Classification of Mycena and Marasmius Species Using Deep Learning Models: An Ecological and Taxonomic Approach
Abstract
1. Introduction
2. Materials and Methods
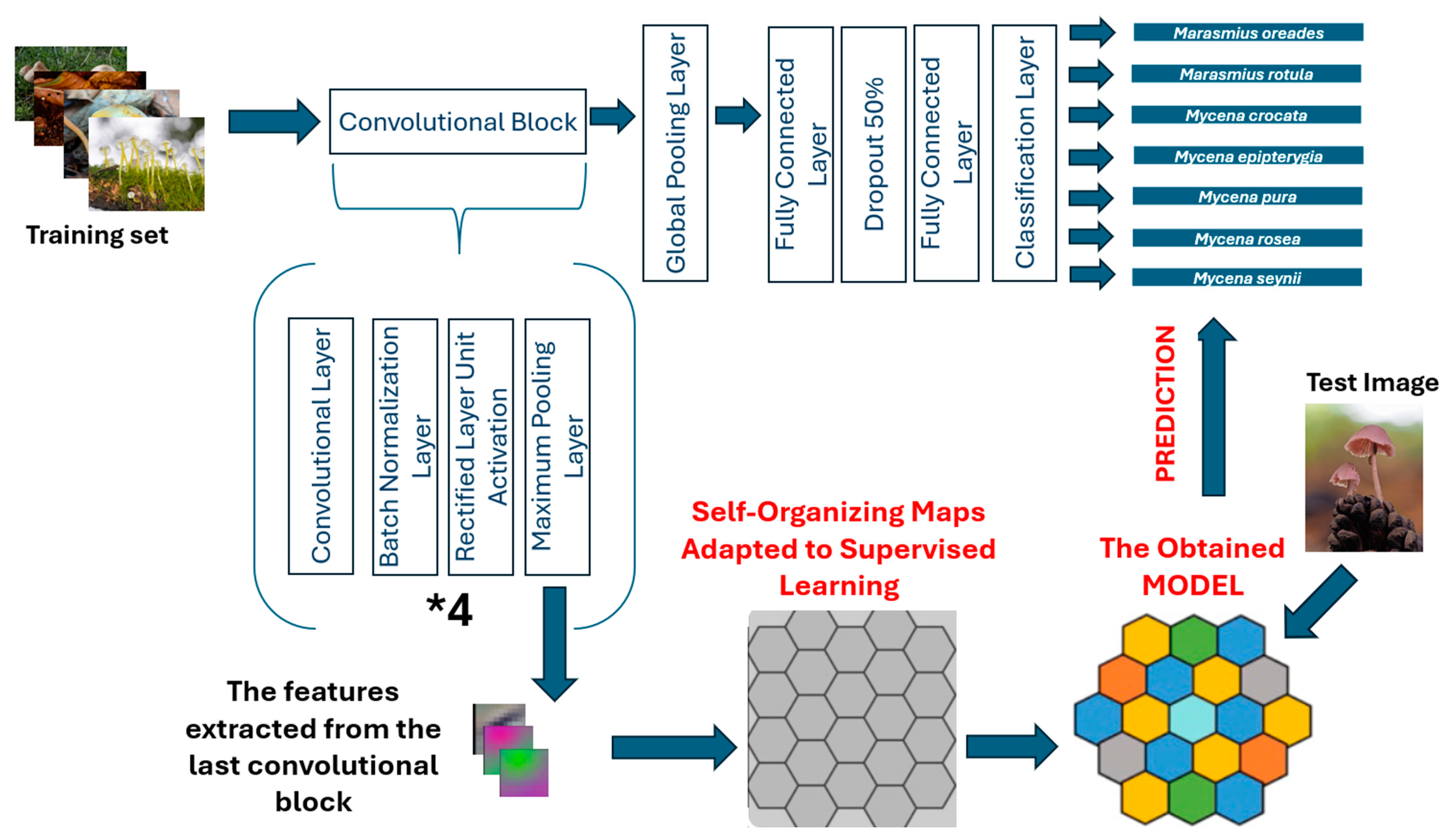
2.1. General Framework
- Discriminative Features Used: The classification relied on the deep features extracted using CNNs. These features include the following:
- Texture and Surface Patterns: Identifying fine-grained details like cap striations, surface roughness, and gill structures.
- Color Variations: Differentiating species based on pigmentation variations influenced by age and environmental conditions.
- Morphological Shapes: Capturing the unique structural attributes of fungi, such as cap, stipe, and gill arrangements.
- Deep Representations: Extracted from CNN layers, enabling the model to learn hierarchical feature relationships that distinguish species.
2.2. Dataset
2.3. Methods
2.4. Experimental Setup
- A total of 4 convolutional blocks, and each has 2D convolutional, batch normalization, ReLU activation, and maximum pooling layers.
- Global Average Pooling (GAP): The feature maps generated by the convolutional blocks are summarized using Adaptive Average Pooling. This technique helps prevent overfitting by reducing the number of parameters.
- Fully Connected Layers: The first fully connected layer consists of 512 neurons and incorporates dropout with a rate of 0.5. The second fully connected layer contains 256 neurons. The final layer is a linear layer with 7 neurons, corresponding to the number of classes.
- Feature Extraction Using a CNN
- A custom CNN architecture is employed to extract high-dimensional features from fungal images.
- The CNN consists of four convolutional blocks, each comprising a convolutional layer, batch normalization, ReLU activation, and max pooling to progressively reduce spatial dimensions while retaining crucial feature information.
- The output of the CNN is a high-dimensional feature vector (14 × 14 × 256 = 50,176 features).
- Feature Clustering and Classification Using SOM
- The self-organizing map (SOM) is a type of neural network used for dimensionality reduction and clustering.
- The extracted CNN features are mapped into a 2D topological grid, where similar fungal species are grouped together.
- Unlike traditional classifiers, SOM provides a structured and interpretable representation of species similarities.
- A Best Matching Unit (BMU) approach is used to classify new images based on the closest neuron in the SOM grid.
- Extract Features from CNN: A test image is passed through CNN to generate a feature vector.
- Find the Best Matching Unit (BMU): The neuron in the SOM grid closest to the input feature vector is identified.
- Assign a Class Label: Each neuron is assigned a fungal species label based on the majority class of training samples mapped to it.
- Final Prediction: The class of the BMU is assigned to the test image.
3. Results
3.1. Evaluation Metrics
3.2. Empirical Results
4. Discussion and Conclusions
- Data Augmentation: Techniques such as random rotations, horizontal flips, and color jittering artificially expand the dataset and help the model learn invariances to image transformations.
- Pretrained Models: By leveraging transfer learning from large-scale image datasets (e.g., ImageNet), our models benefited from prior knowledge, reducing overfitting.
- Regularization Techniques: We employed dropout layers and weight decay to prevent overfitting and enhance generalization.
- Statistical Validation: The chi-square test was applied to assess the reliability of classification results across different models.
- Independent Test Set: We allocated 30% of the dataset for independent testing, ensuring performance metrics reflect real-world applicability.
- Biodiversity Conservation: Automated identification can assist conservationists in mapping fungal distributions and tracking species at risk due to climate change or habitat destruction.
- Agriculture and Forestry: Some fungi are beneficial for soil health, while others are harmful pathogens. Accurate identification supports sustainable agriculture and forest management.
- Citizen Science and Education: Mobile applications utilizing our model can help amateur mushroom pickers and nature enthusiasts correctly identify species, reducing the risk of misidentification, especially for toxic mushrooms.
- Biotechnological Applications: Certain fungi have medicinal and industrial uses. Rapid classification aids in screening potential species for pharmaceutical and environmental applications.
- Autonomous Detection Systems: The model can be integrated into robotic or drone-based systems for real-time fungal identification in forests and agricultural fields.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Souto, M.; Raposeiro, P.M.; Balibrea, A.; Gonçalves, V. Checklist of Basidiomycota and New Records from the Azores Archipelago. Diversity 2024, 16, 170. [Google Scholar] [CrossRef]
- Akata, I.; Kumru, E.; Ediş, G.; Özbey, B.G.; Şahin, E. Three New Records for Turkish Agaricales Inhabiting Ankara University Beşevler 10th Year Campus Area. Kastamonu Univ. J. For. Fac. 2023, 23, 250–263. [Google Scholar] [CrossRef]
- Antonín, V.; Noordeloos, M.E. A Monograph of Marasmioid and Collybioid Fungi in Europe; IHW-Verlag: Eching, Germany, 2010. [Google Scholar]
- Robich, G. Mycena d’Europa; Associazione Micologica Bresadola: Trento, Italy, 2003. [Google Scholar]
- Knudsen, H.; Vesterholt, J. (Eds.) Funga Nordica: Agaricoid, Boletoid, and Cyphelloid Genera; Nordsvamp: Copenhagen, Denmark, 2008. [Google Scholar]
- Aronsen, A.; Læssøe, T. The Genus Mycena s.l. In The Fungi of Northern Europe; The Danish Mycological Society: Copenhagen, Denmark, 2016; Volume 5, 373p. [Google Scholar]
- Breitenbach, J.; Kränzlin, F. Fungi of Switzerland, Volume 3: Boletes and Agarics 1. Part; Verlag Mykologia: Luzern, Switzerland, 1991. [Google Scholar]
- Maas Geesteranus, R. Conspectus of the Mycenas of the Northern Hemisphere—8. Sections Intermediae, Rubromarginatae. Proc. K. Ned. Akad. Wet. C Biol. Med. Sci. 1986, 89, 279–310. [Google Scholar]
- Asif, M.; Maula, F.; Saba, M.; Akram, W.; Raza, M. Taxonomic and Phylogenetic Evidence Reveal a New Species and a New Record of the Genus Marasmius from Pakistan. Phytotaxa 2024, 646, 32–46. [Google Scholar] [CrossRef]
- Ozsari, S.; Kumru, E.; Ekinci, F.; Akata, I.; Guzel, M.S.; Acici, K.; Asuroglu, T. Deep Learning-Based Classification of Macrofungi: Comparative Analysis of Advanced Models for Accurate Fungi Identification. Sensors 2024, 24, 7189. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Liu, H.; Li, J.; Wang, Y. Application of Identification and Evaluation Techniques for Edible Mushrooms: A Review. Crit. Rev. Anal. Chem. 2023, 53, 634–654. [Google Scholar] [CrossRef]
- Picek, L.; Šulc, M.; Matas, J.; Heilmann-Clausen, J.; Jeppesen, T.S.; Lind, E. Automatic Fungi Recognition: Deep Learning Meets Mycology. Sensors 2022, 22, 633. [Google Scholar] [CrossRef]
- Chathurika, K.; Siriwardena, E.; Bandara, H.; Perera, G.; Dilshanka, K. Developing an Identification System for Different Types of Edible Mushrooms in Sri Lanka Using Machine Learning and Image Processing. Int. J. Eng. Manag. Res. 2023, 13, 54–59. [Google Scholar]
- Bartlett, P.; Eberhardt, U.; Schütz, N.; Beker, H.J. Species Determination Using AI Machine-Learning Algorithms: Hebeloma as a Case Study. IMA Fungus 2022, 13, 13. [Google Scholar] [CrossRef]
- Global Core Biodata Resource. Available online: www.gbif.org (accessed on 28 January 2025).
- Macrofungi Species Dataset. Available online: https://tinyurl.com/wp8wefy8 (accessed on 28 January 2025).
- Kohonen, T. The Self-Organizing Map. Proc. IEEE 1990, 78, 1464–1480. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Ono, K.; Hikawa, H. Electroencephalogram-Based Facial Gesture Recognition Using Self-Organizing Map. Sensors 2024, 24, 2741. [Google Scholar] [CrossRef] [PubMed]
- Gholami, V.; Khaleghi, M.R.; Pirasteh, S.; Booij, M.J. Comparison of Self-Organizing Map, Artificial Neural Network, and Co-Active Neuro-Fuzzy Inference System Methods in Simulating Groundwater Quality: Geospatial Artificial Intelligence. Water Resour. Manag. 2022, 36, 451–469. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Vaidya, S.; Ruehle, F.; Halverson, J.; Soljacic, M.; Hou, T.Y.; Tegmark, M. KAN: Kolmogorov-Arnold Networks. arXiv 2024, arXiv:2404.19756. [Google Scholar]
- Firsov, N.; Myasnikov, E.; Lobanov, V.; Khabibullin, R.; Kazanskiy, N.; Khonina, S.; Butt, M.A.; Nikonorov, A. HyperKAN: Kolmogorov–Arnold Networks Make Hyperspectral Image Classifiers Smarter. Sensors 2024, 24, 7683. [Google Scholar] [CrossRef]
- Livieris, I.E. C-KAN: A New Approach for Integrating Convolutional Layers with Kolmogorov–Arnold Networks for Time-Series Forecasting. Mathematics 2024, 12, 3022. [Google Scholar] [CrossRef]
- Hollósi, J.; Ballagi, Á.; Kovács, G.; Fischer, S.; Nagy, V. Detection of Bus Driver Mobile Phone Usage Using Kolmogorov-Arnold Networks. Computers 2024, 13, 218. [Google Scholar] [CrossRef]
- Hollósi, J. Efficiency Analysis of Kolmogorov-Arnold Networks for Visual Data Processing. Eng. Proc. 2024, 79, 68. [Google Scholar] [CrossRef]
- Ibrahum, A.D.M.; Shang, Z.; Hong, J.-E. How Resilient Are Kolmogorov–Arnold Networks in Classification Tasks? A Robustness Investigation. Appl. Sci. 2024, 14, 10173. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Singh, V.; Baral, A.; Kumar, R.; Tummala, S.; Noori, M.; Yadav, S.V.; Kang, S.; Zhao, W. A Hybrid Deep Learning Model for Enhanced Structural Damage Detection: Integrating ResNet50, GoogLeNet, and Attention Mechanisms. Sensors 2024, 24, 7249. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Alshammari, A. Construction of VGG16 Convolution Neural Network (VGG16_CNN) Classifier with NestNet-Based Segmentation Paradigm for Brain Metastasis Classification. Sensors 2022, 22, 8076. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Li, Z.; Tian, X.; Liu, X.; Liu, Y.; Shi, X. A Two-Stage Industrial Defect Detection Framework Based on Improved-YOLOv5 and Optimized-Inception-ResNetV2 Models. Appl. Sci. 2022, 12, 834. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning (PMLR 2019), Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Abd El-Ghany, S.; Mahmood, M.A.; Abd El-Aziz, A.A. Adaptive Dynamic Learning Rate Optimization Technique for Colorectal Cancer Diagnosis Based on Histopathological Image Using EfficientNet-B0 Deep Learning Model. Electronics 2024, 13, 3126. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. EfficientNetV2: Smaller Models and Faster Training. In Proceedings of the International Conference on Machine Learning (PMLR 2021), Virtual, 18–24 July 2021; pp. 10096–10106. [Google Scholar]
- Huang, M.-L.; Liao, Y.-C. Stacking Ensemble and ECA-EfficientNetV2 Convolutional Neural Networks on Classification of Multiple Chest Diseases Including COVID-19. Acad. Radiol. 2023, 30, 1915–1935. [Google Scholar] [CrossRef]
- Tu, Z.; Talebi, H.; Zhang, H.; Yang, F.; Milanfar, P.; Bovik, A.; Li, Y. MaxViT: Multi-Axis Vision Transformer. In Proceedings of the Computer Vision–ECCV 2022: 17th European Conference, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 459–479. [Google Scholar]
- Pacal, I. Enhancing Crop Productivity and Sustainability through Disease Identification in Maize Leaves: Exploiting a Large Dataset with an Advanced Vision Transformer Model. Expert Syst. Appl. 2024, 238, 122099. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Pacal, I. MaxCerVixT: A Novel Lightweight Vision Transformer-Based Approach for Precise Cervical Cancer Detection. Knowl.-Based Syst. 2024, 289, 111482. [Google Scholar] [CrossRef]
- MiniSom Library. Available online: https://github.com/JustGlowing/minisom (accessed on 24 January 2025).
- Harder, C.B.; Hesling, E.; Botnen, S.S.; Lorberau, K.E.; Dima, B.; von Bonsdorff-Salminen, T.; Kauserud, H. Mycena Species Can Be Opportunist-Generalist Plant Root Invaders. Environ. Microbiol. 2023, 25, 1875–1893. [Google Scholar] [CrossRef]
- Niego, A.G.T.; Rapior, S.; Thongklang, N.; Raspé, O.; Hyde, K.D.; Mortimer, P. Reviewing the Contributions of Macrofungi to Forest Ecosystem Processes and Services. Fungal Biol. Rev. 2023, 44, 100294. [Google Scholar] [CrossRef]
- Thoen, E.; Harder, C.B.; Kauserud, H.; Botnen, S.S.; Vik, U.; Taylor, A.F.; Skrede, I. In Vitro Evidence of Root Colonization Suggests Ecological Versatility in the Genus Mycena. New Phytol. 2020, 227, 601–612. [Google Scholar] [CrossRef]
- Koch, R.A.; Liu, J.; Brann, M.; Jumbam, B.; Siegel, N.; Aime, M.C. Marasmioid Rhizomorphs in Bird Nests: Species Diversity, Functional Specificity, and New Species from the Tropics. Mycologia 2020, 112, 1086–1103. [Google Scholar] [CrossRef]
- Bonugli-Santos, R.C.; dos Santos Vasconcelos, M.R.; Passarini, M.R.; Vieira, G.A.; Lopes, V.C.; Mainardi, P.H.; Sette, L.D. Marine-Derived Fungi: Diversity of Enzymes and Biotechnological Applications. Front. Microbiol. 2015, 6, 269. [Google Scholar] [CrossRef]
- Rame, R.; Purwanto, P.; Sudarno, S. Biotechnological Approaches in Utilizing Agro-Waste for Biofuel Production: An Extensive Review on Techniques and Challenges. Bioresour. Technol. Rep. 2023, 24, 101662. [Google Scholar] [CrossRef]
- Dahlberg, A.; Genney, D.R.; Heilmann-Clausen, J. Developing a Comprehensive Strategy for Fungal Conservation in Europe: Current Status and Future Needs. Fungal Ecol. 2010, 3, 50–64. [Google Scholar] [CrossRef]
- Nnadi, N.E.; Carter, D.A. Climate Change and the Emergence of Fungal Pathogens. PLoS Pathog. 2021, 17, e1009503. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hegde, S.; Kumar, A.S.; Zacharias, A.; Mehta, P.; Mukthineni, V.; Acharya, S. Towards Transforming Malaria Vector Surveillance Using VectorBrain: A Novel Convolutional Neural Network for Mosquito Species, Sex, and Abdomen Status Identifications. Sci. Rep. 2024, 14, 23647. [Google Scholar] [CrossRef]
- Khalil, A.F.; Rostam, S. Machine Learning-Based Predictive Maintenance for Fault Detection in Rotating Machinery: A Case Study. Eng. Technol. Appl. Sci. Res. 2024, 14, 13181–13189. [Google Scholar] [CrossRef]
- Picek, L.; Šulc, M.; Matas, J.; Jeppesen, T.S.; Heilmann-Clausen, J.; Læssøe, T.; Frøslev, T. Danish Fungi 2020—Not Just Another Image Recognition Dataset. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 1525–1535. [Google Scholar]
- Jasim, M.A.; Al-Tuwaijari, J.M. Plant Leaf Diseases Detection and Classification Using Image Processing and Deep Learning Techniques. In Proceedings of the 2020 International Conference on Computer Science and Software Engineering (CSASE), Duhok, Iraq, 16–18 April 2020; pp. 259–265. [Google Scholar]
- Shoaib, M.; Hussain, T.; Shah, B.; Ullah, I.; Shah, S.M.; Ali, F.; Park, S.H. Deep Learning-Based Segmentation and Classification of Leaf Images for Detection of Tomato Plant Disease. Front. Plant Sci. 2022, 13, 1031748. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Yan, P.; Li, G.; Jiang, Y.; Luo, H.; Yin, S. Deep Learning Attention Mechanism in Medical Image Analysis: Basics and Beyonds. Int. J. Netw. Dyn. Intell. 2023, 2, 93–116. [Google Scholar] [CrossRef]
- Papa, L.; Proietti Mattia, G.; Russo, P.; Amerini, I.; Beraldi, R. Lightweight and Energy-Aware Monocular Depth Estimation Models for IoT Embedded Devices: Challenges and Performances in Terrestrial and Underwater Scenarios. Sensors 2023, 23, 2223. [Google Scholar] [CrossRef]
- Hernández Medina, R.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine Learning and Deep Learning Applications in Microbiome Research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef] [PubMed]
- Rathnayaka, A.R.; Tennakoon, D.S.; Jones, G.E.; Wanasinghe, D.N.; Bhat, D.J.; Priyashantha, A.H.; Karunarathna, S.C. Significance of Precise Documentation of Hosts and Geospatial Data of Fungal Collections, with an Emphasis on Plant-Associated Fungi. N. Z. J. Bot. 2024, 1–28. [Google Scholar] [CrossRef]
- Pandey, M.; Fernandez, M.; Gentile, F.; Isayev, O.; Tropsha, A.; Stern, A.C.; Cherkasov, A. The Transformational Role of GPU Computing and Deep Learning in Drug Discovery. Nat. Mach. Intell. 2022, 4, 211–221. [Google Scholar] [CrossRef]
- Zhou, L.; Pan, S.; Wang, J.; Vasilakos, A.V. Machine Learning on Big Data: Opportunities and Challenges. Neurocomputing 2017, 237, 350–361. [Google Scholar] [CrossRef]
- Edwards, J.E.; Forster, R.J.; Callaghan, T.M.; Dollhofer, V.; Dagar, S.S.; Cheng, Y.; Smidt, H. PCR and Omics-Based Techniques to Study the Diversity, Ecology and Biology of Anaerobic Fungi: Insights, Challenges and Opportunities. Front. Microbiol. 2017, 8, 1657. [Google Scholar] [CrossRef] [PubMed]
- Pacal, I. Improved Vision Transformer with Lion Optimizer for Lung Diseases Detection. Int. J. Eng. Res. Dev. 2024, 16, 760–776. [Google Scholar] [CrossRef]
- Pouyanfar, S.; Sadiq, S.; Yan, Y.; Tian, H.; Tao, Y.; Reyes, M.P.; Iyengar, S.S. A Survey on Deep Learning: Algorithms, Techniques, and Applications. ACM Comput. Surv. (CSUR) 2018, 51, 92. [Google Scholar] [CrossRef]
- Archana, R.; Jeevaraj, P.E. Deep Learning Models for Digital Image Processing: A Review. Artif. Intell. Rev. 2024, 57, 11. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Dean, J. A Guide to Deep Learning in Healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]


| Mushroom Species | Percentage of Images Received from Source | Number of Samples |
|---|---|---|
| Marasmius oreades | <22% | 222 |
| Marasmius rotula | <20% | 228 |
| Mycena crocata | <25% | 229 |
| Mycena epipterygia | <20% | 243 |
| Mycena pura | <23% | 220 |
| Mycena rosea | <25% | 227 |
| Mycena seynii | <25% | 213 |
| Mushroom Species | Training Set | Independent Test Set |
|---|---|---|
| Marasmius oreades | 154 | 67 |
| Marasmius rotula | 159 | 69 |
| Mycena crocata | 160 | 69 |
| Mycena epipterygia | 171 | 72 |
| Mycena pura | 154 | 66 |
| Mycena rosea | 158 | 69 |
| Mycena seynii | 149 | 64 |
| Name | Attributes |
|---|---|
| Convolutional Block 1 | 3 × 3 kernel size, 32 filters |
| Convolutional Block 2 | 3 × 3 kernel size, 64 filters |
| Convolutional Block 3 | 3 × 3 kernel size, 128 filters |
| Convolutional Block 4 | 3 × 3 kernel size, 256 filters |
| Global Average Pooling | |
| Fully Connected Layers | 512 → 256 → 7 neurons |
| Dropout | 50% |
| Layer | Attribute |
|---|---|
| Linear layer | Input: 2816 Output: 512 |
| Activation | ReLU |
| Dropout | 50% (to prevent overfitting) |
| Linear layer | Input: 512 (256 with dropout) Output: 7 (number of classes) |
| Architecture | Accuracy | Precision | Recall | F1-Score | Specificity | MCC | AUC (OvR) |
|---|---|---|---|---|---|---|---|
| GoogleNet | 0.811 | 0.811 | 0.812 | 0.809 | 0.969 | 0.780 | 0.968 |
| MobileNetV3-L | 0.899 | 0.899 | 0.899 | 0.899 | 0.983 | 0.882 | 0.987 |
| ResNetV2-50 | 0.979 | 0.979 | 0.979 | 0.979 | 0.997 | 0.976 | 0.999 |
| EfficientNet-B0 | 0.958 | 0.959 | 0.958 | 0.958 | 0.993 | 0.951 | 0.998 |
| EfficientNetV2-M | 0.964 | 0.964 | 0.964 | 0.964 | 0.994 | 0.958 | 0.998 |
| VGG19 | 0.876 | 0.877 | 0.876 | 0.876 | 0.979 | 0.856 | 0.987 |
| MaxVit-S | 0.989 | 0.989 | 0.989 | 0.989 | 0.998 | 0.988 | 0.999 |
| CNN | 0.529 | 0.533 | 0.529 | 0.529 | 0.922 | 0.451 | 0.502 |
| CNN-SOM | 0.863 | 0.885 | 0.862 | 0.864 | 0.977 | 0.843 | 0.911 |
| CNN-KAN | 0.763 | 0.762 | 0.763 | 0.759 | 0.961 | 0.724 | 0.861 |
| MaxVit-SOM | 0.977 | 0.977 | 0.977 | 0.977 | 0.996 | 0.973 | 0.986 |
| Ensemble (MaxVit-Small—ResNetV2-50) | 0.935 | 0.936 | 0.935 | 0.935 | 0.989 | 0.925 | 0.96 |
| Models | GoogleNet | MobileNetV3-L | ResNetV2-50 | EfficientNet-B0 | EfficientNetV2-M | VGG19 | MaxVit-S |
|---|---|---|---|---|---|---|---|
| CNN-SOM | 5.147 × 10−4 ← | 2.167 × 10−2 ↑ | 6.107 × 10−3 ↑ | 1.305 × 10−1 | 5.898 × 10−2 | 1.066 × 10−1 | 1.850 × 10−2 ↑ |
| CNN-KAN | 2.195 × 10−1 | 9.431 × 10−3 ↑ | 4.893 × 10−8 ↑ | 9.977 × 10−6 ↑ | 3.629 × 10−6 ↑ | 1.411 × 10−1 | 1.332 × 10−8 ↑ |
| MaxVit-SOM | 1.217 × 10−3 ← | 8.919 × 10−1 | 1.000 | 9.999 × 10−1 | 1.000 | 4.012 × 10−1 | 1.000 |
| Ensemble MaxVit-ResNet | 2.299 × 10−2 ← | 6.475 × 10−1 | 9.999 × 10−1 | 9.588 × 10−1 | 1.000 | 6.569 × 10−1 | 9.874 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekinci, F.; Ugurlu, G.; Ozcan, G.S.; Acici, K.; Asuroglu, T.; Kumru, E.; Guzel, M.S.; Akata, I. Classification of Mycena and Marasmius Species Using Deep Learning Models: An Ecological and Taxonomic Approach. Sensors 2025, 25, 1642. https://doi.org/10.3390/s25061642
Ekinci F, Ugurlu G, Ozcan GS, Acici K, Asuroglu T, Kumru E, Guzel MS, Akata I. Classification of Mycena and Marasmius Species Using Deep Learning Models: An Ecological and Taxonomic Approach. Sensors. 2025; 25(6):1642. https://doi.org/10.3390/s25061642
Chicago/Turabian StyleEkinci, Fatih, Guney Ugurlu, Giray Sercan Ozcan, Koray Acici, Tunc Asuroglu, Eda Kumru, Mehmet Serdar Guzel, and Ilgaz Akata. 2025. "Classification of Mycena and Marasmius Species Using Deep Learning Models: An Ecological and Taxonomic Approach" Sensors 25, no. 6: 1642. https://doi.org/10.3390/s25061642
APA StyleEkinci, F., Ugurlu, G., Ozcan, G. S., Acici, K., Asuroglu, T., Kumru, E., Guzel, M. S., & Akata, I. (2025). Classification of Mycena and Marasmius Species Using Deep Learning Models: An Ecological and Taxonomic Approach. Sensors, 25(6), 1642. https://doi.org/10.3390/s25061642






