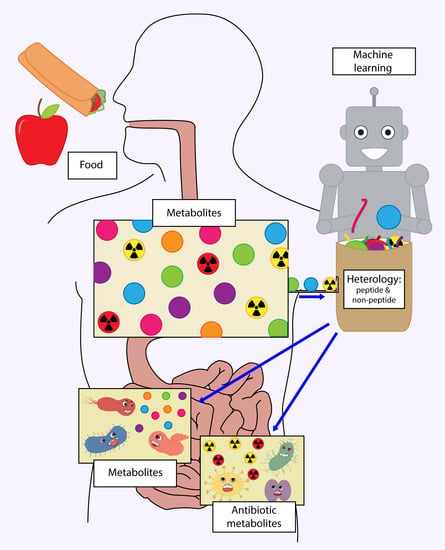
Relevant Features of Polypharmacologic Human-Target Antimicrobials Discovered by Machine-Learning Techniques
Abstract
1. Introduction
2. Results
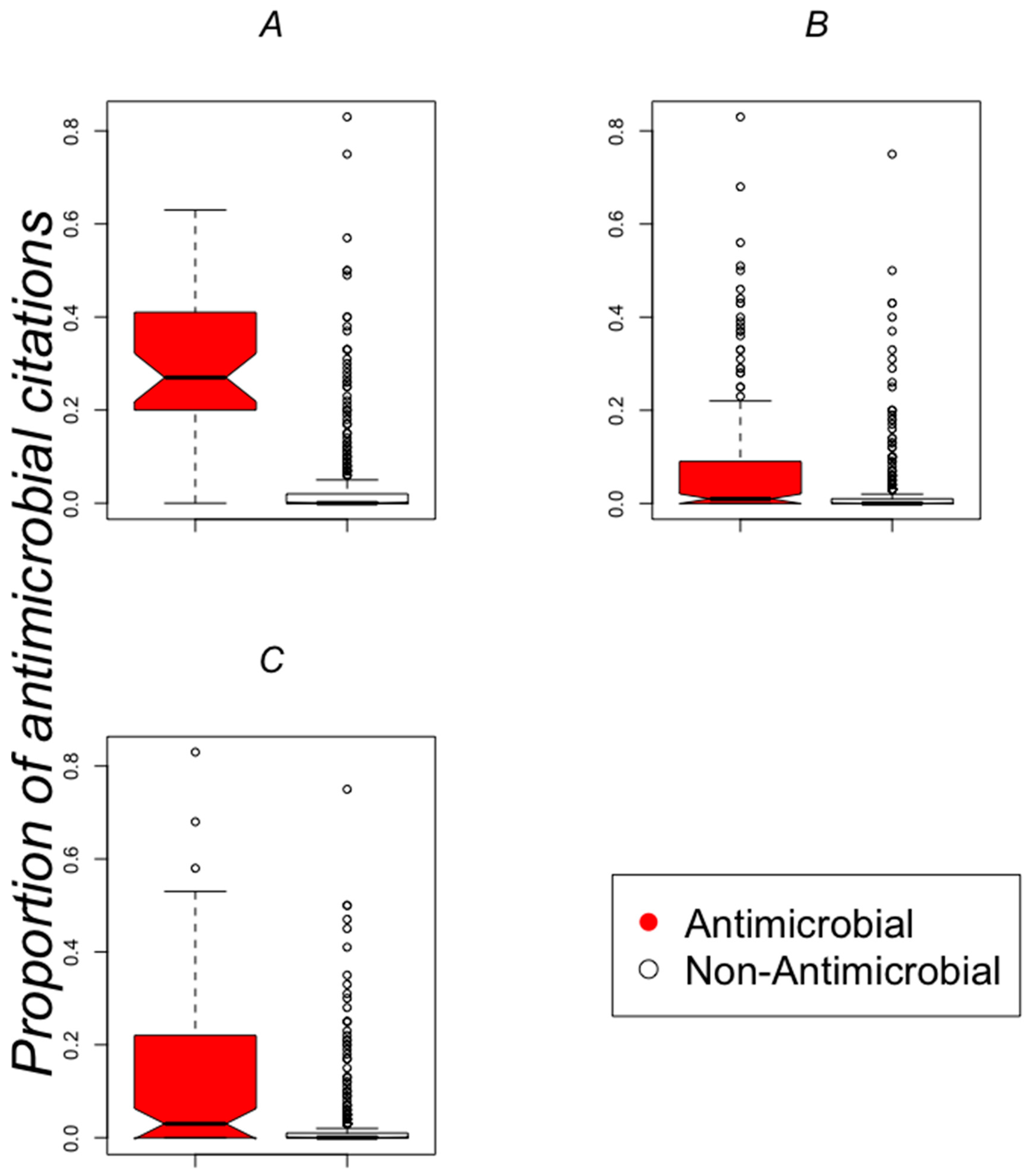
2.1. Publications about Antimicrobial Activity for polyHAM
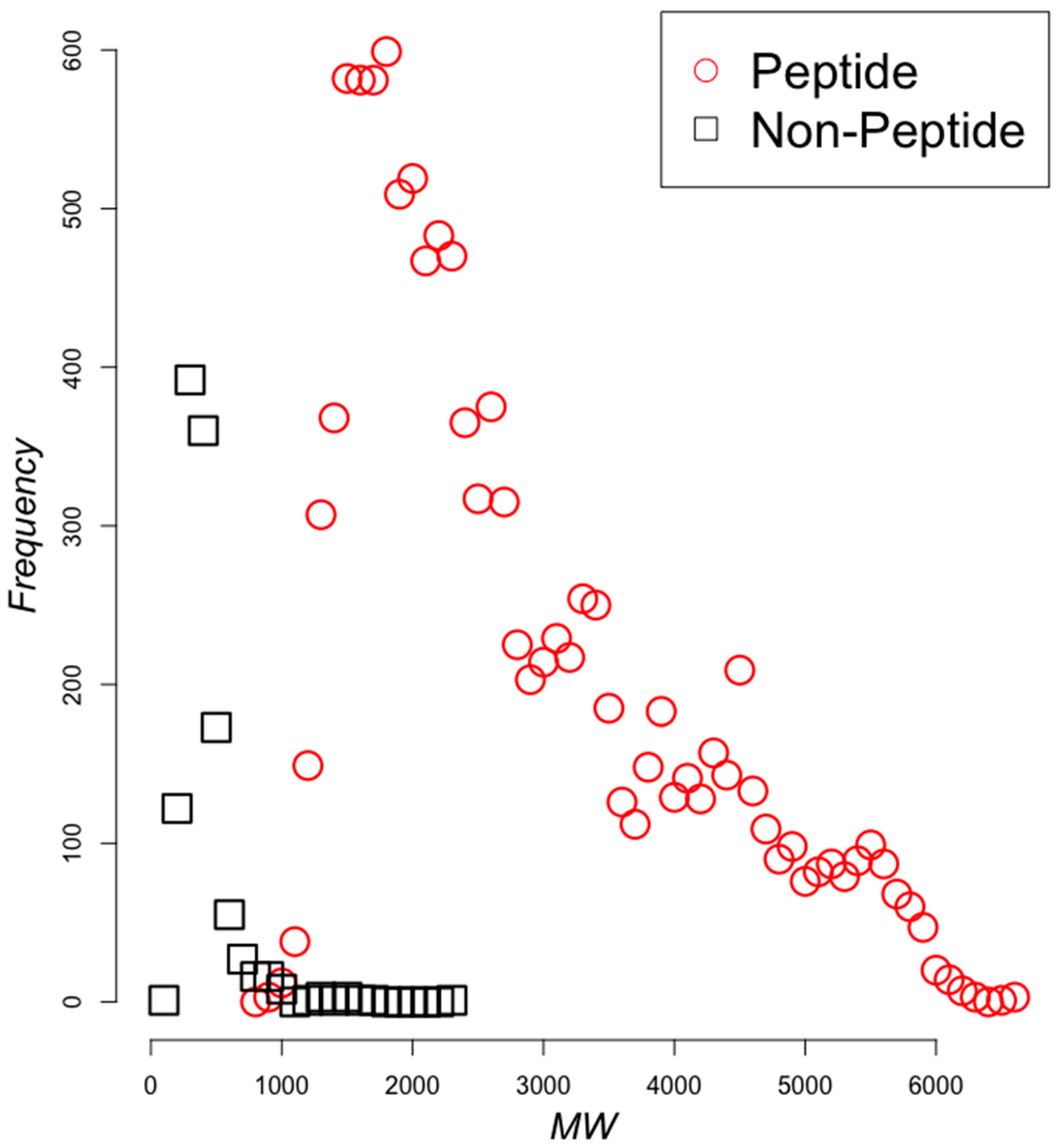
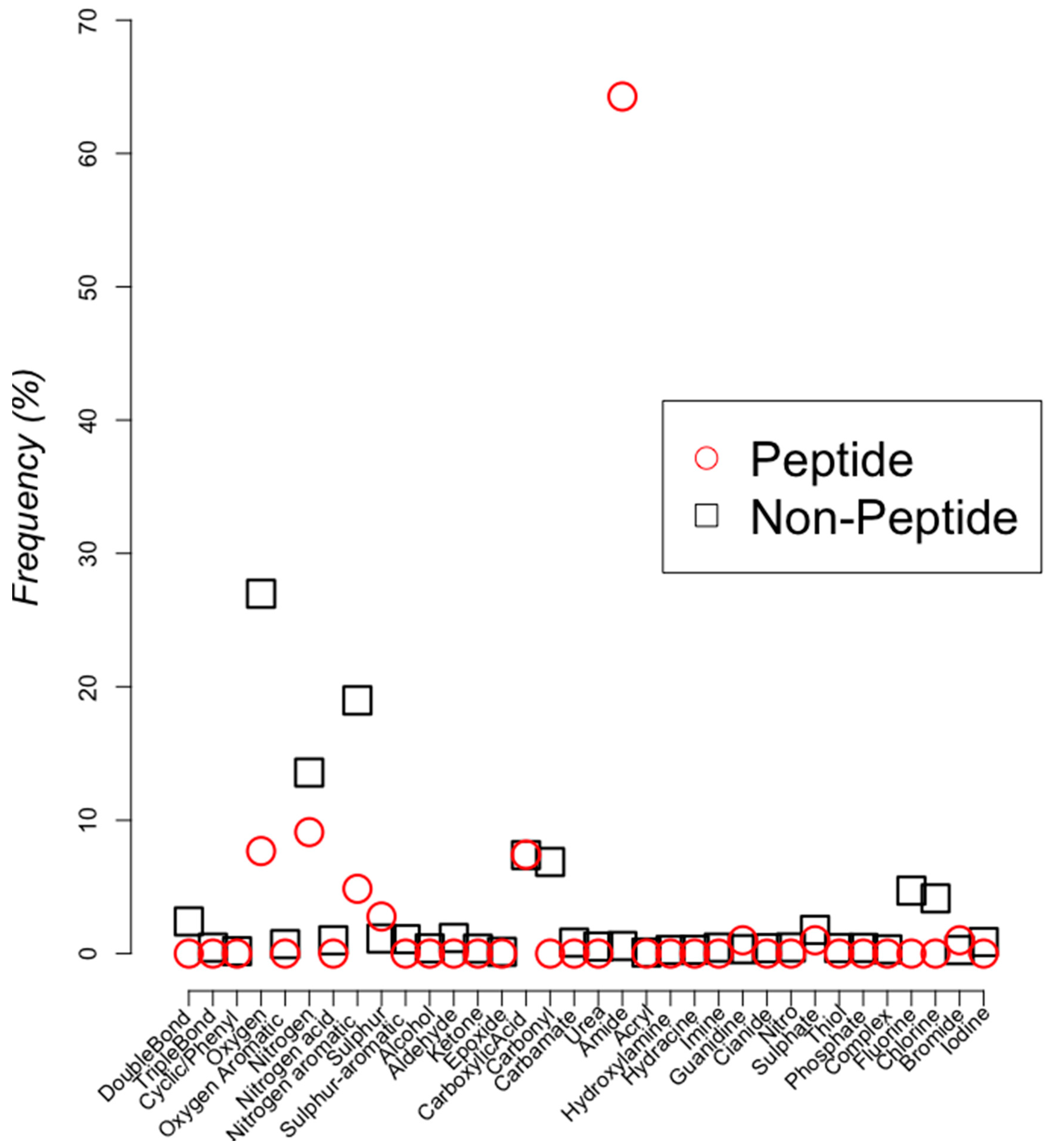
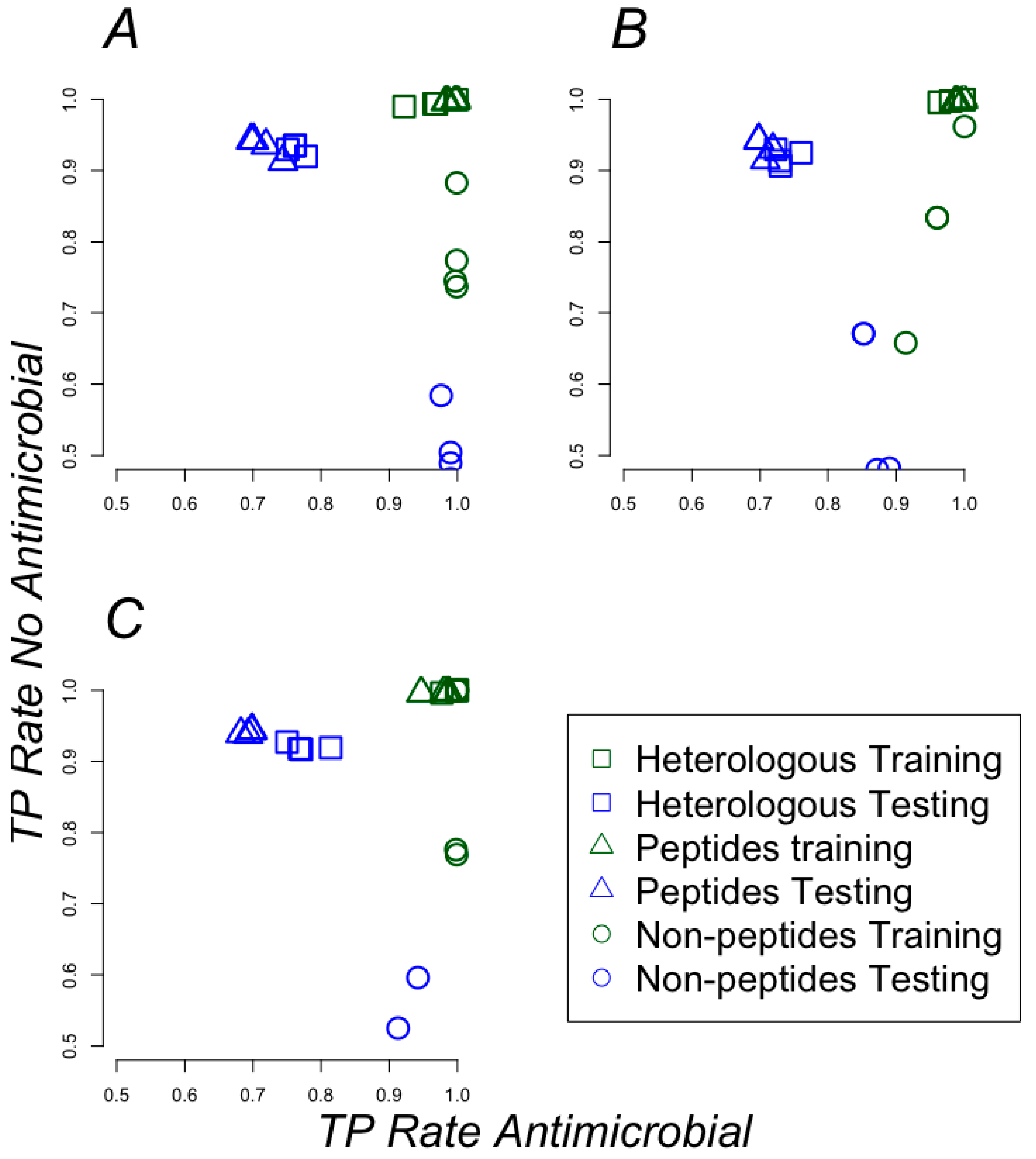
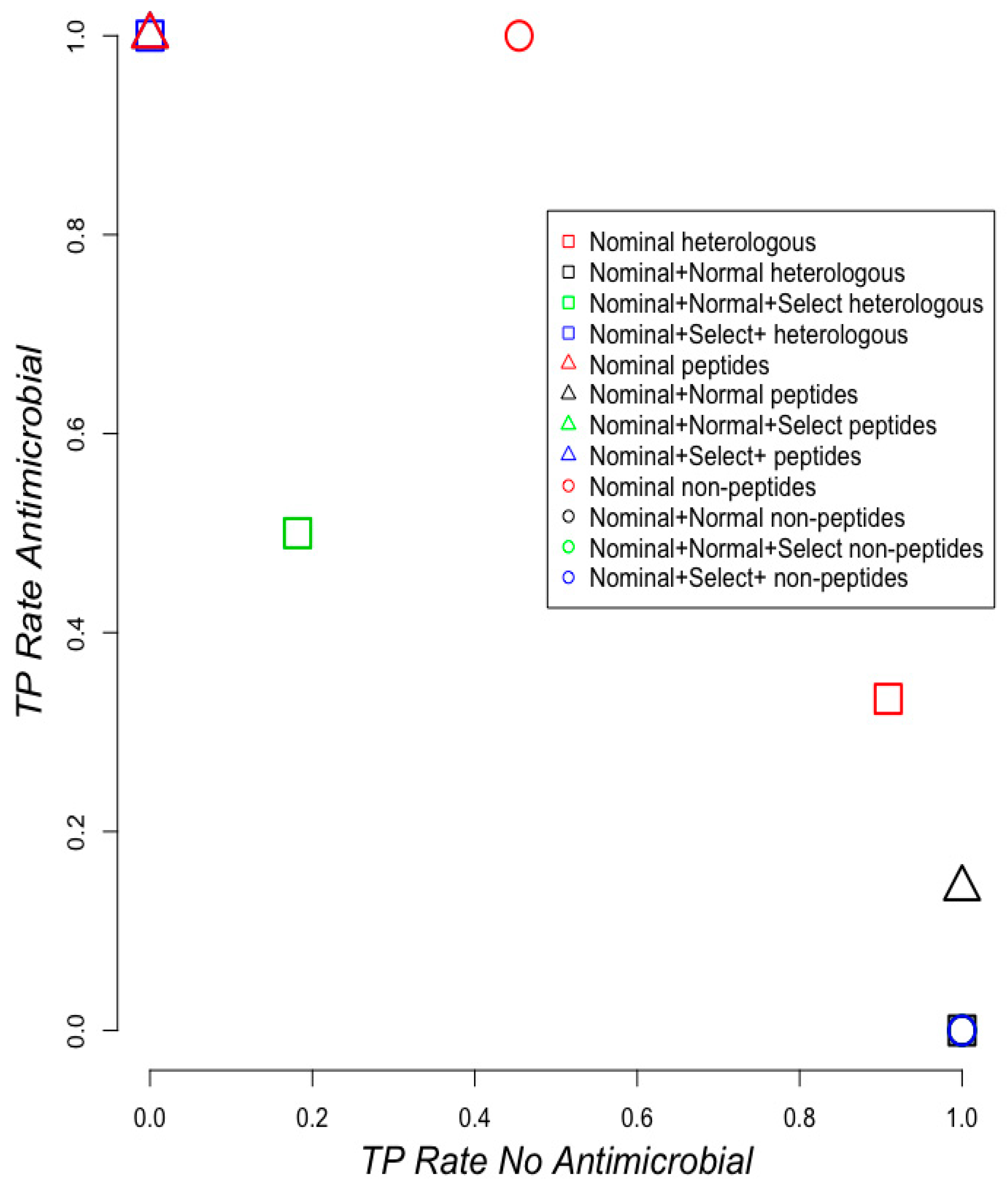
2.2. Classifying Antimicrobials Using Physicochemical Features
3. Discussion
4. Materials and Methods
4.1. Dataset Preparation
4.2. Naïve Text Mining Approach
4.3. Machine-Learning Approach
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fang, Y. Label-free drug discovery. Front. Pharmacol. 2014, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A., Jr.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking Drug Design in the Artificial Intelligence Era. Nat. Rev. Drug Discov. 2019, 19, 353–364. [Google Scholar]
- Sun, X.; Vilar, S.; Tatonetti, N.P. High-Throughput Methods for Combinatorial Drug Discovery. Sci. Transl. Med. 2013, 5, 205rv1. [Google Scholar]
- Oh, D.Y.; Kim, K.; Kwon, H.B.; Seong, J.Y. Cellular and Molecular Biology of Orphan G Protein-Coupled Receptors. Int. Rev. Cytol. 2006, 252, 163–218. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16984818 (accessed on 12 February 2020).
- Vieth, M.; Sutherland, J.J.; Robertson, D.H.; Campbell, R.M. Kinomics: Characterizing the therapeutically validated kinase space. Drug Discov. Today 2005, 10, 839–846. Available online: http://www.ncbi.nlm.nih.gov/pubmed/15970266 (accessed on 12 February 2020). [CrossRef]
- Drawz, S.M.; Bonomo, R.A. Three Decades of β-Lactamase Inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef]
- Amelio, I.; Lisitsa, A.; Knight, R.A.; Melino, G.; Antonov, A.V. Polypharmacology of approved anticancer drugs. Curr. Drug Targets 2016, 18, 534–543. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26926468 (accessed on 12 February 2020). [CrossRef]
- Li, Y.; Wang, P.P.; Li, X.X.; Yu, C.Y.; Yang, H.; Zhou, J.; Xue, W.; Tan, J.; Zhu, F. The Human Kinome Targeted by FDA Approved Multi-Target Drugs and Combination Products: A Comparative Study from the Drug-Target Interaction Network Perspective. PLoS ONE 2016, 11, e0165737. [Google Scholar] [CrossRef]
- Osterloh, I.H. The discovery and development of Viagra® (sildenafil citrate). Sildenafil 2004, 1–13. [Google Scholar] [CrossRef]
- Dutta, S.; Kumar, S.; Hyett, J.; Salomon, C. Molecular Targets of Aspirin and Prevention of Preeclampsia and Their Potential Association with Circulating Extracellular Vesicles during Pregnancy. Int. J. Mol. Sci. 2019, 20, 4370. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57, 1–14. [Google Scholar] [CrossRef]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef] [PubMed]
- Nadal, I.; Donant, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 2007, 56, 1669–1674. Available online: http://www.ncbi.nlm.nih.gov/pubmed/18033837 (accessed on 12 February 2020). [CrossRef]
- Marasco, G.; Di Biase, A.R.; Schiumerini, R.; Eusebi, L.H.; Iughetti, L.; Ravaioli, F.; Scaioli, E.; Colecchia, A.; Festi, D. Gut Microbiota and Celiac Disease. Dig. Dis. Sci. 2016, 61, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G.; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481. Available online: http://www.ncbi.nlm.nih.gov/pubmed/27795980 (accessed on 12 February 2020). [CrossRef]
- Ochoa-Cortes, F.; Turco, F.; Linan-Rico, A.; Soghomonyan, S.; Whitaker, E.; Wehner, S.; Cuomo, R.; Christofi, F.L. Enteric Glial Cells: A New Frontier in Neurogastroenterology and Clinical Target for Inflammatory Bowel Diseases. Inflamm. Bowel. Dis. 2016, 22, 433–449. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26689598 (accessed on 12 February 2020). [CrossRef]
- The Integrative HMP (iHMP) Research Network Consortium; Integrative HMP (iHMP) Research Network Consortium; Buck, G. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Forslund, K.; MetaHIT Consortium; Hildebrand, F.; Nielsen, T.G.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Lara, R.A.N.; Aguilera-Mendoza, L.; Brizuela, C.A.; Peña, A.; Del Rio, G. Heterologous Machine Learning for the Identification of Antimicrobial Activity in Human-Targeted Drugs. Molecules 2019, 24, 1258. Available online: http://www.ncbi.nlm.nih.gov/pubmed/30935109 (accessed on 12 February 2020). [CrossRef]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; Mannhold, R., Kubinyi, H., Timmerman, H., Eds.; Wiley-VCH.: Weinheim, Germany, 2000; p. 667. [Google Scholar]
- Ertl, P. An algorithm to identify functional groups in organic molecules. J. Chemin. 2017, 9, 36. Available online: https://jcheminf.biomedcentral.com/articles/10.1186/s13321-017-0225-z (accessed on 18 February 2020). [CrossRef] [PubMed]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2010, 32, 1466–1474. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21425294 (accessed on 12 February 2020). [CrossRef] [PubMed]
- Zaffiri, L.; Gardner, J.; Toledo-Pereyra, L.H. History of Antibiotics. From Salvarsan to Cephalosporins. J. Investig. Surg. 2012, 25, 67–77. Available online: http://www.ncbi.nlm.nih.gov/pubmed/22439833 (accessed on 12 February 2020). [CrossRef]
- Nuti, R.; Goud, N.S.; Saraswati, A.P.; Alvala, R.; Alvala, M.; Nuti, N.S.G.R. Antimicrobial Peptides: A Promising Therapeutic Strategy in Tackling Antimicrobial Resistance. Curr. Med. Chem. 2017, 24, 4303–4314. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28814242 (accessed on 12 February 2020). [CrossRef] [PubMed]
- Maruvada, P.; Leone, V.; Kaplan, L.M.; Chang, E.B. The Human Microbiome and Obesity: Moving beyond Associations. Cell Host Microbe 2017, 22, 589–599. Available online: http://www.ncbi.nlm.nih.gov/pubmed/29120742 (accessed on 12 February 2020). [CrossRef]
- Tang, W.H.W.; Kitai, T.; Hazen, S.L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 2017, 120, 1183–1196. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28360349 (accessed on 12 February 2020). [CrossRef]
- Wang, J.; Kurilshikov, A.; Radjabzadeh, D.; Turpin, W.; Croitoru, K.; Bonder, M.J.; Jackson, M.A.; Medina-Gomez, C.; Frost, F.; Homuth, G.; et al. Meta-analysis of human genome-microbiome association studies: The MiBioGen consortium initiative. Microbiome 2018, 6, 101. Available online: https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0479-3 (accessed on 17 February 2020). [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Clark, A.G.; Ley, R.E. The Relationship Between the Human Genome and Microbiome Comes into View. Annu. Rev. Genet. 2017, 51, 413–433. [Google Scholar] [CrossRef]
- Gns, H.S.; Gr, S.; Murahari, M.; Krishnamurthy, M. An update on Drug Repurposing: Re-written saga of the drug’s fate. Biomed. Pharmacother. 2019, 110, 700–716. [Google Scholar] [CrossRef]
- Cankaya, S.; Cankaya, B.; Kilic, U.; Kilic, E.; Yulug, B. The therapeutic role of minocycline in Parkinson’s disease. Drugs Context 2019, 8, 1–14. [Google Scholar] [CrossRef]
- Fingerhut, L.C.H.W.; Miller, D.J.; Strugnell, J.M.; Daly, N.L.; Cooke, I.R. OUP accepted manuscript. Bioinformatics 2020, btaa653. [Google Scholar] [CrossRef] [PubMed]
- Khatun, S.; Hasan, M.; Kurata, H.; Khatun, M.S. Efficient computational model for identification of antitubercular peptides by integrating amino acid patterns and properties. FEBS Lett. 2019, 593, 3029–3039. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cai, Y.; Liu, J.; Lin, C.; Liu, X. An advanced approach to identify antimicrobial peptides and their function types for penaeus through machine learning strategies. BMC Bioinform. 2019, 20, 291. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.R.; Tuohy, K.M.; Lindfors, P.; Brown, D.A.; Gibson, G.R.; Wilson, I.D.; Sidaway, J.; Nicholson, J.K.; Holmes, E. Variation in Antibiotic-Induced Microbial Recolonization Impacts on the Host Metabolic Phenotypes of Rats. J. Proteome Res. 2011, 10, 3590–3603. [Google Scholar] [CrossRef] [PubMed]
- Molinos, A.C.; Abriouel, H.; Ben Omar, N.; Lucas, R.; Valdivia, E.; Galvez, A. Inactivation of Listeria monocytogenes in Raw Fruits by Enterocin AS-48. J. Food Prot. 2008, 71, 2460–2467. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A. Correlation-based Feature Selection for Machine Learning The University of Waikato, New Zeland, 1999. Available online: http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.37.4643 (accessed on 12 February 2020).
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Beltran, J.A.; Ibarra, R.T.; Guillen-Ramirez, H.; Brizuela, C.A. Graph-based data integration from bioactive peptide databases of pharmaceutical interest: Toward an organized collection enabling visual network analysis. Bioinformatics 2019, 35, 4739–4747. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2018, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. Available online: https://pubs.acs.org/doi/10.1021/acs.jcim.5b00559 (accessed on 12 February 2020). [CrossRef]
- Wishart, D.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Barton, W.; O’Sullivan, O.; Cotter, P.D. Metabolic phenotyping of the human microbiome. F1000Research 2019, 8, 1956. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. Available online: https://www.kdd.org/exploration_files/p2V11n1.pdf (accessed on 12 February 2020). [CrossRef]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Mach. Learn Res. 2017, 18, 1–5. Available online: http://automl.org/autoweka (accessed on 12 February 2020).


| Antifungal agents pharmacology | Antifungal agents administration and dosage | Antifungal agents administration and dosage therapeutic use | Antifungal agents chemical synthesis chemistry pharmacology |
| Antifungal agents therapeutic use | Drug resistance fungal | Fungi drug effects | Virus replication drug effects |
| Antiviral agents therapeutic use | Anti-bacterial agents analysis | Anti-bacterial agents pharmacology | Anti-bacterial agents therapeutic use |
| Drug resistance bacterial | Drug-resistance multiple bacterial | Mycobacterium tuberculosis drug effects | Anti-bacterial agents |
| Anti-bacterial agents administration and dosage adverse effects | Anti-bacterial agents administration and dosage pharmacology | Anti-bacterial agents administration and dosage therapeutic-use | Anti-bacterial agents adverse-effects |
| Anti-bacterial agents chemistry | Anti-bacterial agents pharmacology therapeutic-use | Anti-bacterial agents toxicity | Bacterial infections drug-therapy |
| DNA-bacterial genetics | Drug-resistance bacterial-genetics | Gram-negative-bacteria drug-effects | Gram-positive-bacteria drug-effects |
| Helicobacter infection drug-therapy microbiology | Helicobacter pylori | Helicobacter pylori drug-effects | Mycobacterium avium complex-drug-effects |
| Attribute Name 1 | Description |
|---|---|
| BCUTw-1h | Eigenvalue based descriptor noted for its utility in chemical diversity |
| AATS5m | Average Broto—Moreau autocorrelation—lag 5/weighted by mass |
| MIC5a | Modified information content index (neighborhood symmetry of 5-order)/ weighted by atoms |
| AATS6m | Average Broto—Moreau autocorrelation—lag 6/weighted by mass |
| AATS7m | Average Broto—Moreau autocorrelation—lag 7/weighted by mass |
| IC5 | Information content index (neighborhood symmetry of 5-order) |
| MIC4 | Modified information content index (neighborhood symmetry of 4-order) |
| AATSC0m | Average centered Broto—Moreau autocorrelation—lag 0/weighted by mass |
| topoDiameter | Topological diameter (maximum atom eccentricity) |
| AATS0m | Average Broto—Moreau autocorrelation—lag 0/weighted by mass |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nava Lara, R.A.; Beltrán, J.A.; Brizuela, C.A.; Del Rio, G. Relevant Features of Polypharmacologic Human-Target Antimicrobials Discovered by Machine-Learning Techniques. Pharmaceuticals 2020, 13, 204. https://doi.org/10.3390/ph13090204
Nava Lara RA, Beltrán JA, Brizuela CA, Del Rio G. Relevant Features of Polypharmacologic Human-Target Antimicrobials Discovered by Machine-Learning Techniques. Pharmaceuticals. 2020; 13(9):204. https://doi.org/10.3390/ph13090204
Chicago/Turabian StyleNava Lara, Rodrigo A., Jesús A. Beltrán, Carlos A. Brizuela, and Gabriel Del Rio. 2020. "Relevant Features of Polypharmacologic Human-Target Antimicrobials Discovered by Machine-Learning Techniques" Pharmaceuticals 13, no. 9: 204. https://doi.org/10.3390/ph13090204
APA StyleNava Lara, R. A., Beltrán, J. A., Brizuela, C. A., & Del Rio, G. (2020). Relevant Features of Polypharmacologic Human-Target Antimicrobials Discovered by Machine-Learning Techniques. Pharmaceuticals, 13(9), 204. https://doi.org/10.3390/ph13090204