Abstract
Panax ginseng C.A.Mey. is an adaptogenic plant traditionally used to enhance mental and physical capacities in cases of weakness, exhaustion, tiredness, or loss of concentration, and during recovery. According to ancient records, red ginseng root preparations enhance longevity with long-term intake. Recent pharmacokinetic studies of ginsenosides in humans and our in vitro study in neuronal cells suggest that ginsenosides are effective when their levels in blood is low—at concentrations from 10−6 to 10−18 M. In the present study, we compared the effects of red ginseng root preparation HRG80TM(HRG) at concentrations from 0.01 to 10,000 ng/mL with effects of white ginseng (WG) and purified ginsenosides Rb1, Rg3, Rg5 and Rk1 on gene expression in isolated hippocampal neurons. The aim of this study was to predict the effects of differently expressed genes on cellular and physiological functions in organismal disorders and diseases. Gene expression profiling was performed by transcriptome-wide mRNA microarray analyses in murine HT22 cells after treatment with ginseng preparations. Ingenuity pathway downstream/upstream analysis (IPA) was performed with datasets of significantly up- or downregulated genes, and expected effects on cellular function and disease were identified by IPA software. Ginsenosides Rb1, Rg3, Rg5, and Rk1 have substantially varied effects on gene expression profiles (signatures) and are different from signatures of HRG and WG. Furthermore, the signature of HRG is changed significantly with dilution from 10,000 to 0.01 ng/mL. Network pharmacological analyses of gene expression profiles showed that HRG exhibits predictable positive effects in neuroinflammation, senescence, apoptosis, and immune response, suggesting beneficial soft-acting effects in cancer, gastrointestinal, and endocrine systems diseases and disorders in a wide range of low concentrations in blood.
1. Introduction
Panax ginseng C.A.Mey. is likely one of the most widely used botanicals in the world [1,2]. This adaptogenic plant [2,3,4,5,6,7,8] is approved in Europe and other countries as an herbal medicinal product to enhance cognitive functions, physical capacities in weakness, exhaustion, tiredness, loss of concentration, and during convalescence [9,10]. According to ancient records, red ginseng root enhances longevity with long-term intake [11]. Overall, ginseng is a promising treatment for aging-related diseases, including neurodegenerative, cardiovascular diseases, diabetes, and cancer [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15].
A challenge in ginseng research and herbal therapy, in general, is the consistency and reproducibility of the results obtained from various studies [10]. One of the critical solutions to this problem is the reproducible quality of herbal preparations, which is possible to achieve by implementation of GMP and Good Agriculture Practices in the production of herbal preparations, including their cultivation in well-controlled conditions.
Recently, we have demonstrated that both hydroponically cultivated red ginseng root powder HRG80TM (HRG) and white ginseng (WG) preparations were effective in preventing and mitigating the stress-induced deterioration of cognitive functions in healthy subjects [16] and elderly patients with mild cognitive disorders [17] at daily doses of 418 mg HRG (64 mg of total ginsenosides, 1.2 mg Rg5) and 764 mg WG (19.6 mg of total ginsenosides, 0.16 mg Rg5). Both HRG and WG preparations substantially impacted brain activity, affecting various brain regions depending on the mental load during relaxation and cognitive tasks associated with memory, attention, and mental performance. Both ginseng preparations activated electroencephalogram (EEG) spectral powers compared to placebo [17]. The spectral changes in the quantitative EEG induced by HRG indicated an improvement in mood and calming effects evidenced by the modulation of β2 waves, representing changes in GABA-ergic neurotransmission. HRG attenuated δ/θ wave power, which is increased in aging [17]. Red ginseng HRG preparation was more active than WG in humans [17] and in an animal study, where HRG induced higher excitability of pyramidal cells by modulation of ionotropic glutamate NMDA and kainate receptor-mediated transmission at daily doses of 10 to 50 mg/kg body weight corresponding to human doses of 100–500 mg [18].
We had several aims in this study, intending to cover several gaps in ginseng research and specifically in hydroponically cultivated roots used to produce red ginseng preparations. These are related to the following issues:
Numerous studies of purified ginsenosides show their potential efficacy in treating cancer and other diseases [19,20,21,22,23,24,25,26,27,28], suggesting that the total extract or powdered roots have similar or probably better therapeutic effectiveness [23,24,25,26]. However, clinical evidence supporting this suggestion in comparative studies is missing [27,28].
As a matter of fact, blood cells and tissues are continuously exposed to varying concentrations of ginsenosides after oral administration of ginseng in daily therapeutic doses of 0.6–9.0 g [10,29,30]. They range from maximal detected concentrations of 35 ng/mL to the limit of detection 0.5 ng/mL and less within 12–48 h after oral intake [29]. A recent study demonstrated that ginsenoside Rg5 exhibits soft-acting effects in a wide range of physiological and sub-physiological concentrations from 1 μM to 1 aM. However, low-dose studies in corresponding concentrations are not available in the scientific literature.
Pharmacological effects of ginseng preparations at various levels of regulation of homeostasis have been studied [26,31,32,33,34,35,36,37,38]; however, network pharmacology studies [26,33,34,35,36,37,38] at the transcriptomic level of regulation of cellular homeostasis are limited [38].
Therefore, the first aim of this study was to compare the effects of red ginseng root preparation (HRG80TM) at concentrations from 0.01 to 10,000 ng/mL with effects of white ginseng (WG) and purified ginsenosides Rb1, Rg3, Rg5, and Rk1 on gene expression of isolated hippocampal neurons. The study’s second aim was to predict the effects of differently expressed genes on cellular and physiological functions in organismal disorders and diseases using a network pharmacology approach to transcriptomics.
2. Results
2.1. Effect of Ginsenosides Rb1, Rg3, Rg5 and Rk1, WG and HRG80TM at Different Concentrations on Gene Expression Profile in the Hippocampal Neuronal Cell Line HT22
Table 1 shows the number of genes deregulated (>20 fold compared to control) by ginsenosides Rb1, Rg3, Rg5 and Rk1, WG and HRG80TM at different concentrations in the hippocampal neuronal cell line HT22, which in the same range—from 283 to 461 at all tested concentrations of HRG80TM and within 20% RSD of the mean value 383 ± 77 (Supplementary Table S1 in Supplement S1).

Table 1.
The number of genes deregulated (>20 fold compared to controls) by ginsenosides Rb1, Rg3, Rg5 and Rk1, WG and HRG80TM at different concentrations in the hippocampal neuronal cell line HT22.
The number of genes deregulated by WG and purified ginsenosides is also within this range, despite the difference in the content of ginsenosides Rb1, Rg3, Rg5, and Rk1 in WG and HRG80TM (Table 1). However, the gene expression profile (signature) was substance-specific and concentration-specific, containing many genes that are not deregulated in other concentrations or other test samples alone (Figure 1, Table 1, and Supplementary Table S2 in Supplement S2).
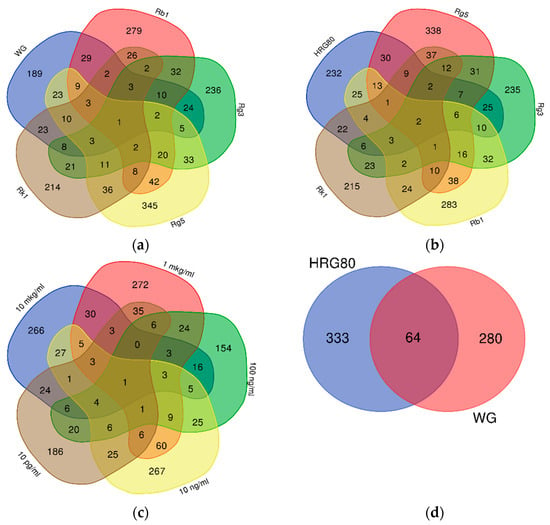
Figure 1.
Venn diagrams of: (a) the number of genes deregulated by WG and ginsenosides Rb1, Rg3, Rg5 and Rk1, (b) the number of genes deregulated by HRG and ginsenosides Rb1, Rg3, Rg5 and Rk1, (c) the number of genes deregulated by HRG at concentrations of 10 μg/mL, 1 μg/mL, 100 ng/mL, 10 ng/mL, and 0.01 ng/mL; (d) the number of genes deregulated by WG and HRG in the hippocampal neuronal cell line HT22.
Only one gene, SUOX, encoding mitochondrial sulfite oxidase, was commonly deregulated by all purified ginsenosides, WG, and HRG80TM (Figure 1a,b), and the KRTAP 10-7 gene in all tested concentrations of HRG80TM and total extract HRG80TM (Figure 1c).
Deregulated genes expression profiles (signatures) of WG and HRG80TM were also significantly different; only 64 of 398 and 344 genes deregulated by HRG80TM (16% of total) and WG (18% of total), respectively, were the same (Figure 1d).
These observations demonstrate that red ginseng preparation HRG80TM significantly impacts the gene expression of hippocampal neurons in a wide range of concentrations, from 1 μg/mL to 0.01 ng/mL.
These observations also reveal that the gene expression profile of hippocampal neurons is specific for every purified ginsenoside and is quite different from red ginseng HRG80TM and WG preparations.
2.2. Effect of Ginsenoside Rg5 on Signaling Canonical Pathways
Figure 2 show the predicted effects (−log p-value > 1.3, z-score > 2) of red and white ginseng extracts (HRG80TM and WG) and their major constituents ginsenosides Rb1, Rg3, Rg5, and Rk1 on canonical signaling pathways, including inhibition of:
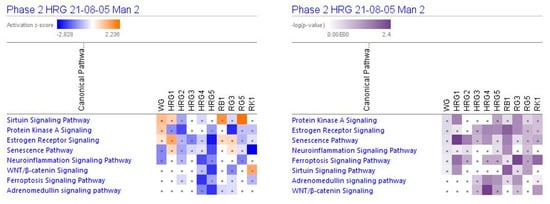
Figure 2.
Effects of ginseng extracts WG (10 μg/mL), HRG at concentrations of 10 μg/mL (HRG1), 1 μg/mL (HRG2), 100 ng/mL (HRG3), 10 ng/mL (HRG4), and 10 pg/mL (HRG5), and ginsenosides Rb1, Rg3, Rg5, Rk1 at a concentration 100 nM on selected canonical pathways. The brown color shows the predicted activation and the blue color the predicted inhibition of diseases; symbol · shows that the activation z-score was < 2 and a -log p-value < 1.3 = p < 0.05. An absolute z-score ≥ 2 is considered significant activation (+) or inhibition (−). The activation z-score predicts the activation state of the canonical pathway, using the gene expression patterns of the genes within the pathway. The p-value calculated using a right-tailed Fisher’s exact test indicates the statistical significance of the overlap of analyzed dataset genes that are within a pathway.
- neurotransmitters and nervous system signaling involved in neuroinflammation signaling (Figure 3);
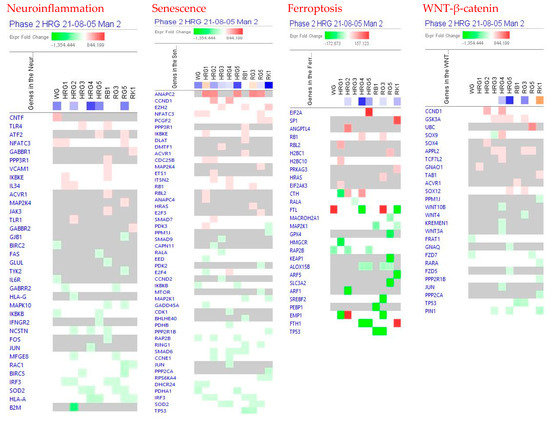 Figure 3. Effects of WG at a concentration of 10 μg/mL, HRG at concentrations of 10 μg/mL (HRG1), 1 μg/mL (HRG2), 100 ng/mL (HRG3), 10 ng/mL (HRG4), and 10 pg/mL (HRG5), and ginsenosides Rb1, Rg3, Rg5 and Rk1 at a concentration of 100 nM on neuroinflammation, senescence, ferroptosis, and WNT—β-catenin signaling pathways. The heatmap of gene expression (in fold-changes compared to controls; red—upregulation, green—downregulation), after exposure of cells with test samples at different concentrations; the column represents signatures of test samples with solid red or green squares indicating genes that are expected to be upregulated or downregulated, respectively; color intensity indicates the actual log-fold changes.
Figure 3. Effects of WG at a concentration of 10 μg/mL, HRG at concentrations of 10 μg/mL (HRG1), 1 μg/mL (HRG2), 100 ng/mL (HRG3), 10 ng/mL (HRG4), and 10 pg/mL (HRG5), and ginsenosides Rb1, Rg3, Rg5 and Rk1 at a concentration of 100 nM on neuroinflammation, senescence, ferroptosis, and WNT—β-catenin signaling pathways. The heatmap of gene expression (in fold-changes compared to controls; red—upregulation, green—downregulation), after exposure of cells with test samples at different concentrations; the column represents signatures of test samples with solid red or green squares indicating genes that are expected to be upregulated or downregulated, respectively; color intensity indicates the actual log-fold changes. - senescence, ferroptosis, adrenomedullin, and WNT/β-catenin signaling—canonical pathways involved in cellular stress response, injury, and cancer progression (Figure 3 and Figure 4);
 Figure 4. Effects of WG at a concentration of 10 μg/mL, HRG at concentrations of 10 μg/mL (HRG1), 1 μg/mL (HRG2), 100 ng/mL (HRG3), 10 ng/mL (HRG4), and 10 pg/mL (HRG5), and ginsenosides Rb1, Rg3, Rg5 and Rk1 at a concentration of 100 nM on adrenomedullin, PKA, sirtuin and estrogen receptor-signaling pathways. The heatmap of gene expression (in fold-changes compared to control, red—upregulation, green—downregulation), after exposure of cells with test samples at different concentrations; the column represents signatures of test samples with solid red or green squares indicating genes that are expected to be upregulated or downregulated, respectively; color intensity indicates the actual log-fold changes.
Figure 4. Effects of WG at a concentration of 10 μg/mL, HRG at concentrations of 10 μg/mL (HRG1), 1 μg/mL (HRG2), 100 ng/mL (HRG3), 10 ng/mL (HRG4), and 10 pg/mL (HRG5), and ginsenosides Rb1, Rg3, Rg5 and Rk1 at a concentration of 100 nM on adrenomedullin, PKA, sirtuin and estrogen receptor-signaling pathways. The heatmap of gene expression (in fold-changes compared to control, red—upregulation, green—downregulation), after exposure of cells with test samples at different concentrations; the column represents signatures of test samples with solid red or green squares indicating genes that are expected to be upregulated or downregulated, respectively; color intensity indicates the actual log-fold changes. - intracellular second messenger c-AMP mediated protein kinase A signaling (Figure 4);
- transcriptional regulation of sirtuin and nuclear receptors, and estrogen receptor-mediated signaling (Figure 4).
The effects of ginseng extracts WG and HRG, and ginsenosides Rb1, Rg3, Rg5, and Rk1 on gene expression involved in these signaling pathways are shown in Supplements S3–S10 in detail.
2.3. Predicted Effects of Ginsenosides and Ginseng Extracts HRG80TM and WG
A large proportion (about 75%) of deregulated genes consisted of networks that are significantly associated with cancer, gastrointestinal, and endocrine systems diseases and disorders (Table 2 and Figure 5).

Table 2.
The number of genes associated with various diseases and deregulated by ginseng extracts HRG80TM and WG, and ginsenosides Rb1, Rg3, Rg5.
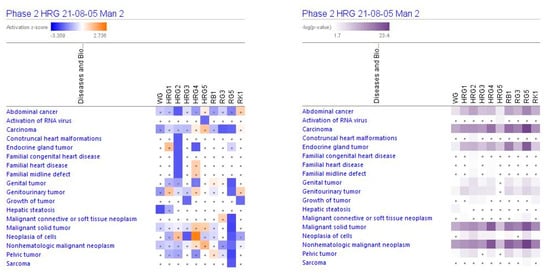
Figure 5.
Effects of ginseng extracts WG (10 μg/mL), HRG80TM at concentrations of 10 μg/mL (HRG1), 1 μg/mL (HRG2), 100 ng/mL (HRG3), 10 ng/mL (HRG4), and 10 pg/mL (HRG5), and ginsenosides Rb1, Rg3, Rg5, Rk1 at a concentration 100 nM on diseases. Disease scores are presented using a gradient from dark blue to brown for predicted activation and light to dark blue for predicted inhibition of diseases; symbol · shows that the activation z-score was <2 and a -log p-value < 1.3 = p < 0.05. An absolute z-score ≥ 2 is considered significant activation (+) or inhibition (−).
IPA analysis shows diseases and bio functions (Table 3) that are expected to be significantly (activation score z > [±2],−log p-value) > 1.5) correlated to ginseng extracts HRG80TM and WG, and ginsenosides Rb1, Rg3, Rg5. HRG80TM is expected to inhibit endocrine gland tumors, abdominal cancer and neoplasm, necrosis, connective tissue cell death, and heart disease at a concentration of 1 μM.

Table 3.
Predicted effects of ginseng extracts HRG80TM and WG, and ginsenosides Rb1, Rg3, Rg5 in various diseases and biofunctions expressed by activation z-score values. Positive values show predicted activation; negative values showpredicted inhibition.
At lower concentrations (100 or 10 ng/mL), HRG80TM may inhibit neoplasia of cells, invasion of breast cancer cell lines, and growth of tumors and endocrine gland tumors, while the highest concentration of 10 μg/mL may inhibit necrosis and proliferation of leukemia cell lines.
WG is expected to inhibit hepatic stenosis and abdominal neoplasm at the same concentration of 10 μg/mL. Antitumor activity profiles of purified ginsenosides are different from ginseng total extracts, where the most polyvalent is Rg5, which is expected to inhibit genital and genitourinary tumors, malignant neoplasm and solid tumors, neoplasia of cells, pelvic tumors, carcinomas, and sarcomas (Table 3 and Figure 5).
Expected inhibition of endocrine gland tumors at two different concentrations of 1 μg/mL and 10 pg/mL (Figure 6) is associated with two distinct networks (Figure 6a,b) including quite different sets of molecules, where 72 genes are commonly deregulated at these concentrations, while 250 and 251 are unique for each of them, respectively (Figure 6c and Supplementary Table S3 in Supplement S2). IPA analysis of gene expression of neurons shows predicted inhibition of endocrine tumors at both a concentration of HRG80 of 10 ng/mL (HRG4) and of 1000 ng/mL (HRG2). The same canonical pathways were inhibited in both concentrations; however, these effects were mediated by quite different sets of deregulated genes. Figure 5 shows only 72 genes involved in inhibiting endocrine tumors deregulated at both concentrations of HRG. In addition, 260 genes contribute to the overall anti-tumor effect at a concentration of 1000 ng/mL, and 251 other genes at a concentration of 10 ng/mL. Since the content of Rg5 comprises 7.534% of the extract, the concentration of Rg5 in cell culture incubation media is 75.3 ng/mL (98 nM) and 0.75 ng/mL (0.98 nM), respectively (Table 1). This is in line with the results of pharmacokinetic studies in humans, where the max level of Rg5 in blood was found to be 8 ng/mL.
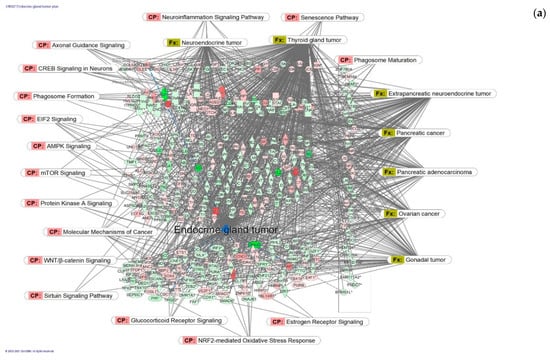
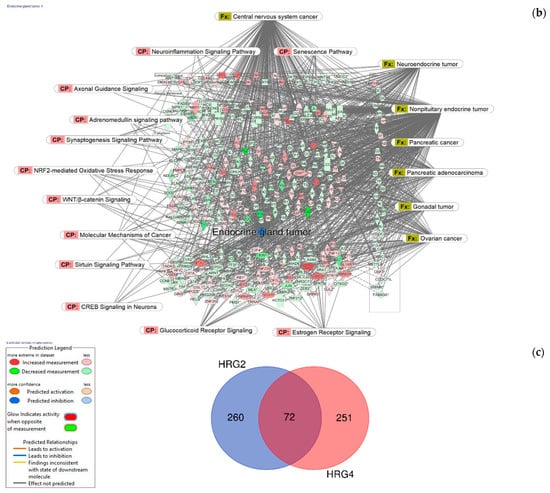
Figure 6.
Molecular network shows predicted inhibition of exocrine gland tumor by HRG80TM at concentrations of: (a) 1 μg/mL and (b) 10 pg/mL. Solid red or green color nodes indicate genes that are up-regulated and down-regulated, respectively; color intensity indicates the actual log-fold changes. The tags labeled with purple display the canonical pathways related to particular genes of the network. The tags labeled with khaki show various types of tumors associated with the molecules in subnetworks. (c) Venn diagram showing the number of concentration-specific and commonly deregulated (72) genes at concentrations of 1 μg/mL and 10 pg/mL.
Similarly, expected inhibition of abdominal neoplasm by red HRG2 and white WG ginseng is associated with two distinct networks (Figure 7a,b) including quite different sets of molecules, where 54 genes are commonly deregulated by HRG or WG, while 407 and 290 are unique for each ginseng preparation, respectively (Figure 7c).
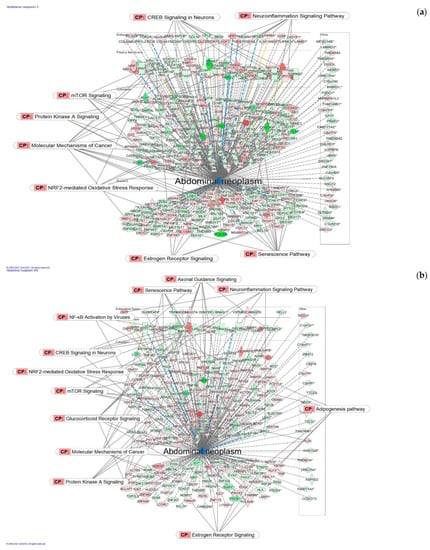
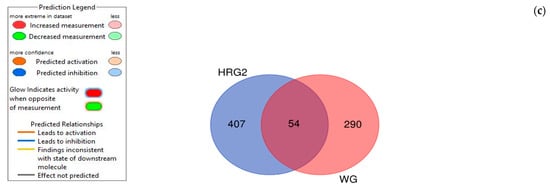
Figure 7.
Molecular network shows predicted inhibition of abdominal neoplasm by: (a) HRG80TM at a concentration of 1 μg/mL and (b) WG at a concentration of 10 μg/mL. Solid red or green color nodes indicate upregulated and downregulated genes, respectively; color intensity indicates the actual log-fold changes. The tags labeled with purple display the canonical pathways related to particular genes of the network. The tags labeled with khaki show various types of tumors associated with the molecules in subnetworks. (c) Venn diagram showing the number of product-specific and commonly deregulated (54) genes.
3. Discussion
This study’s primary aim was to utilize transcriptomics of neuronal cells to uncover potential pharmacological activities and indications for use in medicine hydroponically cultivated Panax ginseng root preparation HRG80TM (HRG). This task is closely associated with exploring HRG at reasonable in vitro concentrations matching the concentrations of ginsenosides found in in vivo studies on human subjects.
Our study provides evidence that HRG is pharmacologically active at the concentrations 10, 100, 1000, and 10,000 ng/mL, corresponding to the concentrations of ginsenosides (from 0.05 to 35 nM, equivalent to 5–7000 ng/mL, assuming that the content of ginsenoside Rg5 is 2% in the HRG80TM, Supplement S1) detected in the blood of human subjects orally administered with red ginseng preparations in therapeutic doses [29].
At all tested concentrations, HRG deregulated genes significantly associated with cancer, gastrointestinal, and endocrine system diseases and disorders (Table 2 and Figure 4).
However, the expected (based on IPA analysis results) pharmacological profile and possible indications for use are different for both WG and HRG, as well as for various doses of HRG in blood. HRG80TM has the potential to inhibit endocrine gland tumors, abdominal cancer and neoplasm, necrosis, connective tissue cell death, and heart disease at blood concentrations of 1 μM, while at other concentrations, HRG80TM has the potential to inhibit neoplasia of cells, invasion of breast cancer cell lines, growth of tumors and endocrine gland tumors, and necrosis and proliferation of leukemia cell lines. On the other hand, WG can presumably inhibit hepatic stenosis, which is not expected with HRG.
These conclusions are in line with other reports on the antitumor activity of ginseng and purified ginsenosides [19,23,25,26,36,39,40,41,42,43,44].
Meanwhile, antitumor activity profiles of purified ginsenosides are different from ginseng total extracts (Table 3). The results obtained for purified ginsenosides cannot be simply extrapolated on the total extract or powdered roots, which exhibit quite different pharmacological and therapeutic activities.
The mechanism of action of HRG80TM is multitargeted and associated with stress-induced abnormalities, neuroinflammation, and senescence, and other signaling pathways (Figure 3, Figure 4, Figure 5 and Figure 6) playing an important role in these physiological and cellular processes, including apoptosis, tumorigenesis, and progression of cancer, which is typical for adaptogens [8,45,46].
Overall, the results of this study are in line both with the traditional use of ginseng in aging and the theory in which low-grade chronic inflammation (inflammaging) that develops with senescence plays a crucial role in the progression of aging-related diseases—primarily cancer [47,48,49]—, and in line with the adaptogen concept, suggesting efficacy of adaptogens in stress-induced aging-related diseases [8,50,51] (Figure 8).
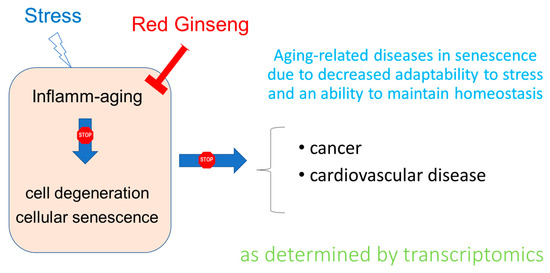
Figure 8.
Hypothetical scheme of the effect of Ginseng on aging-related diseases as determined by transcriptomics.
There are several constraints in this study. One of them is the lack of scientific literature about the direction (positive or negative) of correlations between gene expression and physiological function or disease for predicting effects of some experimental findings used in silico analysis. The second is related to the number of concentration points in the dose–response correlation study; more intermediate points over the 10-fold difference range will show smooth changes from point to point. Based on the results of the IPA analysis, we do not have yet explanations for observations of differential regulation of gene expression profiles at different concentrations of ginseng extracts or individual ginsenosides. More mechanistic studies at multiple concentration points within narrower concentration ranges are required to elucidate the regulatory mechanisms, which, clearly, are multi-targeted, multilevel, upstream, and downstream feedback-regulated, resulting in maintaining of intracellular and extracellular homeostasis.
Overall, the effects of total extracts on gene expression profiles are different from the effects of purified ginsenosides in all tested concentrations; however, the predicted effects on signaling pathways, biological functions, and diseases are similar, probably due to adaptive compensatory regulation of alternative signaling molecules of molecular networks.
Further preclinical and clinical studies are required to assess the therapeutic efficacy of HRG80TM in different types of endocrine tumors and abdominal cancers.
Overall, this is the first evidence of pharmacological activity of ginseng preparations at concentrations found in the blood of human subjects after oral administration in therapeutic doses.
4. Materials and Methods
All materials and methods used in the present study have been described in detail in our previously published studies [52,53]. Therefore, only a short description of herbal extracts, mRNA microarray hybridization, and ingenuity pathway analysis (IPA) is provided below.
4.1. Test Samples and Reference Standard
Powdered red ginseng preparation HRG80TM and its extract (HRG) were obtained at Botalys S. A. (Ath, Belgium). Harvested roots were air-dried and steamed to red ginseng. The red ginseng HRG80TM preparation was standardized for the content of the ginsenosides Rg1, Re, Rf, Rb1, Rg2, Rc, Rh1, Rb2, F1, Rd, Rg6, F2, Rh4, Rg3-(S-R), PPT (20-R), Rk1, C(k), Rg5, Rh2, Rh3, 20S-PPT, and PPD (Supplement S11).
Powdered red ginseng preparation HRG80TM and its extract (HRG) were obtained at Botalys S. A. (Ath, Belgium). Korean Ginseng (P. ginseng Meyer) root was hydroponically cultivated in controlled conditions, air-dried, steamed to red ginseng, which was powdered, and standardized for the content of ginsenosides Rg1, Re, Rf, Rb1, Rg2, Rc, Rh1, Rb2, F1, Rd, Rg6, F2, Rh4, Rg3-(S-R), PPT (20-R), Rk1, C(k), Rg5, Rh2, Rh3, 20S-PPT, and PPD to obtain red ginseng HRG80TM preparation containing 7.6% of total ginsenosides (Supplemental Table S4 in Supplement S11). HRG80TM preparation was exhaustively extracted by 40% ethanol and evaporated to dryness to obtain HRG extract (DER 4: 1) used for in vitro experiments. The content of ginsenosides in the HRG extract was 30.32% (Supplement S11).
The reference standard, P. ginseng Meyer powdered root preparation, and the extract contained 5.57% and 22.15% total ginsenosides, respectively (Supplemental Table S4 in Supplement S11). All herbal preparations were analyzed and certified by Botalys S. A.
Purified reference standards of ginsenosides were purchased from Merck https://www.sigmaaldrich.com/, accessed on 27 September 2021.
4.2. mRNA Microarray Hybridization
After seeding, we let hippocampal neuronal HT22 cells of mouse origin grow for 24 h before using them for treatment with test compounds. Following this, the cells were subjected to different drug concentrations and combinations or DMSO as a solvent control (0.5%) for another 24 h. An InviTrap® Spin Universal RNA mini kit (250 preps; Stratec Molecular) was used for RNA isolation. A total RNA Nanochip assay was applied on an Agilent 2100 bioanalyzer (Agilent Technologies GmbH) for quality control of RNA, and an RNA index value of >8.5 served as a threshold for further processing of RNA samples. For every two independent experiments, quality control of RNA was performed. The mRNA microarray hybridization was performed at the Genomics and Proteomics Core Facility, German Cancer Research Center (Heidelberg, Germany) using Affymetrix GeneChips® with mouse Clariom S assays. The measurements were normalized by a quantile normalization algorithm without background subtraction. The standard deviation differences were calculated in one-by-one comparisons to identify differentially regulated genes. Chipster software (The Finnish IT Center for Science CSC) was used for further evaluation of results.
4.3. Ingenuity Pathway Analysis (IPA)
The interpretation of microarray data and gene expression changes was performed using ingenuity pathway analysis (IPA) software, summer release 2021 (QIAGEN Bioinfor-matics, Aarhus C, Denmark), which relies on the Ingenuity Knowledge Base, a continuously updated database that gathers research findings, with more than 8.1 million findings manually curated from the biomedical literature or integrated from 45 third-party databases. The IPA network contains 40,000 nodes representing mammalian genes, molecules, and biologic functions, linked by 1,480,000 edges representing experimentally observed cause–effect relationships (either activating or inhibiting) related to gene expression, transcription, activation, molecular metabolism, and binding [54].
Using the IPA Core Analysis tool for all tested transcriptomic datasets, we performed predictive analyses of the impact of test samples on canonical signaling and metabolic pathways, which displayed the molecules of interest within well-established pathways; and diseases, disorders, molecular and cellular functions that are activated or inhibited downstream and upstream of the genes, whose expression has been altered.
4.4. Statistical Analysis
Two statistical methods of analysis of gene expression datasets were used in IPA: (i) the gene-set-enrichment method, where differentially expressed genes are intersected with sets of genes that are associated with a particular biological function or pathway, providing an ‘enrichment’ score (Fisher’s exact test p-value) that measures overlap of observed and predicted regulated gene sets [55,56], (ii) a method based on cause–effect relationships related to the direction of effects reported in the literature, that provides a so-called z-score that measures the match of observed and predicted up/down-regulation [55,56,57,58]. The predicted effects are based on gene expression changes in the experimental samples relative to the control; z-score > 2, -log p-value > 1.3.
5. Conclusions
In this study, we, for the first time, have demonstrated that the gene expression profile of murine hippocampal neuronal HT22 cells is changed significantly in response to exposure of hydroponically cultivated red ginseng preparation HRG80TM in concentrations ranging from 10 μg/mL to 0.01 ng/mL. This is in line with concentrations of ginsenosides Rb1, Rg3, Rg5, and Rk1 found in in vivo studies of human subjects taking orally therapeutic doses of red ginseng. Purified ginsenosides Rb1, Rg3, Rg5, and Rk1 have substantially varied effects on gene expression profiles (signatures) and predicted pharmacological activities, as determined by in silico analysis of transcriptomics. Their signatures are different from the signatures and expected therapeutic indications of red ginseng HRG80TM and white ginseng preparations, which are also quite varied.
Comparative in silico transcriptome analysis of microarray-based gene expression profiles of neuronal cells exposed to red and white ginseng extracts and their major constituents ginsenosides Rb1, Rg3, Rg5, and Rk1 predicts a potential beneficial effect in neuroinflammation, senescence, and cancer, gastrointestinal, and endocrine system diseases and disorders. HRG80TM has the potential to inhibit endocrine gland tumors, abdominal cancers and neoplasm, necrosis, connective tissue cell death, and heart disease at blood concentrations of 1 μg/mL.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph14101010/s1, Supplementary data S1–S11.
Author Contributions
S.A. contributed to the execution of experiments, data management, and manuscript preparation. T.E. contributed to the overall management of the study and manuscript preparation. A.P. contributed to the study conception, design and evaluation, data analysis, interpretation of the results, and drafting and preparation of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Europharma USA (Green Bay, WI, USA) and Botalys SA (Ath, Belgium). The research sponsors were Terrence Lemerond, Europharma USA Inc., USA; Pierre-Antoine Mariage and Paul-Evence Coppee, Botalys SA, Belgium (grant number 2019-03).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained in the article and Supplementary Materials.
Acknowledgments
The author acknowledges the support of Botalys SA for their supply of investigational products and their characterization. Quality control was performed by Pierre-Antoine Mariage and Sylvie Defrère, Botalys SA, Belgium. We thank the Microarray Unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing excellent expression profiling services.The authors are grateful to Professor Rer. Nat. Reiner Kunze (Institute of Physiology and Pathophysiology, Department of Medicine, Heidelberg University) for the generous gift of the murine neuronal cell line HT-22.
Conflicts of Interest
A.P. has an independent contractor agreement with EuroPharmaUSA Inc. All other authors declare no competing interests. The funders had no role in the study’s design, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Lee, S.M.; Bae, B.-S.; Park, H.-W.; Ahn, N.-G.; Cho, B.-G.; Cho, Y.-L.; Kwak, Y.-S. Characterization of Korean red ginseng (Panax ginseng Meyer): History, preparation method, and chemical composition. J. Ginseng Res. 2015, 39, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Flagg, A.J. Traditional and current use of ginseng. Nurs. Clin. N. Am. 2021, 56, 109–121. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Youn, S.H.; Kwak, Y.-S.; Han, C.-K.; Haidere, M.F.; Kim, J.K.; Min, H.; Jung, Y.-J.; Hosseinzadeh, H.; Hyun, S.H.; et al. Adaptogenic effects of Panax ginseng on modulation of immune functions. J. Ginseng Res. 2020, 45, 32–40. [Google Scholar] [CrossRef]
- Panossian, A.; Brendler, T. The Role of Adaptogens in Prophylaxis and Treatment of Viral Respiratory Infections. Pharmaceuticals 2020, 13, 236. [Google Scholar] [CrossRef]
- Irfan, M.; Kwak, Y.-S.; Han, C.-K.; Hyun, S.H.; Rhee, M.H. Adaptogenic effects of Panax ginseng on modulation of cardiovascular functions. J. Ginseng Res. 2020, 44, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.-Y.; He, Y.-F.; Li, L.; Meng, H.; Dong, Y.-M.; Yi, F.; Xiao, P.-G. A preliminary review of studies on adaptogens: Comparison of their bioactivity in TCM with that of ginseng-like herbs used worldwide. Chin. Med. 2018, 13, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.; Rauf, A. Adaptogenic herb ginseng (Panax) as medical food: Status quo and future prospects. Biomed. Pharmacother. 2016, 85, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Panossian, A.G.; Efferth, T.; Shikov, A.N.; Pozharitskaya, O.N.; Kuchta, K.; Mukherjee, P.K.; Banerjee, S.; Heinrich, M.; Wu, W.; Guo, D.; et al. Evolution of the adaptogenic concept from traditional use to medical systems: Pharmacology of stress- and aging-related diseases. Med. Res. Rev. 2020, 41, 630–703. [Google Scholar] [CrossRef] [PubMed]
- EMA/HMPC/321232/2012. Assessment Report on Panax Ginseng C.A. Meyer, Radix. Based on Article 16d (1), Article 16f and Article 16h of Directive 2001/83/EC as Amended (Traditional Use); European Medicines Agency: Amsterdam, The Netherlands, 2014. [Google Scholar]
- EMA/HMPC/321232/2012. Community Herbal Monograph on Panax Ginseng C.A. Meyer; Radix European Medicines Agency: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Kim, H.J.; Jung, S.W.; Kim, S.Y.; Cho, I.H.; Kim, H.C.; Rhim, H.; Kim, M.; Nah, S.Y. Panax ginseng as an adjuvant treatment for Alzheimer’s disease. J. Ginseng Res. 2018, 42, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.J.; Kim, S.K.; Lee, N.Y.; Choi, Y.R.; Kim, H.S.; Gupta, H.; Youn, G.S.; Sung, H.; Shin, M.J.; Suk, K.T. Effect of Korean red ginseng on metabolic syndrome. J. Ginseng Res. 2020, 45, 380–389. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kim, A.K. Anti-breast cancer activity of Fine Black ginseng (Panax ginseng Meyer) and ginsenoside Rg5. J. Ginseng Res. 2014, 39, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Bilia, A.R.; Bergonzi, M.C. The G115 standardized ginseng extract: An example for safety, efficacy, and quality of an herbal medicine. J. Ginseng Res. 2019, 44, 179–193. [Google Scholar] [CrossRef]
- Hou, W.; Wang, Y.; Zheng, P.; Cui, R. Effects of Ginseng on Neurological Disorders. Front. Cell. Neurosci. 2020, 14, 55. [Google Scholar] [CrossRef] [Green Version]
- Mariage, P.A.; Hovhannisyan, A.; Panossian, A.G. Efficacy of Panax ginseng Meyer herbal preparation HRG80 in preventing and mitigating stress-induced failure of cognitive functions in healthy subjects: A pilot, randomized, double-blind, placebo-controlled crossover trial. Pharmaceuticals 2020, 13, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimpfel, W.; Mariage, P.-A.; Panossian, A. Effects of Red and white ginseng Preparations on Electrical Activity of the Brain in Elderly Subjects: A Randomized, Double-Blind, Placebo-Controlled, Three-Armed Cross-Over Study. Pharmaceuticals 2021, 14, 182. [Google Scholar] [CrossRef] [PubMed]
- Dimpfel, W.; Schombert, L.; Panossian, A.G. Panax ginseng preparations enhance long term potentiation in rat hippocampal slices by glutamatergic NMDA and kainate receptor mediated transmission. J. Altern. Complementary Integr. Med. 2020, 6, 106. [Google Scholar]
- Chen, C.; Lv, Q.; Li, Y.; Jin, Y.-H. The Anti-Tumor Effect and Underlying Apoptotic Mechanism of Ginsenoside Rk1 and Rg5 in Human Liver Cancer Cells. Molecules 2021, 26, 3926. [Google Scholar] [CrossRef]
- Zhu, G.-X.; Zuo, J.-L.; Xu, L.; Li, S.-Q. Ginsenosides in vascular remodeling: Cellular and molecular mechanisms of their therapeutic action. Pharmacol. Res. 2021, 169, 105647. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, M.; Ling, C.; Zhu, Y.; Ren, H.; Hong, C.; Qin, J.; Liu, T.; Wang, J. Neuroprotective Effects of Ginsenosides against Cerebral Ischemia. Molecules 2019, 24, 1102. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.Y.; Kim, K.J.; Song, J.H.; Lee, B.Y. Ginsenoside Rg5 prevents apoptosis by modulating heme-oxygenase-1/nuclear factor E2-related factor 2 signaling and alters the expression of cognitive impairment-associated genes in thermal stress-exposed HT22 cells. J Ginseng Res. 2018, 42, 225–228. [Google Scholar] [CrossRef]
- Ahuja, A.; Kim, J.H.; Kim, J.-H.; Yi, Y.-S.; Cho, J.Y. Functional role of ginseng-derived compounds in cancer. J. Ginseng Res. 2017, 42, 248–254. [Google Scholar] [CrossRef]
- Park, S.K.; Hyun, S.H.; In, G.; Park, C.-K.; Kwak, Y.-S.; Jang, Y.-J.; Kim, B.; Kim, J.-H.; Han, C.-K. The antioxidant activities of Korean red ginseng (Panax ginseng) and ginsenosides: A systemic review through in vivo and clinical trials. J. Ginseng Res. 2020, 45, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Z.; Anderson, S.; Du, W.; He, T.-C.; Yuan, C.-S. Red ginseng and cancer treatment. Chin. J. Nat. Med. 2016, 14, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, P.; Subramaniyam, S.; Mathiyalagan, R.; Yang, D.-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J. Ginseng Res. 2017, 42, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Kim, T.-H.; Choi, T.-Y.; Lee, M.S. Ginseng for Health Care: A Systematic Review of Randomized Controlled Trials in Korean Literature. PLoS ONE 2013, 8, e59978. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.P.; Behm, B. Ginseng: A Qualitative Review of Benefits for Palliative Clinicians. Am. J. Hosp. Palliat. Med. 2019, 36, 630–659. [Google Scholar] [CrossRef]
- Yoo, S.; Park, B.-I.; Kim, D.-H.; Lee, S.; Lee, S.-H.; Shim, W.-S.; Seo, Y.; Kang, K.; Lee, K.-T.; Yim, S.-V.; et al. Ginsenoside Absorption Rate and Extent Enhancement of Black Ginseng (CJ EnerG) over red ginseng in Healthy Adults. Pharmaceutics 2021, 13, 487. [Google Scholar] [CrossRef]
- Kim, H.-K. Pharmacokinetics of ginsenoside Rb1 and its metabolite compound K after oral administration of Korean red ginseng extract. J. Ginseng Res. 2013, 37, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Cui, R.; Zhao, L.; Fan, J.; Li, B. Mechanisms of Panax ginseng action as an antidepressant. Cell Prolif. 2019, 52, e12696. [Google Scholar] [CrossRef] [Green Version]
- Nocerino, E.; Amato, M.; Izzo, A. The aphrodisiac and adaptogenic properties of ginseng. Fitoterapia 2000, 71, S1–S5. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Park, J.-H.; Kim, H.-S.; Lee, C.-Y.; Lee, H.-J.; Kang, K.S.; Kim, C.-E. Systems-level mechanisms of action of Panax ginseng: A network pharmacological approach. J. Ginseng Res. 2017, 42, 98–106. [Google Scholar] [CrossRef]
- Ding, Q.; Zhu, W.; Diao, Y.; Xu, G.; Wang, L.; Qu, S.; Shi, Y. Elucidation of the Mechanism of Action of Ginseng Against Acute Lung Injury/Acute Respiratory Distress Syndrome by a Network Pharmacology-Based Strategy. Front. Pharmacol. 2021, 11, 611794. [Google Scholar] [CrossRef]
- Li, Q.-Y.; Hou, C.-Z.; Yang, L.-P.; Chu, X.-L.; Wang, Y.; Zhang, P.; Zhao, Y. Study on the Mechanism of Ginseng in the Treatment of Lung Adenocarcinoma Based on Network Pharmacology. Evid. Based Complement. Altern. Med. 2020, 2020, 2658795. [Google Scholar] [CrossRef]
- Jeong, E.; Lim, Y.; Kim, K.J.; Ki, H.-H.; Lee, D.; Suh, J.; So, S.-H.; Kwon, O.; Kim, J.Y. A Systems Biological Approach to Understanding the Mechanisms Underlying the Therapeutic Potential of red ginseng Supplements against Metabolic Diseases. Molecules 2020, 25, 1967. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wang, P.; Yang, W.; Zhao, C.; Zhang, L.; Zhang, J.; Qin, Y.; Xu, H.; Huang, L. Characterization of the Components and Pharmacological Effects of Mountain-Cultivated Ginseng and Garden Ginseng Based on the Integrative Pharmacology Strategy. Front. Pharmacol. 2021, 12, 659954. [Google Scholar] [CrossRef]
- Park, J.-M.; Lee, H.-J.; Yoo, J.H.; Ko, W.J.; Cho, J.Y.; Hahm, K.B. Overview of gastrointestinal cancer prevention in Asia. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 855–867. [Google Scholar] [CrossRef]
- Kim, J.W.; Han, S.W.; Cho, J.Y.; Chung, I.-J.; Kim, J.G.; Lee, K.H.; Park, K.U.; Baek, S.K.; Oh, S.C.; Lee, M.A.; et al. Korean red ginseng for cancer-related fatigue in colorectal cancer patients with chemotherapy: A randomised phase III trial. Eur. J. Cancer 2020, 130, 51–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bespalov, V.G.; Alexandrov, V.A.; Semenov, A.L.; Kovan’Ko, E.G.; Ivanov, S.D. Anticarcinogenic activity of alpha-difluoromethylornithine, ginseng, eleutherococcus, and leuzea on radiation-induced carcinogenesis in female rats. Int. J. Radiat. Biol. 2014, 90, 1191–1200. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, M.-K.; Lee, M.; Kwon, B.-S.; Suh, D.H.; Song, Y.S. Effect of red ginseng on Genotoxicity and Health-Related Quality of Life after Adjuvant Chemotherapy in Patients with Epithelial Ovarian Cancer: A Randomized, Double Blind, Placebo-Controlled Trial. Nutrients 2017, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, I.-K.; Lee, K.Y.; Kang, J.; Park, J.S.; Jeong, J. Immune-modulating Effect of Korean red ginseng by Balancing the Ratio of Peripheral T Lymphocytes in Bile Duct or Pancreatic Cancer Patients With Adjuvant Chemotherapy. In Vivo 2021, 35, 1895–1900. [Google Scholar] [CrossRef]
- Oh, J.; Bin Jeon, S.; Lee, Y.; Lee, H.; Kim, J.; Kwon, B.R.; Kyung-Min, C.; Cha, J.-D.; Hwang, S.-M.; Choi, K.-M.; et al. Fermented red ginseng Extract Inhibits Cancer Cell Proliferation and Viability. J. Med. Food 2015, 18, 421–428. [Google Scholar] [CrossRef]
- Panossian, A.; Seo, E.-J.; Efferth, T. Novel molecular mechanisms for the adaptogenic effects of herbal extracts on isolated brain cells using systems biology. Phytomedicine 2018, 50, 257–284. [Google Scholar] [CrossRef]
- Panossian, A. Understanding adaptogenic activity: Specificity of the pharmacological action of adaptogens and other phytochemicals. Ann. N. Y. Acad. Sci. 2017, 1401, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Fulop, T.; Witkowski, J.M.; Olivieri, F.; Larbi, A. The integration of inflammaging in age-related diseases. Semin. Immunol. 2018, 40, 17–35. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Accardi, G.; Monastero, R.; Nicoletti, F.; Libra, M. Ageing: From inflammation to cancer. Immun. Ageing 2018, 15, 1. [Google Scholar] [CrossRef]
- Zuo, L.; Prather, E.R.; Stetskiv, M.; Garrison, D.E.; Meade, J.R.; Peace, T.I.; Zhou, T. Inflammaging and Oxidative Stress in Human Diseases: From Molecular Mechanisms to Novel Treatments. Int. J. Mol. Sci. 2019, 20, 4472. [Google Scholar] [CrossRef] [Green Version]
- Panossian, A.; Gerbarg, P. Potential use of plant adaptogens in age-related disorders. In Complementary, Alternative, and Integrative Interventions in Mental Health and Aging; Lavretsky, H., Sajatovic, M., Reynolds, C.F., III, Eds.; Oxford University Press: New York, NY, USA, 2016; pp. 197–211. [Google Scholar]
- Panossian, A.; Wikman, G. Evidence based efficacy and effectiveness of Rhodiola SHR-5 extract in treating stress- and age-associated disorders. In Rhodiola rosea, Series: Traditional Herbal Medicines for Modern Times; Cuerrier, A., Ampong-Nyarko, K., Eds.; CRC Press Taylor; Francis Group: Boca-Raton, FL, USA; London, UK; New York, NY, USA, 2014; pp. 203–221. [Google Scholar]
- Seo, E.-J.; Klauck, S.M.; Efferth, T.; Panossian, A. Adaptogens in chemobrain (Part I): Plant extracts attenuate cancer chemotherapy-induced cognitive impairment—Transcriptome-wide microarray profiles of neuroglia cells. Phytomedicine 2018, 55, 80–91. [Google Scholar] [CrossRef]
- Panossian, A.; Seo, E.-J.; Klauck, S.M.; Efferth, T. Adaptogens in chemobrain (part IV): Adaptogenic plants prevent the chemotherapeutics-induced imbalance of redox homeostasis by modulation of expression of genes encoding Nrf2-mediated signaling proteins and antioxidant, metabolizing, detoxifying enzymes in neuroglia cells. Longhua Chin. Med. 2020, 3, 4. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Abatangelo, L.; Maglietta, R.; Distaso, A.; D’Addabbo, A.; Creanza, T.M.; Mukherjee, S.; Ancona, N. Comparative study of gene set enrichment methods. BMC Bioinform. 2009, 10, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihnatova, I.; Popovici, V.; Budinska, E. A critical comparison of topology-based pathway analysis methods. PLoS ONE 2018, 13, e0191154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakhry, C.T.; Choudhary, P.; Gutteridge, A.; Sidders, B.; Chen, P.; Ziemek, D.; Zarringhalam, K. Interpreting transcriptional changes using causal graphs: New methods and their practical utility on public networks. BMC Bioinform. 2016, 17, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chindelevitch, L.; Ziemek, D.; Enayetallah, A.; Randhawa, R.; Sidders, B.; Brockel, C.; Huang, E.S. Causal reasoning on biological networks: Interpreting transcriptional changes. Bioinformatics 2012, 28, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).