Integrative Genomic–Epigenomic Analysis of Clozapine-Treated Patients with Refractory Psychosis
Abstract
1. Introduction
2. Results
2.1. Clinical and Demographic Characteristics of Patients
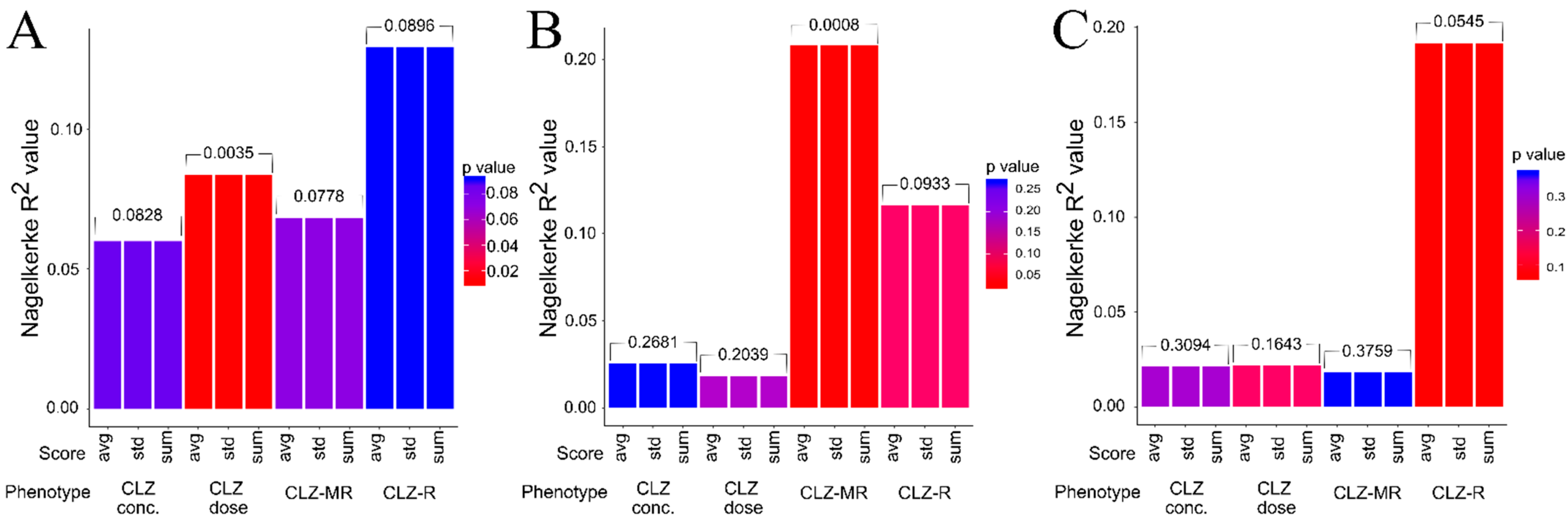
2.2. Association Between Genetic Risk Scores and Clozapine-Associated Phenotypes
2.3. Functional Prediction of the SNPs Included in the BD-PRS Associated with CLZ Metabolic Ratio
2.4. Differentially Methylated Sites Between Patients Grouped by BD-PRS and CLZ Metabolic Ratios
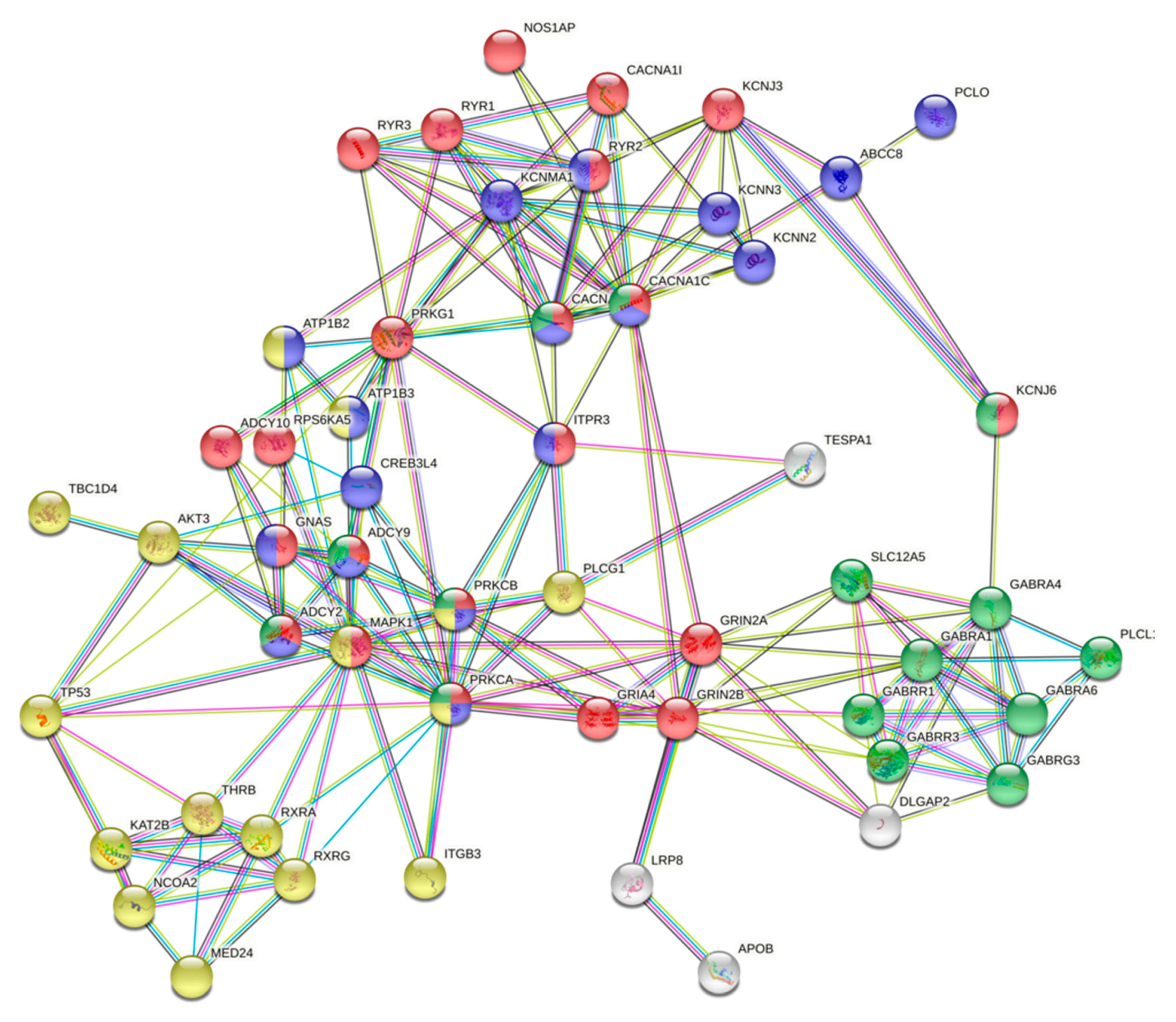
2.5. Protein–Protein Interactions Between Gene Products with High Impact Variants in the Top Enriched Pathways and Differentially Methylated Sites
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Clozapine and Norclozapine Plasma Concentrations.
4.3. Analysis and Quality Control of Microarrays
4.4. Analysis of Polygenic Risk Score
4.5. Analysis of Differentially Methylated Regions (DMRs)
4.6. Functional Annotation and Pathway Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siskind, D.J.; Lee, M.; Ravindran, A.; Zhang, Q.; Ma, E.; Motamarri, B.; Kisely, S. Augmentation strategies for clozapine refractory schizophrenia: A systematic review and meta-analysis. Aust. N. Z. J. Psychiatry 2018, 52, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Ifteni, P.; Teodorescu, A.; Dima, L.; Burtea, V. Rapid Titration of Clozapine in Schizophrenia and Bipolar Disorder. Am. J. Ther. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-García, C.; Iglesias-Alonso, A.; Bobes, J. Concentrations in plasma clozapine levels in schizophrenic and schizoaffective patients. Rev. Psiquiatr. Salud Ment. 2017, 10, 192–196. [Google Scholar] [CrossRef]
- Scheepers, F.E.; de Mul, J.; Boer, F.; Hoogendijk, W.J. Psychosis as an Evolutionary Adaptive Mechanism to Changing Environments. Front Psychiatry 2018, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Burton, C.Z.; Ryan, K.A.; Kamali, M.; Marshall, D.F.; Harrington, G.; McInnis, M.G.; Tso, I.F. Psychosis in bipolar disorder: Does it represent a more “severe” illness? Bipolar Disord. 2018, 20, 18–26. [Google Scholar] [CrossRef]
- Rothschild, A.J. Challenges in the Treatment of Major Depressive Disorder with Psychotic Features. Schizophr. Bull. 2013, 39, 787–796. [Google Scholar] [CrossRef]
- Costa-Dookhan, K.A.; Agarwal, S.M.; Chintoh, A.; Tran, V.N.; Stogios, N.; Ebdrup, B.H.; Sockalingam, S.; Rajji, T.K.; Remington, G.J.; Siskind, D.; et al. The clozapine to norclozapine ratio: A narrative review of the clinical utility to minimize metabolic risk and enhance clozapine efficacy. Expert Opin. Drug Saf. 2020, 19, 43–57. [Google Scholar] [CrossRef]
- Numata, S.; Umehara, H.; Ohmori, T.; Hashimoto, R. Clozapine Pharmacogenetic Studies in Schizophrenia: Efficacy and Agranulocytosis. Front. Pharmacol. 2018, 9, 1049. [Google Scholar] [CrossRef]
- Ameer, M.A.; Saadabadi, A. Neuroleptic Medications. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: http://www.ncbi.nlm.nih.gov/books/NBK459150/ (accessed on 12 November 2020).
- Fehr, B.S.; Ozcan, M.E.; Suppes, T. Low doses of clozapine may stabilize treatment-resistant bipolar patients. Eur. Arch. Psychiatry Clin. Neurosci. 2005, 255, 10–14. [Google Scholar] [CrossRef]
- Haidary, H.A.; Padhy, R.K. Clozapine. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: http://www.ncbi.nlm.nih.gov/books/NBK535399/ (accessed on 29 November 2020).
- Schenone, M.; Dančík, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef]
- Gregori-Puigjané, E.; Setola, V.; Hert, J.; Crews, B.A.; Irwin, J.J.; Lounkine, E.; Marnett, L.; Roth, B.L.; Shoichet, B.K. Identifying mechanism-of-action targets for drugs and probes. Proc. Natl. Acad. Sci. USA 2012, 109, 11178–11183. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, A.B.; Wang, Q.Q.; Joseph, P.; Zheng, C.; Chen, Y.; Kovalenko, O.; Singh, S.; Armstrong, A.; Resnick, K.; Zanotti, K.; et al. Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment. Oncogene 2018, 37, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bauman, J.E.; Bologa, C.G.; Buranda, T.; Chigaev, A.; Edwards, B.S.; Jarvik, J.W.; Gresham, H.D.; Haynes, M.K.; Hjelle, B.; et al. Drug repurposing from an academic perspective. Drug Discov. Today Ther. Strategy 2011, 8, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Henkel, N.D.; Alganem, K.; Hamoud, A.; Reigle, J.; Alnafisah, R.S.; Eby, H.M.; Imami, A.S.; Creeden, J.F.; Miruzzi, S.A.; et al. Signature-based approaches for informed drug repurposing: Targeting CNS disorders. Neuropsychopharmacology 2021, 46, 116–130. [Google Scholar] [CrossRef]
- Alaimo, S.; Pulvirenti, A. Network-Based Drug Repositioning: Approaches, Resources, and Research Directions. Methods Mol. Biol. 2019, 1903, 97–113. [Google Scholar] [CrossRef]
- Sahu, N.U.; Kharkar, P.S. Computational Drug Repositioning: A Lateral Approach to Traditional Drug Discovery? Curr. Top. Med. Chem. 2016, 16, 2069–2077. [Google Scholar] [CrossRef]
- González-Esquivel, D.F.; Castro, N.; Ramírez-Bermúdez, J.; Custodio, V.; Rojas-Tomé, S.; Castro-Román, R.; Jung-Cook, H. Plasma levels of clozapine and norclozapine in Mexican schizophrenia patients. Arzneimittelforschung 2011, 61, 335–339. [Google Scholar] [CrossRef]
- Couchman, L.; Morgan, P.E.; Spencer, E.P.; Flanagan, R.J. Plasma Clozapine, Norclozapine, and the Clozapine:Norclozapine Ratio in Relation to Prescribed Dose and Other Factors: Data from a Therapeutic Drug Monitoring Service, 1993–2007. Ther. Drug Monit. 2010, 32, 438–447. [Google Scholar] [CrossRef]
- Subramanian, S.; Völlm, B.A.; Huband, N. Clozapine dose for schizophrenia. Cochrane Database Syst. Rev. 2017, 6, CD009555. [Google Scholar] [CrossRef]
- Simpson, G.M.; Josiassen, R.C.; Stanilla, J.K.; de Leon, J.; Nair, C.; Abraham, G.; Odom-White, A.; Turner, R.M. Double-blind study of clozapine dose response in chronic schizophrenia. Am. J. Psychiatry 1999, 156, 1744–1750. [Google Scholar] [CrossRef]
- Suzuki, T.; Uchida, H.; Watanabe, K.; Kashima, H. Factors Associated with Response to Clozapine in Schizophrenia: A Review. Psychopharmacol. Bull. 2011, 44, 32–60. [Google Scholar]
- Alessandro, G.; Erbo, D.; Grayson, D.R. Epigenetic Basis of Clozapine Action. J. Drug Des. Res. 2017, 4, 1055. [Google Scholar] [PubMed]
- O’Donovan, M.C.; Owen, M.J. The implications of the shared genetics of psychiatric disorders. Nat. Med. 2016, 22, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Hannon, E.; Dempster, E.; Viana, J.; Burrage, J.; Smith, A.R.; Macdonald, R.; St Clair, D.; Mustard, C.; Breen, G.; Therman, S.; et al. An integrated genetic-epigenetic analysis of schizophrenia: Evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016, 17, 176. [Google Scholar] [CrossRef] [PubMed]
- Vaquero-Baez, M.; Díaz-Ruíz, A.; Tristán-López, L.; Aviña-Cervantes, C.; Torner, C.; Ramírez-Bermúdez, J.; Montes, S.; Ríos, C. Clozapine and desmethylclozapine: Correlation with neutrophils and leucocytes counting in Mexican patients with schizophrenia. BMC Psychiatry 2019, 19, 295. [Google Scholar] [CrossRef]
- Nucifora, F.C.; Mihaljevic, M.; Lee, B.J.; Sawa, A. Clozapine as a Model for Antipsychotic Development. Neurotherapeutics 2017, 14, 750–761. [Google Scholar] [CrossRef]
- Wilkowska, A.; Cubała, W.J. Clozapine as Transformative Treatment in Bipolar Patients. Neuropsychiatr. Dis. Treat. 2019, 15, 2901–2905. [Google Scholar] [CrossRef]
- Marx, C.E.; Shampine, L.J.; Duncan, G.E.; VanDoren, M.J.; Grobin, A.C.; Massing, M.W.; Madison, R.D.; Bradford, D.W.; Butterfield, M.I.; Lieberman, J.A.; et al. Clozapine markedly elevates pregnenolone in rat hippocampus, cerebral cortex, and serum: Candidate mechanism for superior efficacy? Pharmacol. Biochem. Behav. 2006, 84, 598–608. [Google Scholar] [CrossRef]
- Leveque, J.-C.; Macías, W.; Rajadhyaksha, A.; Carlson, R.R.; Barczak, A.; Kang, S.; Li, X.M.; Coyle, J.T.; Huganir, R.L.; Heckers, S.; et al. Intracellular Modulation of NMDA Receptor Function by Antipsychotic Drugs. J. Neurosci. 2000, 20, 4011–4020. [Google Scholar] [CrossRef]
- Spivak, B.; Shabash, E.; Sheitman, B.; Weizman, A.; Mester, R. The effects of clozapine versus haloperidol on measures of impulsive aggression and suicidality in chronic schizophrenia patients: An open, nonrandomized, 6-month study. J. Clin. Psychiatry 2003, 64, 755–760. [Google Scholar] [CrossRef]
- Klemen, M.S.; Dolenšek, J.; Rupnik, M.S.; Stožer, A. The triggering pathway to insulin secretion: Functional similarities and differences between the human and the mouse β cells and their translational relevance. Islets 2017, 9, 109–139. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.L.; Haan, N.; Wilkinson, L.S.; Thomas, K.L.; Hall, J. CACNA1C: Association with Psychiatric Disorders, Behavior, and Neurogenesis. Schizophr. Bull. 2018, 44, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Gedvilaite, E.; Badner, J.A.; Erdman, C.; Baird, L.; Matsunami, N.; Leppert, M.; Xing, J.; Byerley, W. A Rare Variant in CACNA1D Segregates with 7 Bipolar I Disorder Cases in a Large Pedigree. Mol. Neuropsychiatry 2016, 2, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Hu, Y.; Xin, J.; Zhao, M.; Wang, J. Analyzing the genes and pathways related to major depressive disorder via a systems biology approach. Brain Behav. 2020, 10, e01502. [Google Scholar] [CrossRef] [PubMed]
- Charles, E.F.; Lambert, C.G.; Kerner, B. Bipolar disorder and diabetes mellitus: Evidence for disease-modifying effects and treatment implications. Int. J. Bipolar Disord. 2016, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Calkin, C.V.; Gardner, D.M.; Ransom, T.; Alda, M. The relationship between bipolar disorder and type 2 diabetes: More than just co-morbid disorders. Ann. Med. 2013, 45, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, J.L.; Nanda, P.; Tandon, N.; Mothi, S.S.; Bolo, N.; McCarroll, S.; Clementz, B.A.; Gershon, E.S.; Pearlson, G.D.; Sweeney, J.A.; et al. Polygenic risk for type 2 diabetes mellitus among individuals with psychosis and their relatives. J. Psychiatr. Res. 2016, 77, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Hackinger, S.; Prins, B.; Mamakou, V.; Zengini, E.; Marouli, E.; Brčić, L.; Serafetinidis, I.; Lamnissou, K.; Kontaxakis, V.; Dedoussis, G.; et al. Evidence for genetic contribution to the increased risk of type 2 diabetes in schizophrenia. Transl. Psychiatry 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Postolache, T.T.; Bosque-Plata L del Jabbour, S.; Vergare, M.; Wu, R.; Gragnoli, C. Co-shared genetics and possible risk gene pathway partially explain the comorbidity of schizophrenia, major depressive disorder, type 2 diabetes, and metabolic syndrome. Am. J. Med. Genet B Neuropsychiatr. Genet. 2019, 180, 186–203. [Google Scholar] [CrossRef]
- Reay, W.R.; Atkins, J.R.; Carr, V.J.; Green, M.J.; Cairns, M.J. Pharmacological enrichment of polygenic risk for precision medicine in complex disorders. Sci. Rep. 2020, 10, 879. [Google Scholar] [CrossRef]
- Łojko, D.; Owecki, M.; Suwalska, A. Impaired Glucose Metabolism in Bipolar Patients: The Role of Psychiatrists in Its Detection and Management. Int. J. Environ. Res. Public Health 2019, 16, 1132. [Google Scholar] [CrossRef] [PubMed]
- Davidson, M.D.; Ballinger, K.R.; Khetani, S.R. Long-term exposure to abnormal glucose levels alters drug metabolism pathways and insulin sensitivity in primary human hepatocytes. Sci. Rep. 2016, 6, 28178. [Google Scholar] [CrossRef]
- Yau, L.S.; Strother, A.; Buchholz, J.; Abu-el-Haj, S. Glucose effect on drug action, metabolism, and pharmacokinetic parameters in mice. Drug Nutr. Interact. 1987, 5, 9–20. [Google Scholar] [PubMed]
- Imam, M.U.; Ismail, M. Effects of Brown Rice and White Rice on Expression of Xenobiotic Metabolism Genes in Type 2 Diabetic Rats. Int. J. Mol. Sci. 2012, 13, 8597–8608. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, K.; Wan, Y.-J.Y. Retinoids activate RXR/CAR-mediated pathway and induce CYP3A. Biochem. Pharmacol. 2010, 79, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Konishi, T.; Han, G.; Campwala, K.H.; French, S.W.; Wan, Y.-J.Y. The role of hepatocyte RXRα in xenobiotic-sensing nuclear receptor-mediated pathways. Eur. J. Pharm. Sci. 2002, 15, 89–96. [Google Scholar] [CrossRef]
- Wan, Y.J.; Cai, Y.; Lungo, W.; Fu, P.; Locker, J.; French, S.; Sucov, H.M. Peroxisome proliferator-activated receptor alpha-mediated pathways are altered in hepatocyte-specific retinoid X receptor alpha-deficient mice. J. Biol. Chem. 2000, 275, 28285–28290. [Google Scholar] [CrossRef]
- Mak, P.A.; Laffitte, B.A.; Desrumaux, C.; Joseph, S.B.; Curtiss, L.K.; Mangelsdorf, D.J.; Tontonoz, P.; Edwards, P.A. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J. Biol. Chem. 2002, 277, 31900–31908. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Li, R.; Chen, G. Transcriptional Factors Mediating Retinoic Acid Signals in the Control of Energy Metabolism. Int. J. Mol. Sci. 2015, 16, 14210–14244. [Google Scholar] [CrossRef]
- Vu-Dac, N.; Gervois, P.; Torra, I.P.; Fruchart, J.C.; Kosykh, V.; Kooistra, T.; Princen, H.M.; Dallongeville, J.; Staels, B. Retinoids increase human apo C-III expression at the transcriptional level via the retinoid X receptor. Contribution to the hypertriglyceridemic action of retinoids. J. Clin. Investig. 1998, 102, 625–632. [Google Scholar] [CrossRef]
- Dursun, S.M.; Szemis, A.; Andrews, H.; Reveley, M.A. The effects of clozapine on levels of total cholesterol and related lipids in serum of patients with schizophrenia: A prospective study. J. Psychiatry Neurosci. 1999, 24, 453–455. [Google Scholar] [PubMed]
- Kumar, M.; Sidana, A. Clozapine-induced Acute Hypertriglyceridemia. Indian J. Psychol. Med. 2017, 39, 682–684. [Google Scholar] [CrossRef] [PubMed]
- Procyshyn, R.M.; Wasan, K.M.; Thornton, A.E.; Barr, A.M.; Chen, E.Y.H.; Pomarol-Clotet, E.; Stip, E.; Williams, R.; Macewan, G.W.; Birmingham, C.L.; et al. Changes in serum lipids, independent of weight, are associated with changes in symptoms during long-term clozapine treatment. J. Psychiatry Neurosci. 2007, 32, 331–338. [Google Scholar] [PubMed]
- Li, M.; Huang, L.; Grigoroiu-Serbanescu, M.; Bergen, S.E.; Landén, M.; Hultman, C.M.; Forstner, A.J.; Strohmaier, J.; Hecker, J.; Schulze, T.G.; et al. Convergent Lines of Evidence Support LRP8 as a Susceptibility Gene for Psychosis. Mol. Neurobiol. 2016, 53, 6608–6619. [Google Scholar] [CrossRef]
- Guidotti, A.; Grayson, D.R.; Caruncho, H.J. Epigenetic RELN Dysfunction in Schizophrenia and Related Neuropsychiatric Disorders. Front. Cell. Neurosci. 2016, 10, 89. [Google Scholar] [CrossRef]
- Vik-Mo, A.O.; Fernø, J.; Skrede, S.; Steen, V.M. Psychotropic drugs up-regulate the expression of cholesterol transport proteins including ApoE in cultured human CNS- and liver cells. BMC Pharmacol. 2009, 9, 10. [Google Scholar] [CrossRef]
- Samanaite, R.; Gillespie, A.; Sendt, K.-V.; McQueen, G.; MacCabe, J.H.; Egerton, A. Biological Predictors of Clozapine Response: A Systematic Review. Front Psychiatry 2018, 9, 327. [Google Scholar] [CrossRef]
- Van Eck, M.; Van Dijk, K.W.; Herijgers, N.; Hofker, M.H.; Groot, P.H.E.; Van Berkel, T.J.C. Essential role for the (hepatic) LDL receptor in macrophage apolipoprotein E-induced reduction in serum cholesterol levels and atherosclerosis. Atherosclerosis 2001, 154, 103–112. [Google Scholar] [CrossRef]
- Peloso, G.M.; Nomura, A.; Khera, A.V.; Chaffin, M.; Won, H.-H.; Ardissino, D.; Danesh, J.; Schunkert, H.; Wilson, J.G.; Samani, N.; et al. Rare Protein-Truncating Variants in APOB, Lower Low-Density Lipoprotein Cholesterol, and Protection Against Coronary Heart Disease. Circ. Genom. Precis. Med. 2019, 12, e002376. [Google Scholar] [CrossRef]
- Lee, G.H.; D’Arcangelo, G. New Insights into Reelin-Mediated Signaling Pathways. Front. Cell. Neurosci. 2016, 10, 122. [Google Scholar] [CrossRef]
- Gill, I.; Droubi, S.; Giovedi, S.; Fedder, K.N.; Bury, L.A.D.; Bosco, F.; Sceniak, M.P.; Benfenati, F.; Sabo, S.L. Presynaptic NMDA receptors—Dynamics and distribution in developing axons in vitro and in vivo. J. Cell Sci. 2015, 128, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, P.; Perez, J.; Barale, F.; Schettini, G.; Soares, J.C. GABAergic dysfunction in mood disorders. Mol. Psychiatry 2003, 8, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Ifteni, P.; Teodorescu, A.; Moga, M.A.; Pascu, A.M.; Miclaus, R.S. Switching bipolar disorder patients treated with clozapine to another antipsychotic medication: A mirror image study. Neuropsychiatr. Dis. Treat. 2017, 13, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-B.; Tang, Y.-L.; Wang, C.-Y.; de Leon, J. Clozapine for treatment-resistant bipolar disorder: A systematic review. Bipolar Disord. 2015, 17, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Tueting, P.; Matrisciano, F.; Grayson, D.R.; Guidotti, A. Behavioral and molecular neuroepigenetic alterations in prenatally stressed mice: Relevance for the study of chromatin remodeling properties of antipsychotic drugs. Transl. Psychiatry 2016, 6, e711. [Google Scholar] [CrossRef]
- Dong, E.; Nelson, M.; Grayson, D.R.; Costa, E.; Guidotti, A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc. Natl. Acad. Sci. USA 2008, 105, 13614–13619. [Google Scholar] [CrossRef]
- Nair, P.C.; McKinnon, R.A.; Miners, J.O.; Bastiampillai, T. Binding of clozapine to the GABA B receptor: Clinical and structural insights. Mol. Psychiatry 2020, 25, 1910–1919. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Wang, T.; Luo, X. Aetiology of bipolar disorder: Contribution of the L-type voltage-gated calcium channels. Gen. Psychiatry 2019, 32, e100009. [Google Scholar] [CrossRef]
- Starnawska, A.; Demontis, D.; Pen, A.; Hedemand, A.; Nielsen, A.L.; Staunstrup, N.H.; Grove, J.; Als, T.D.; Jarram, A.; O’Brien, N.L.; et al. CACNA1C hypermethylation is associated with bipolar disorder. Transl. Psychiatry 2016, 6, e831. [Google Scholar] [CrossRef]
- Bigos, K.L.; Mattay, V.S.; Callicott, J.H.; Straub, R.E.; Vakkalanka, R.; Kolachana, B.; Hyde, T.M.; Lipska, B.K.; Kleinman, J.E.; Weinberger, D.R. Genetic Variation in CACNA1C Affects Brain Circuitries Related to Mental Illness. Arch. Gen. Psychiatry 2010, 67, 939. [Google Scholar] [CrossRef]
- Smedler, E.; Abé, C.; Pålsson, E.; Ingvar, M.; Landén, M. CACNA1C polymorphism and brain cortical structure in bipolar disorder. J. Psychiatry Neurosci. 2020, 45, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.I.; Boehning, D. Cardiac inositol 1,4,5-trisphosphate receptors. Biochim. Biophys. Acta BBA Mol. Cell Res. 2017, 1864, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Martinez, J.A. Dopamine gene methylation patterns are associated with obesity markers and carbohydrate intake. Brain Behav. 2018, 8, e01017. [Google Scholar] [CrossRef] [PubMed]
- Prole, D.L.; Taylor, C.W. Inositol 1,4,5-trisphosphate receptors and their protein partners as signalling hubs. J. Physiol. 2016, 594, 2849–2866. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, H.; Fujimoto, T.; Tanaka, M.; Shirasawa, S. Tespa1 is a novel component of mitochondria-associated endoplasmic reticulum membranes and affects mitochondrial calcium flux. Biochem. Biophys. Res. Commun. 2013, 433, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zheng, M.; Qiu, Y.; Guo, C.; Ji, J.; Lei, L.; Zhang, X.; Liang, J.; Lou, J.; Huang, W.; et al. Tespa1 negatively regulates FcεRI-mediated signaling and the mast cell–mediated allergic response. J. Exp. Med. 2014, 211, 2635–2649. [Google Scholar] [CrossRef]
- Matsuzaki, H.; Fujimoto, T.; Ota, T.; Ogawa, M.; Tsunoda, T.; Doi, K.; Hamabashiri, M.; Tanaka, M.; Shirasawa, S. Tespa1 is a novel inositol 1,4,5-trisphosphate receptor binding protein in T and B lymphocytes. FEBS Open Biol. 2012, 2, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, E.; Kroken, R.A. Drug treatment developments in schizophrenia and bipolar mania: Latest evidence and clinical usefulness. Ther. Adv. Chronic Dis. 2012, 3, 287–300. [Google Scholar] [CrossRef]
- Calafato, M.S.; Thygesen, J.H.; Ranlund, S.; Zartaloudi, E.; Cahn, W.; Crespo-Facorro, B.; Díez-Revuelta, A.; Forti, M.D.; Hall, M.H.; Iyegbe, C.; et al. Use of schizophrenia and bipolar disorder polygenic risk scores to identify psychotic disorders. Br. J. Psychiatry 2018, 213, 535–541. [Google Scholar] [CrossRef]
- Cardno, A.G.; Owen, M.J. Genetic Relationships between Schizophrenia, Bipolar Disorder, and Schizoaffective Disorder. Schizophr. Bull. 2014, 40, 504–515. [Google Scholar] [CrossRef]
- Santoro, M.L.; Ota, V.; de Jong, S.; Noto, C.; Spindola, L.M.; Talarico, F.; Gouvea, E.; Lee, S.H.; Moretti, P.; Curtis, C.; et al. Polygenic risk score analyses of symptoms and treatment response in an antipsychotic-naive first episode of psychosis cohort. Transl. Psychiatry 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-P.; Robinson, D.; Yu, J.; Gallego, J.; Fleischhacker, W.W.; Kahn, R.S.; Crespo-Facorro, B.; Vazquez-Bourgon, J.; Kane, J.M.; Malhotra, A.K.; et al. Schizophrenia Polygenic Risk Score as a Predictor of Antipsychotic Efficacy in First-Episode Psychosis. Am. J. Psychiatry 2018, 176, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Graw, S.; Henn, R.; Thompson, J.A.; Koestler, D.C. pwrEWAS: A user-friendly tool for comprehensive power estimation for epigenome wide association studies (EWAS). BMC Bioinform. 2019, 20, 218. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, DSM-5, 5th ed.; American Psychiatric Publishing: Washington, DC, USA, 2013; pp. 87–123. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [PubMed]
- Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.R.; Farh, K.-H.; Holmans, P.A.; Lee, P.; Bulik-Sullivan, B.; Collier, D.A.; Huang, H.; et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Howard, D.M.; Adams, M.J.; Clarke, T.-K.; Hafferty, J.D.; Gibson, J.; Shirali, M.; Coleman, J.R.I.; Hagenaars, S.P.; Ward, J.; Wigmore, E.M.; et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019, 22, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 Loci Associated with Bipolar Disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Euesden, J.; Lewis, C.M.; O’Reilly, P.F. PRSice: Polygenic Risk Score software. Bioinformatics 2015, 31, 1466–1468. [Google Scholar] [CrossRef]
- Conomos, M.P.; Miller, M.B.; Thornton, T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015, 39, 276–293. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Number (%) or Mean ± Standard Deviation |
|---|---|
| Clinical diagnosis | |
| Schizophrenia | 31 (70.45%) |
| Schizoaffective disorder | 9 (20.45%) |
| Bipolar disorder | 4 (9.09%) |
| Number of Male Patients (%) | 28 (63.60%) |
| Age (years) | 37.40 ± 11.30 |
| Age at onset | 18.50 ± 9.80 |
| School (Years) | 13.30 ± 2.90 |
| Number of patients who are smokers (%) | 22 (50.00%) |
| Number of patients who are drinkers (%) | 13 (29.50%) |
| CLZ Dose (mg/day) | 202.60 ± 138.02 |
| CLZ responders | 36 (81.80%) |
| CLZ and its metabolite determinations | |
| * Plasma concentrations of CLZ (ng/mL) | 154.03 ± 191.97 |
| Location † | Gene Symbol | Gene Name | Genetic Variant ID | Minor Allele Frequency | Protein ID | Variant Location in Coding Region |
|---|---|---|---|---|---|---|
| chr1:53712727 | LRP8 | LDL receptor-related protein 8 | rs5174 | T = 0.204 | NP_004622.2:p.Arg952Gln | Missense variant |
| chr1:151374025 | PSMB4 | Proteasome 20S subunit beta 4 | rs4603 | C = 0.273 | NP_002787.2:p.Ile234Asn | Missense variant |
| chr1:151733335 | MRPL9 | Mitochondrial ribosomal protein L9 | rs8480 | G = 0.443 | NP_113608.1:p.Glu210Val | Missense variant |
| chr4:162307312 | FSTL5 | Follistatin-like 5 | rs3749598 | A = 0.216 | NP_064501.2:p.Asp711Tyr | Missense variant |
| chr5:7520768 | ADCY2 | Adenylate cyclase 2 | rs13166360 | T = 0.057 | NP_065433.2:p.Val147Met | Missense variant |
| chr5:898209847 | LYSMD3 | LysM domain containing 3 | rs10069050 | C = 0.375 | NP_938014.1:p.Glu41Asp | Missense variant |
| chr6:142396790 | NMBR | Neuromedin B receptor | rs7453944 | T = 0.307 | NP_002502.2:p.Leu390Met | Missense variant |
| chr7:64439701 | ZNF117 | Zinc finger protein 117 | rs3807069 | T = 0.307 | NP_056936.2:p.Cys83Tyr | Missense variant |
| chr7: 92733766 | SAMD9 | Sterile alpha motif domain-containing 9 | rs10279499 | A = 0.091 | NP_001180236.1:p.Val549Leu | Missense variant |
| chr7:104717517 | KMT2E | Lysine methyltransferase 2E (inactive) | rs2240455 | T = 0.216 | NP_061152.3:p.Tyr292Ter | * Stop_gained |
| chr7:129663496 | ZC3HC1 | Zinc finger C3HC-type containing 1 | rs11556924 | T = 0.148 | NP_057562.3:p.Arg363His | Missense variant |
| chr8:1514009 | DLGAP2 | DLG associated protein 2 | rs2301963 | C = 0.284 | NP_001333739.1:p.Pro464Gln | Missense variant |
| chr12:108618630 | WSCD2 | WSC domain containing 2 | rs3764002 | T = 0.125 | NP_055468.2:p.Thr266Ile | Missense variant |
| chr15:84639350 | ADAMTSL3 | ADAMTS-like 3 | rs2277849 | T = 0.189 | NP_997400.2:p.Leu869Phe | Missense variant |
| chr16:3639827 | SLX4 | SLX4 structure-specific endonuclease subunit | rs3810813 | A = 0.079 | NP_115820.2:p.Ser1271Phe | Missense variant |
| chr17:35988672 | DDX52 | DExD-box helicase 52 | rs7224513 | C = 0.239 | NP_008941.3:p.Arg264Ser | Missense variant |
| chr17:73513677 | TSEN54 | tRNA splicing endonuclease subunit 54 | rs11559205 | C = 0.091 | NP_997229.2:p.Ile137Leu | Missense variant |
| Location † | Gene Symbol | CpG Site | Feature | Location Relative to cgi | LogFC | Avg Methylation | p-Value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| High PRS | Medium PRS | Low PRS | High-Medium PRS | Medium-Low PRS | ||||||
| TESPA1 | cg23612423 | 3’UTR | Open sea | −0.14346761 | 0.5651155 | 0.42164789 | 0.52374574 | 9.06 × 10−7 | 4.01 × 10−2 | |
| chr2:21266669-21266961 | APOB | cg16723488 | TSS200 | Island | 0.09815776 | 0.37368773 | 0.4718455 | 0.39797591 | 8.38 × 10−6 | 2.42 × 10−5 |
| chr2:21266669-21266961 | APOB | cg05337441 | Body | Shore | 0.08863618 | 0.15337978 | 0.2555618 | 0.16692562 | 2.46 × 10−5 | 3.02 × 10−6 |
| chr8:58055960-58056244 | - | cg11062466 | IGR | Shore | 0.27464151 | 0.30018264 | 0.57482415 | 0.36696224 | 8.92 × 10−6 | 6.11 × 10−3 |
| chr10:135170645-135171954 | C10orf125 | cg05456948 | TSS200 | Island | −0.04000716 | 0.19107189 | 0.15466524 | 0.1946724 | 3.04 × 10−04 | 1.54 × 10−06 |
| STAG1 | cg16760310 | Body | Open sea | 0.02946489 | 0.932180391 | 0.96574714 | 0.93628225 | 1.09 × 10−3 | 7.21 × 10−6 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayén-Lobo, Y.G.; Martínez-Magaña, J.J.; Pérez-Aldana, B.E.; Ortega-Vázquez, A.; Genis-Mendoza, A.D.; Dávila-Ortiz de Montellano, D.J.; Soto-Reyes, E.; Nicolini, H.; López-López, M.; Monroy-Jaramillo, N. Integrative Genomic–Epigenomic Analysis of Clozapine-Treated Patients with Refractory Psychosis. Pharmaceuticals 2021, 14, 118. https://doi.org/10.3390/ph14020118
Mayén-Lobo YG, Martínez-Magaña JJ, Pérez-Aldana BE, Ortega-Vázquez A, Genis-Mendoza AD, Dávila-Ortiz de Montellano DJ, Soto-Reyes E, Nicolini H, López-López M, Monroy-Jaramillo N. Integrative Genomic–Epigenomic Analysis of Clozapine-Treated Patients with Refractory Psychosis. Pharmaceuticals. 2021; 14(2):118. https://doi.org/10.3390/ph14020118
Chicago/Turabian StyleMayén-Lobo, Yerye Gibrán, José Jaime Martínez-Magaña, Blanca Estela Pérez-Aldana, Alberto Ortega-Vázquez, Alma Delia Genis-Mendoza, David José Dávila-Ortiz de Montellano, Ernesto Soto-Reyes, Humberto Nicolini, Marisol López-López, and Nancy Monroy-Jaramillo. 2021. "Integrative Genomic–Epigenomic Analysis of Clozapine-Treated Patients with Refractory Psychosis" Pharmaceuticals 14, no. 2: 118. https://doi.org/10.3390/ph14020118
APA StyleMayén-Lobo, Y. G., Martínez-Magaña, J. J., Pérez-Aldana, B. E., Ortega-Vázquez, A., Genis-Mendoza, A. D., Dávila-Ortiz de Montellano, D. J., Soto-Reyes, E., Nicolini, H., López-López, M., & Monroy-Jaramillo, N. (2021). Integrative Genomic–Epigenomic Analysis of Clozapine-Treated Patients with Refractory Psychosis. Pharmaceuticals, 14(2), 118. https://doi.org/10.3390/ph14020118








