 42, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Badia, M.; Moussaoui, M.; Sanchez, D.; Nogués, M.V.; Boix, E. Comparison of Human RNase 3 and RNase 7 Bactericidal Action at the Gram-Negative and Gram-Positive Bacterial Cell Wall. FEBS J. 2010, 277, 1713–1725. [Google Scholar] [CrossRef]
- Arispe, N.; Rojas, E.; Pollard, H.B. Alzheimer Disease Amyloid β Protein Forms Calcium Channels in Bilayer Membranes: Blockade by Tromethamine and Aluminum. Proc. Natl. Acad. Sci. USA 1993, 90, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Arispe, N.; Pollard, H.B.; Rojas, E. Zn2+ Interaction with Alzheimer Amyloid β Protein Calcium Channels. Proc. Natl. Acad. Sci. USA 1996, 90, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Bhatia, R.; Lal, R. Amyloid β Protein Forms Ion Channels: Implications for Alzheimer’s Disease Pathophysiology. FASEB J. 2001, 15, 2433–2444. [Google Scholar] [CrossRef]
- Quist, A.; Doudevski, I.; Lin, H.; Azimova, R.; Ng, D.; Frangione, B.; Kagan, B.; Ghiso, J.; Lal, R. Amyloid Ion Channels: A Common Structural Link for Protein-Misfolding Disease. Proc. Natl. Acad. Sci. USA 2005, 4, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Lashuel, H.A.; Hartley, D.; Petre, B.M.; Walz, T.; Lansbury, P.T. Neurodegenerative Disease: Amyloid Pores from Pathogenic Mutations. Nature 2002, 418, 291. [Google Scholar] [CrossRef]
- Jang, H.; Zheng, J.; Nussinov, R. Models of β-Amyloid Ion Channels in the Membrane Suggest That Channel Formation in the Bilayer Is a Dynamic Process. Biophys. J. 2007, 93, 1938–1949. [Google Scholar] [CrossRef]
- Jang, H.; Arce, F.T.; Ramachandran, S.; Capone, R.; Azimova, R.; Kagan, B.L.; Nussinov, R.; Lal, R. Truncated β-Amyloid Peptide Channels Provide an Alternative Mechanism for Alzheimer’s Disease and Down Syndrome. Proc. Natl. Acad. Sci. USA 2010, 107, 6538–6543. [Google Scholar] [CrossRef]
- Moir, R.D.; Lathe, R.; Tanzi, R.E. The Antimicrobial Protection Hypothesis of Alzheimer’s Disease. Alzheimer Dement. 2018, 14, 1602–1614. [Google Scholar] [CrossRef]
- Bishop, G.M.; Robinson, S.R. The Amyloid Hypothesis: Let Sleeping Dogmas Lie? Neurobiol. Aging 2002, 23, 1101–1105. [Google Scholar] [CrossRef]
- Robinson, S.R.; Bishop, G.M. Aβ as a Bioflocculant: Implications for the Amyloid Hypothesis of Alzheimer’s Disease. Neurobiol. Aging 2002, 23, 1051–1072. [Google Scholar] [CrossRef]
- Miklossy, J. Bacterial Amyloid and DNA Are Important Constituents of Senile Plaques: Further Evidence of the Spirochetal and Biofilm Nature of Senile Plaques. J. Alzheimer Dis. 2016, 53, 1459–1473. [Google Scholar] [CrossRef]
- Pulliam, L. HIV Regulation of Amyloid Beta Production. J. Neuroimmune Pharmacol. 2009, 4, 213–217. [Google Scholar] [CrossRef]
- Rempel, H.C.; Pulliam, L. HIV-1 Tat Inhibits Neprilysin and Elevates Amyloid β. AIDS 2005, 4, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Soscia, S.J.; Kirby, J.E.; Washicosky, K.J.; Tucker, S.M.; Ingelsson, M.; Hyman, B.; Burton, M.A.; Goldstein, L.E.; Duong, S.; Tanzi, R.E.; et al. The Alzheimer’s Disease-Associated Amyloid β-Protein Is an Antimicrobial Peptide. PLoS ONE 2010, 5, e9505. [Google Scholar] [CrossRef]
- Boix, E.; Salazar, V.A.; Torrent, M.; Pulido, D.; Nogués, M.V.; Moussaoui, M. Structural Determinants of the Eosinophil Cationic Protein Antimicrobial Activity. Biol. Chem. 2012, 393, 801–815. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Lin, Y.-M.; Chang, T.-W.; Wu, S.-J.; Lee, Y.-S.; Chang, M.D.-T.; Chen, C.; Wu, S.-H.; Liao, Y.-D. The Flexible and Clustered Lysine Residues of Human Ribonuclease 7 Are Critical for Membrane Permeability and Antimicrobial Activity. J. Biol. Chem. 2007, 282, 4626–4633. [Google Scholar] [CrossRef] [PubMed]
- Samir, P.; Link, A.J. Analyzing the Cryptome: Uncovering Secret Sequences. AAPS J. 2011, 13, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Pizzo, E.; Notomista, E.; de la Fuente-Nunez, C.; Arciello, A. Host Defence Cryptides from Human Apolipoproteins: Applications in Medicinal Chemistry. Curr. Top. Med. Chem. 2020, 20, 1324–1337. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.A.; Harrison, I.; McKnight, Á.; Dobson, C.B. Anti-Infective Activity of Apolipoprotein Domain Derived Peptides in Vitro: Identification of Novel Antimicrobial Peptides Related to Apolipoprotein B with Anti-HIV Activity. BMC Immunol. 2010, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.B.; Sales, S.D.; Hoggard, P.; Wozniak, M.A.; Crutcher, K.A. The Receptor-Binding Region of Human Apolipoprotein E Has Direct Anti-Infective Activity. J. Infect. Dis. 2006, 193, 442–450. [Google Scholar] [CrossRef]
- Arciello, A. Host Defence Peptides in Medicinal Chemistry: Identification, Engineering, Characterization and Beyond. Curr. Top. Med. Chem. 2020, 20, 1235–1237. [Google Scholar] [CrossRef]
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a Histone-Derived Antimicrobial Peptide in Atlantic Halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta Proteins Proteom. 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Groß, R.; Bauer, R.; Krüger, F.; Rücker-Braun, E.; Olari, L.R.; Ständker, L.; Preising, N.; Rodríguez, A.A.; Conzelmann, C.; Gerbl, F.; et al. A Placenta Derived C-Terminal Fragment of β-Hemoglobin with Combined Antibacterial and Antiviral Activity. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Kohn, E.M.; Shirley, D.J.; Arotsky, L.; Picciano, A.M.; Ridgway, Z.; Urban, M.W.; Carone, B.R.; Caputo, G.A. Role of Cationic Side Chains in the Antimicrobial Activity of C18G. Molecules 2018, 23, 329. [Google Scholar] [CrossRef]
- Athira, P.P.; Anju, M.V.; Anooja, V.V.; Archana, K.; Neelima, S.; Rosamma, P. A Histone H2A-Derived Antimicrobial Peptide, Hipposin from Mangrove Whip Ray, Himantura Walga: Molecular and Functional Characterisation. 3 Biotech 2020, 10. [Google Scholar] [CrossRef]
- Dell’Olmo, E.; Gaglione, R.; Cesaro, A.; Cafaro, V.; Teertstra, W.R.; de Cock, H.; Notomista, E.; Haagsman, H.P.; Veldhuizen, E.J.A.; Arciello, A. Host Defence Peptides Identified in Human Apolipoprotein B as Promising Antifungal Agents. Appl. Microbiol. Biotechnol. 2021, 105, 953–1964. [Google Scholar] [CrossRef]
- Dell’Olmo, E.; Gaglione, R.; Sabbah, M.; Schibeci, M.; Cesaro, A.; di Girolamo, R.; Porta, R.; Arciello, A. Host Defense Peptides Identified in Human Apolipoprotein B as Novel Food Biopreservatives and Active Coating Components. Food Microbiol. 2021, 99. [Google Scholar] [CrossRef] [PubMed]
- Pane, K.; Durante, L.; Crescenzi, O.; Cafaro, V.; Pizzo, E.; Varcamonti, M.; Zanfardino, A.; Izzo, V.; di Donato, A.; Notomista, E. Antimicrobial Potency of Cationic Antimicrobial Peptides Can Be Predicted from Their Amino Acid Composition: Application to the Detection of “Cryptic” Antimicrobial Peptides. J. Theor. Biol. 2017, 419, 254–265. [Google Scholar] [CrossRef]
- Gaglione, R.; Pane, K.; Dell’Olmo, E.; Cafaro, V.; Pizzo, E.; Olivieri, G.; Notomista, E.; Arciello, A. Cost-Effective Production of Recombinant Peptides in Escherichia coli. New Biotechnol. 2019, 51, 39–48. [Google Scholar] [CrossRef]
- Brancaccio, D.; Pizzo, E.; Cafaro, V.; Notomista, E.; De Lise, F.; Bosso, A.; Gaglione, R.; Merlino, F.; Novellino, E.; Ungaro, F.; et al. Antimicrobial Peptide Temporin-L Complexed with Anionic Cyclodextrins Results in a Potent and Safe Agent against Sessile Bacteria. Int. J. Pharm. 2020, 584. [Google Scholar] [CrossRef]
- Na, Y.J.; Han, S.B.; Kang, J.S.; Yoon, Y.D.; Park, S.-K.; Kim, H.M.; Yang, K.-H.; Joe, C.O. Lactoferrin Works as a New LPS-Binding Protein in Inflammatory Activation of Macrophages. Int. Immunopharmacol. 2004, 4, 1187–1199. [Google Scholar] [CrossRef]
- MacGowan, A.P.; Wootton, M.; Hedges, A.J.; Bowker, K.E.; Holt, H.A.; Reeves, D.S. A New Time-Kill Method of Assessing the Relative Efficacy of Antimicrobial Agents Alone and in Combination Developed Using a Representative β-Lactam, Aminoglycoside and Fluoroquinolone. J. Antimicrob. Chemother. 1996, 38, 193–203. [Google Scholar] [CrossRef][Green Version]
- Garidel, P.; Blume, A. Miscibility of Phosphatidylethanolamine-Phosphatidylglycerol Mixtures as a Function of PH and Acyl Chain Length. Eur. Biophys. J. 2000, 28, 629–638. [Google Scholar] [CrossRef]
- Arouri, A.; Dathe, M.; Blume, A. Peptide Induced Demixing in PG/PE Lipid Mixtures: A Mechanism for the Specificity of Antimicrobial Peptides towards Bacterial Membranes? Biochim. Biophys. Acta Biomembr. 2009, 1788, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; del Vecchio, P.; Grimaldi, A.; Notomista, E.; Cafaro, V.; Pane, K.; Schuabb, V.; Winter, R.; Petraccone, L. Membrane Disintegration by the Antimicrobial Peptide (P)GKY20: Lipid Segregation and Domain Formation. Phys. Chem. Chem. Phys. 2019, 21, 3989–3998. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Chino, M.; Pane, K.; Pistorio, V.; de Santis, A.; Pizzo, E.; D’Errico, G.; Pavone, V.; Lombardi, A.; del Vecchio, P.; et al. Exploring the Role of Unnatural Amino Acids in Antimicrobial Peptides. Sci. Rep. 2018, 8, 8888. [Google Scholar] [CrossRef] [PubMed]
- Cañadas, O.; Casals, C. Differential Scanning Calorimetry of Protein–Lipid Interactions. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 974, pp. 55–71. [Google Scholar]
- Gong, Z.; Ikonomova, S.P.; Karlsson, A.J. Secondary Structure of Cell-Penetrating Peptides during Interaction with Fungal Cells. Protein Sci. 2018, 27, 702–713. [Google Scholar] [CrossRef]
- Malgieri, G.; Avitabile, C.; Palmieri, M.; D’Andrea, L.D.; Isernia, C.; Romanelli, A.; Fattorusso, R. Structural Basis of a Temporin 1b Analogue Antimicrobial Activity against Gram Negative Bacteria Determined by CD and NMR Techniques in Cellular Environment. ACS Chem. Biol. 2015, 10, 965–969. [Google Scholar] [CrossRef]
- Avitabile, C.; D’Andrea, L.D.; Romanelli, A. Circular Dichroism Studies on the Interactions of Antimicrobial Peptides with Bacterial Cells. Sci. Rep. 2014, 4, 4293. [Google Scholar] [CrossRef]
- Kagan, B.L.; Jang, H.; Capone, R.; Teran Arce, F.; Ramachandran, S.; Lal, R.; Nussinov, R. Antimicrobial Properties of Amyloid Peptides. Mol. Pharm. 2012, 9, 708–717. [Google Scholar] [CrossRef]
- LeVine, H. Thioflavine T Interaction with Amyloid β-Sheet Structures. Amyloid 1995, 2, 1–6. [Google Scholar] [CrossRef]
- Biancalana, M.; Makabe, K.; Koide, A.; Koide, S. Molecular Mechanism of Thioflavin-T Binding to the Surface of β-Rich Peptide Self-Assemblies. J. Mol. Biol. 2009, 385, 1052–1063. [Google Scholar] [CrossRef]
- Wu, C.; Biancalana, M.; Koide, S.; Shea, J.-E. Binding Modes of Thioflavin-T to the Single-Layer β-Sheet of the Peptide Self-Assembly Mimics. J. Mol. Biol. 2009, 394, 627–633. [Google Scholar] [CrossRef]
- Han, D.; Sherman, S.; Filocamo, S.; Steckl, A.J. Long-Term Antimicrobial Effect of Nisin Released from Electrospun Triaxial Fiber Membranes. Acta Biomater. 2017, 53, 242–249. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K. The Roles of Antimicrobial Peptides in Innate Host Defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [PubMed]
- Song, D.W.; Kim, S.H.; Kim, H.H.; Lee, K.H.; Ki, C.S.; Park, Y.H. Multi-Biofunction of Antimicrobial Peptide-Immobilized Silk Fibroin Nanofiber Membrane: Implications for Wound Healing. Acta Biomater. 2016, 39, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Echalier, C.; Jebors, S.; Laconde, G.; Brunel, L.; Verdié, P.; Causse, L.; Bethry, A.; Legrand, B.; van den Berghe, H.; Garric, X.; et al. Sol–Gel Synthesis of Collagen-Inspired Peptide Hydrogel. Mater. Today 2017, 20, 59–66. [Google Scholar] [CrossRef]
- Hacke, M.; Gruber, T.; Schulenburg, C.; Balbach, J.; Arnold, U. Consequences of Proline-to-Alanine Substitutions for the Stability and Refolding of Onconase. FEBS J. 2013, 280, 4454–4462. [Google Scholar] [CrossRef]
- Sigel, S.; Bunk, S.; Meergans, T.; Doninger, B.; Stich, K.; Stulnig, T.; Derfler, K.; Hoffmann, J.; Deininger, S.; von Aulock, S.; et al. Apolipoprotein B100 Is a Suppressor of Staphylococcus aureus-Induced Innate Immune Responses in Humans and Mice. Eur. J. Immunol. 2012, 42, 2983–2989. [Google Scholar] [CrossRef]
- Stöckl, M.; Fischer, P.; Wanker, E.; Herrmann, A. α-Synuclein Selectively Binds to Anionic Phospholipids Embedded in Liquid-Disordered Domains. J. Mol. Biol. 2008, 375, 1394–1404. [Google Scholar] [CrossRef]
- Bradford, A.M.; Bowie, J.H.; Tyler, M.J.; Wallace, J.C. New Antibiotic Uperin Peptides from the Dorsal Glands of the Australian Toadlet Uperoleia Mjobergii. Aust. J. Chem. 1996, 1861, 2342–2353. [Google Scholar] [CrossRef]
- Martin, L.L.; Kubeil, C.; Piantavigna, S.; Tikkoo, T.; Gray, N.P.; John, T.; Calabrese, A.N.; Liu, Y.; Hong, Y.; Hossain, M.A.; et al. Amyloid Aggregation and Membrane Activity of the Antimicrobial Peptide Uperin 3.5. Pept. Sci. 2018, 110, e24052. [Google Scholar] [CrossRef]
- Branch, T.; Girvan, P.; Barahona, M.; Ying, L. Introduction of a Fluorescent Probe to Amyloid-β to Reveal Kinetic Insights into Its Interactions with Copper(II). Angew. Chem. 2015, 127, 1243–1246. [Google Scholar] [CrossRef]
- Berthelot, K.; Cullin, C.; Lecomte, S. What Does Make an Amyloid Toxic: Morphology, Structure or Interaction with Membrane? Biochimie 2013, 95, 12–19. [Google Scholar] [CrossRef]
- Steinborner, S.T.; Currie, G.J.; Bowie, J.H.; Wallace, J.C.; Tyler, M.J. New Antibiotic Caerin 1 Peptides from the Skin Secretion of the Australian Tree Frog Litoria Chloris. Comparison of the Activities of the Caerin 1 Peptides from the Genus Litoria. J. Pept. Res. 1998, 51, 121–126. [Google Scholar] [CrossRef]
- Bucciantini, M.; Nosi, D.; Forzan, M.; Russo, E.; Calamai, M.; Pieri, L.; Formigli, L.; Quercioli, F.; Soria, S.; Pavone, F.; et al. Toxic Effects of Amyloid Fibrils on Cell Membranes: The Importance of Ganglioside GM1. FASEB J. 2012, 26, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of Action of Membrane Active Antimicrobial Peptides. Biopolym. Pept. Sci. Sect. 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Lorenzen, K.; Olia, A.S.; Uetrecht, C.; Cingolani, G.; Heck, A.J.R. Determination of Stoichiometry and Conformational Changes in the First Step of the P22 Tail Assembly. J. Mol. Biol. 2008, 379, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Ehrnhoefer, D.E.; Bieschke, J.; Boeddrich, A.; Herbst, M.; Masino, L.; Lurz, R.; Engemann, S.; Pastore, A.; Wanker, E.E. EGCG Redirects Amyloidogenic Polypeptides into Unstructured, off-Pathway Oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Srinivasan, Y.; de Anda, J.; Nicastro, L.K.; Tükel, Ç.; Wong, G.C.L. Functional Reciprocity of Amyloids and Antimicrobial Peptides: Rethinking the Role of Supramolecular Assembly in Host Defense, Immune Activation, and Inflammation. Front. Immunol. 2020, 11, 1629. [Google Scholar] [CrossRef]
- Pane, K.; Durante, L.; Pizzo, E.; Varcamonti, M.; Zanfardino, A.; Sgambati, V.; di Maro, A.; Carpentieri, A.; Izzo, V.; di Donato, A.; et al. Rational Design of a Carrier Protein for the Production of Recombinant Toxic Peptides in Escherichia coli. PLoS ONE 2016, 11, e0146552. [Google Scholar] [CrossRef]
- Biltonen, R.L.; Lichtenberg, D. The Use of Differential Scanning Calorimetry as a Tool to Characterize Liposome Preparations. Chem. Phys. Lipids 1993, 64, 129–142. [Google Scholar] [CrossRef]
- Andrushchenko, V.V.; Vogel, H.J.; Prenner, E.J. Interactions of Tryptophan-Rich Cathelicidin Antimicrobial Peptides with Model Membranes Studied by Differential Scanning Calorimetry. Biochim. Biophys. Acta Biomembr. 2007, 1768, 2447–2458. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Pane, K.; Bosso, A.; Landi, N.; Ragucci, S.; Russo, R.; Gaglione, R.; Torres, M.D.T.; de la Fuente-Nunez, C.; Arciello, A.; et al. Novel Bioactive Peptides from PD-L1/2, a Type 1 Ribosome Inactivating Protein from Phytolacca dioica L. Evaluation of Their Antimicrobial Properties and Anti-Biofilm Activities. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Smaldone, G.; Di Girolamo, R.; Piccoli, R.; Pedone, E.; Arciello, A. Cell Milieu Significantly Affects the Fate of AApoAI Amyloidogenic Variants: Predestination or Serendipity? Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 377–384. [Google Scholar] [CrossRef] [PubMed]



| Peptide Name | Sequence | Molecular Weight | Net Charge at Neutral pH | Isoelectric Point | Grand Average of Hydropathicity |
|---|---|---|---|---|---|
| r(P)ApoBLPro | PHVALKPGKLKFIIPSPKRPVKLLSGGNTLHLVSTTKT | 4074.96 Da | 7.2 | 11.43 | 0.005 |
| r(P)ApoBLAla | PHVALKAGKLKFIIPSPKRPVKLLSGGNTLHLVSTTKT | 4048.92 Da | 7.2 | 11.43 | 0.005 |
| Gram-Negative Strains | r(P)ApoBLPro | r(P)ApoBLAla |
|---|---|---|
| Escherichia coli ATCC 35218 | 2.5 | 2.5 |
| Pseudomonas aeruginosa PAO1 | 5–10 | 2.5–10 |
| Bulkolderia cenocepacia J2315 | 40 | 40 |
| Gram-positive strains | ||
| Staphylococcus aureus MRSA WKZ-2 | 5–10 | 2.5 |
| Bacillus subtlis subsp. spizizenii ATCC 6633 | 5 | 5 |
| Staphylococcus aureus ATCC 12600 | 20 | 20 |
| System | ΔHm (kJ mol−1) a,b,c | Tm (°C) b,d |
|---|---|---|
| DPPE/ DPPG | 39.7 | 60.5 |
| + r(P) ApoBLPro | 37.5 | 62.0 |
| + r(P) ApoBLAla | 34.8 | 62.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaglione, R.; Smaldone, G.; Cesaro, A.; Rumolo, M.; De Luca, M.; Di Girolamo, R.; Petraccone, L.; Del Vecchio, P.; Oliva, R.; Notomista, E.; et al. Impact of a Single Point Mutation on the Antimicrobial and Fibrillogenic Properties of Cryptides from Human Apolipoprotein B. Pharmaceuticals 2021, 14, 631. https://doi.org/10.3390/ph14070631
Gaglione R, Smaldone G, Cesaro A, Rumolo M, De Luca M, Di Girolamo R, Petraccone L, Del Vecchio P, Oliva R, Notomista E, et al. Impact of a Single Point Mutation on the Antimicrobial and Fibrillogenic Properties of Cryptides from Human Apolipoprotein B. Pharmaceuticals. 2021; 14(7):631. https://doi.org/10.3390/ph14070631
Chicago/Turabian StyleGaglione, Rosa, Giovanni Smaldone, Angela Cesaro, Mariano Rumolo, Maria De Luca, Rocco Di Girolamo, Luigi Petraccone, Pompea Del Vecchio, Rosario Oliva, Eugenio Notomista, and et al. 2021. "Impact of a Single Point Mutation on the Antimicrobial and Fibrillogenic Properties of Cryptides from Human Apolipoprotein B" Pharmaceuticals 14, no. 7: 631. https://doi.org/10.3390/ph14070631
APA StyleGaglione, R., Smaldone, G., Cesaro, A., Rumolo, M., De Luca, M., Di Girolamo, R., Petraccone, L., Del Vecchio, P., Oliva, R., Notomista, E., Pedone, E., & Arciello, A. (2021). Impact of a Single Point Mutation on the Antimicrobial and Fibrillogenic Properties of Cryptides from Human Apolipoprotein B. Pharmaceuticals, 14(7), 631. https://doi.org/10.3390/ph14070631