MRI-Based Meningioma Firmness Classification Using an Adversarial Feature Learning Approach
Abstract
1. Introduction
2. Materials and Methods
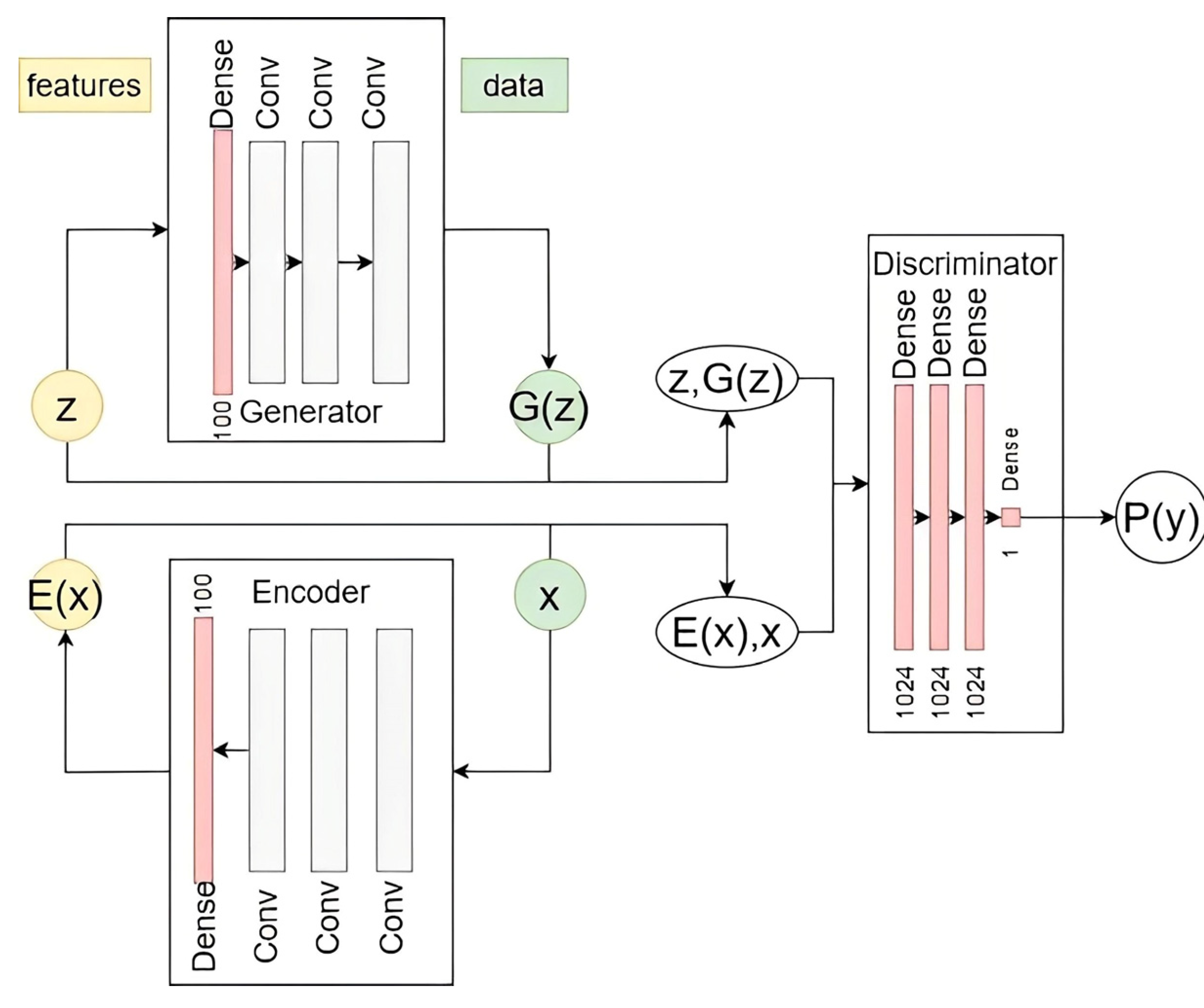
2.1. Adversarial Learning-Based Feature Extraction
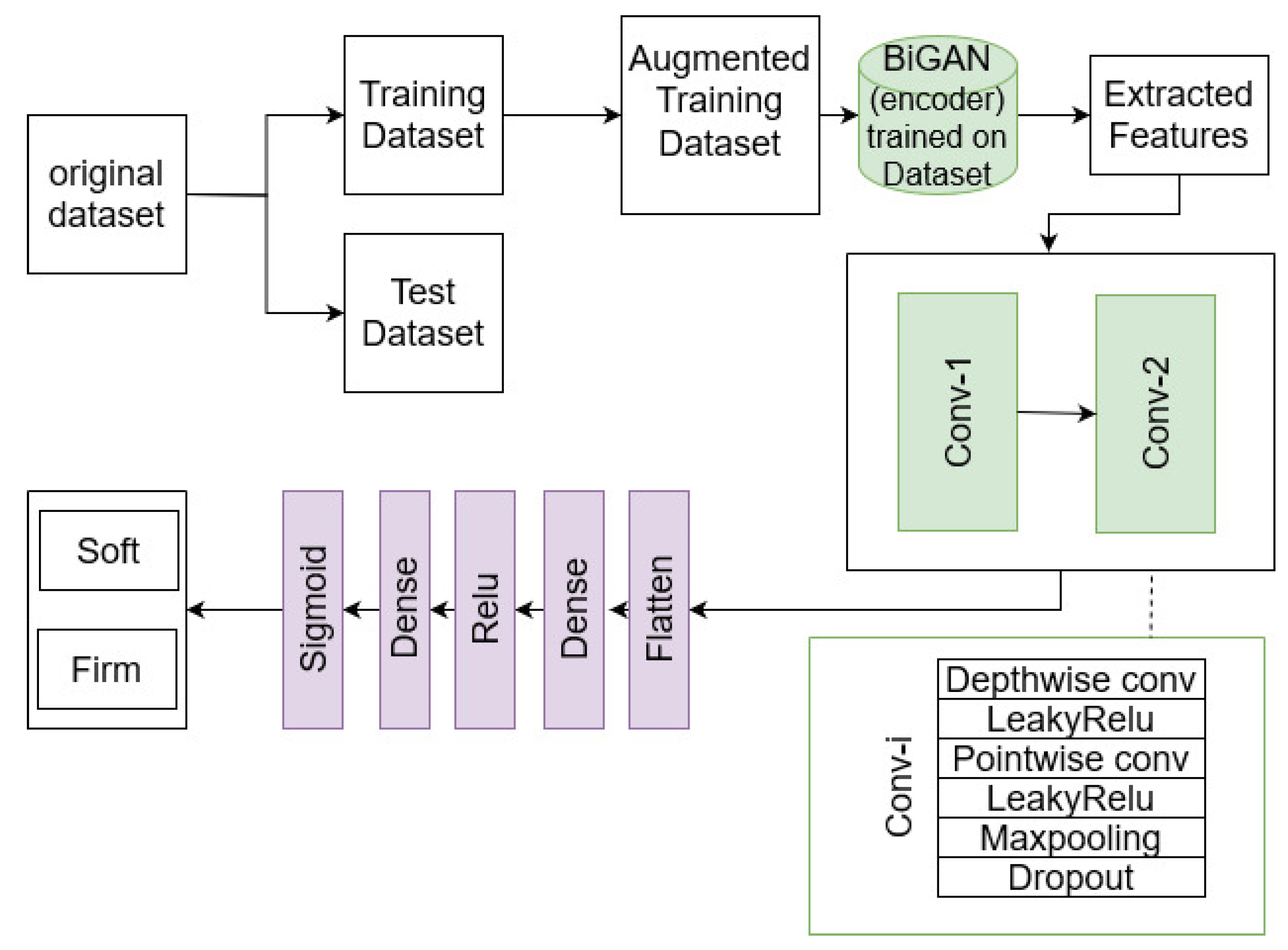
2.2. Depth-Wise Separable Deep Learning
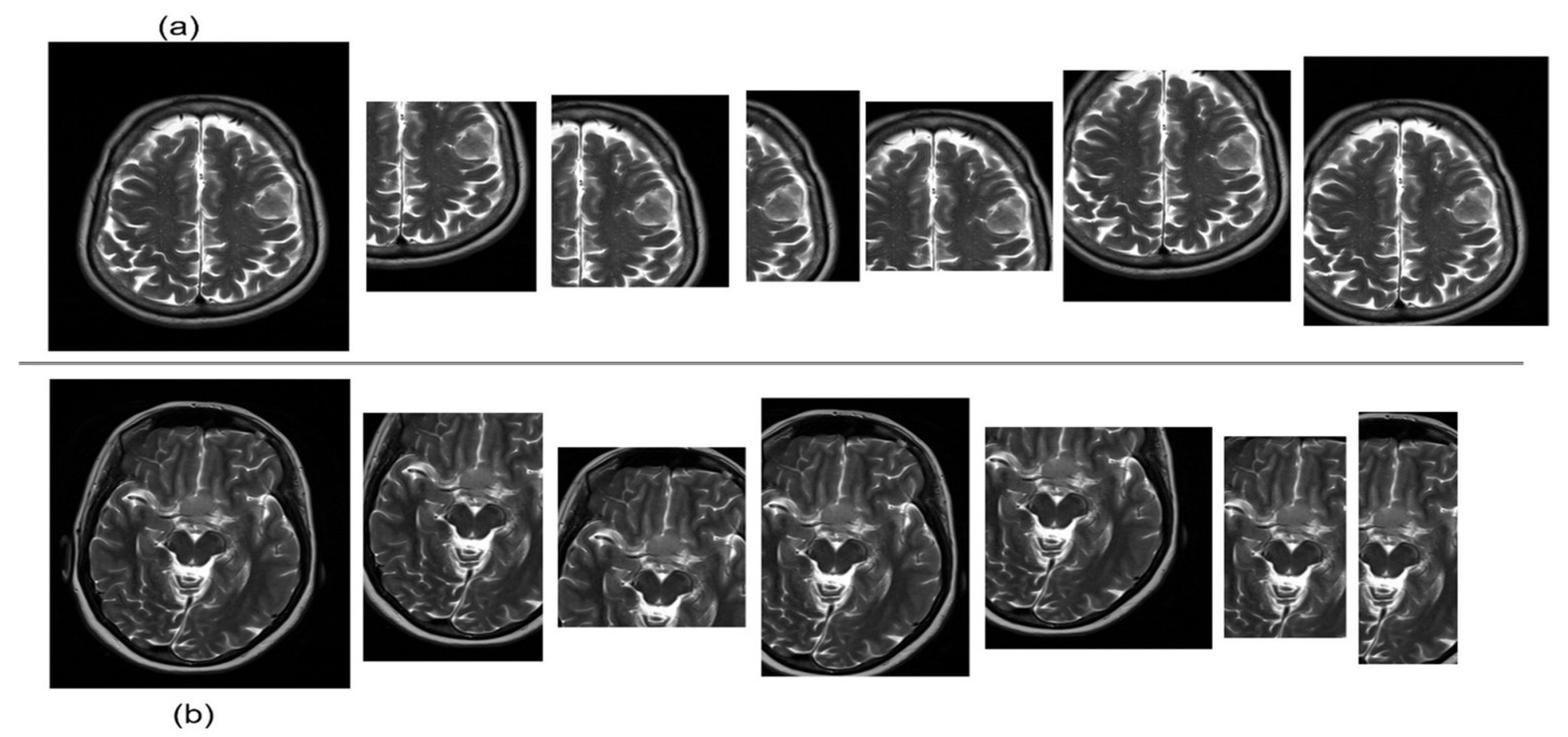
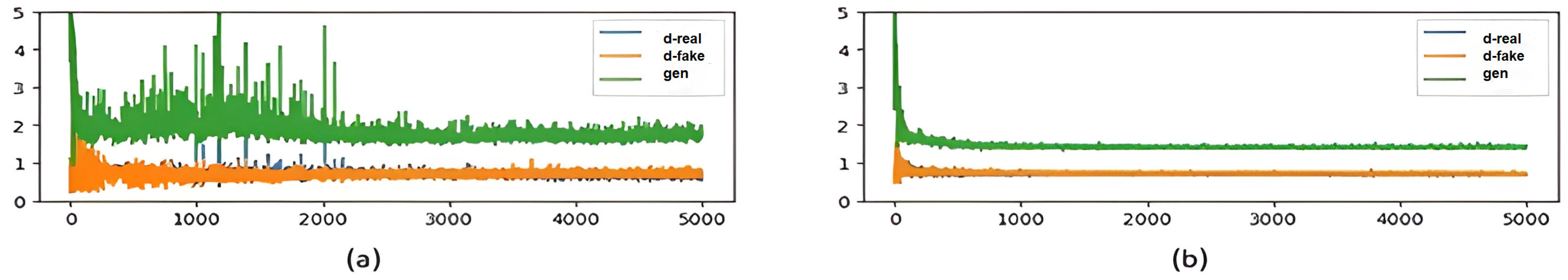
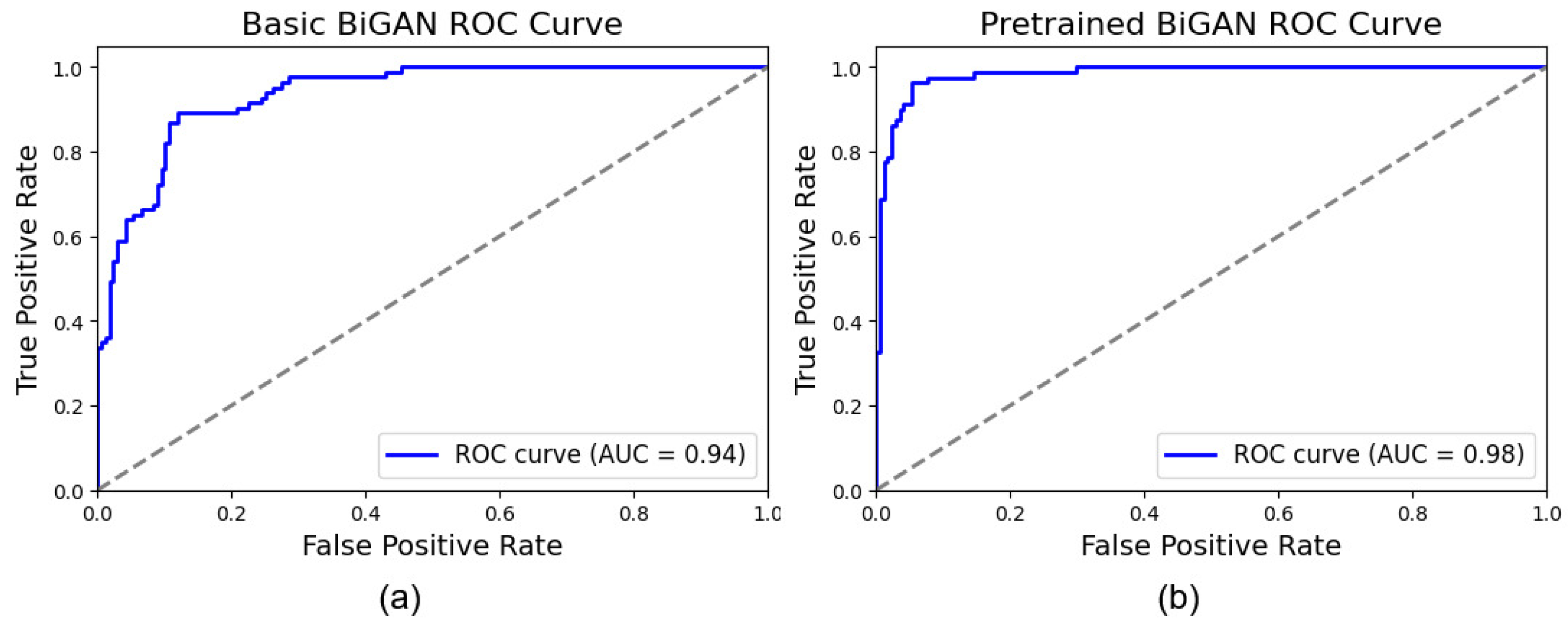
3. Experiments
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alqahtani, W.S.; Almufareh, N.A.; Domiaty, D.M.; Albasher, G.; Alduwish, M.A.; Alkhalaf, H.; Almuzzaini, B.; Al-Marshidy, S.S.; Alfraihi, R.; Elasbali, A.M.; et al. Epidemiology of cancer in Saudi Arabia thru 2010–2019: A systematic review with constrained meta-analysis. AIMS Public Health 2020, 7, 679–696. [Google Scholar] [PubMed]
- Sahoo, A.K.; Parida, P. A clustering based approach for meningioma tumors extraction from brain MRI images. In Proceedings of the 2020 IEEE International Symposium on Sustainable Energy, Signal Processing and Cyber Security (iSSSC), Gunupur, Odisha, 16–17 December 2020. [Google Scholar]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic Brain Tumor Detection and Segmentation Using U-Net Based Fully Convolutional Networks. In Proceedings of the Medical Image Understanding and Analysis, Edinburgh, UK, 11–13 July 2017. [Google Scholar]
- Firmino, M.; Morais, A.H.; Mendoça, R.M.; Dantas, M.R.; Hekis, H.R.; Valentim, R. Computer-aided detection system for lung cancer in computed tomography scans: Review and future prospects. Biomed. Eng. Online 2014, 13, 41. [Google Scholar] [CrossRef]
- Cepeda, S.; Arrese, I.; García-García, S.; Velasco-Casares, M.; Escudero-Caro, T.; Zamora, T.; Sarabia, R. Meningioma Consistency Can Be Defined by Combining the Radiomic Features of Magnetic Resonance Imaging and Ultrasound Elastography. A Pilot Study Using Machine Learning Classifiers. World Neurosurg. 2021, 146, e1147–e1159. [Google Scholar] [CrossRef] [PubMed]
- Ben Ismail, M.M. Local Ensemble Approach for Meningioma Tumor Firmness Detection in MRI Images. In Proceedings of the 2020 2nd International Conference on Image, Video and Signal Processing, New York, NY, USA, 20–22 March 2020. [Google Scholar]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef]
- Ranjbarzadeh, R.; Bagherian Kasgari, A.; Jafarzadeh Ghoushchi, S.; Anari, S.; Naseri, M.; Bendechache, M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci. Rep. 2021, 11, 10930. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Coarse Tumor Boundaries. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019. [Google Scholar]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N. Brain Tumor Type Classification via Capsule Networks. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018. [Google Scholar]
- Irmak, E. Multi-Classification of Brain Tumor MRI Images Using Deep Convolutional Neural Network with Fully Optimized Framework. Iran. J. Sci. Technol. Trans. Electr. Eng. 2021, 45, 1015–1036. [Google Scholar] [CrossRef]
- Xue, Y.; Yang, Y.; Farhat, F.G.; Shih, F.Y.; Boukrina, O.; Barrett, A.M.; Binder, J.R.; Graves, W.W.; Roshan, U.W. Brain Tumor Classification with Tumor Segmentations and a Dual Path Residual Convolutional Neural Network from MRI and Pathology Images. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Virtual, 25–28 October 2020. [Google Scholar]
- Cheng, Y.; Qin, G.; Zhao, R.; Liang, Y.; Sun, M. ConvCaps: Multi-input Capsule Network for Brain Tumor Classification. In Proceedings of the Neural Information Processing: 26th International Conference, ICONIP 2019, Sydney, NSW, Australia, 12–15 December 2019. [Google Scholar]
- Kumar, R.L.; Kakarla, J.; Isunuri, B.V.; Singh, M. Multi-class brain tumor classification using residual network and global average pooling. Multimed. Tools Appl. 2021, 80, 13429–13438. [Google Scholar] [CrossRef]
- Abiwinanda, N.; Hanif, M.; Hesaputra, S.T.; Handayani, A.; Mengko, T.R. Brain Tumor Classification Using Convolutional Neural Network. In World Congress on Medical Physics and Biomedical Engineering 2018; Springer: Singapore, 2019. [Google Scholar]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Brain Tumor Dataset. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427/8 (accessed on 23 January 2025).
- Pashaei, A.; Sajedi, H.; Jazayeri, N. Brain tumor classification via convolutional neural network and extreme learning machines. In Proceedings of the 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 25–26 October 2018. [Google Scholar]
- Luo, F.; Liu, G.; Guo, W.; Chen, G.; Xiong, N. ML-KELM: A kernel extreme learning machine scheme for multi-label classification of real time data stream in SIoT. IEEE Trans. Netw. Sci. Eng. 2021, 9, 1044–1055. [Google Scholar] [CrossRef]
- Sadoon, T.A.; Ali, M.H. Deep learning model for glioma, meningioma and pituitary classification. Int. J. Adv. Appl. Sci. ISSN 2021, 2252, 8814. [Google Scholar] [CrossRef]
- Ge, C.; Qu, Q.; Gu, I.Y.-H.; Store Jakola, A. 3D Multi-Scale Convolutional Networks for Glioma Grading Using MR Images. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018. [Google Scholar]
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018. [Google Scholar]
- Anaraki, A.K.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74. [Google Scholar] [CrossRef]
- Banzato, T.; Cherubini, G.B.; Atzori, M.; Zotti, A. Development of a deep convolutional neural network to predict grading of canine meningiomas from magnetic resonance images. Vet. J. 2018, 235, 90–92. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- AlKubeyyer, A.; Ben Ismail, M.M.; Bchir, O.; Alkubeyyer, M. Automatic detection of the meningioma tumor firmness in MRI images. J. Xray Sci. Technol. 2020, 28, 659–682. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Sebastian, V.B.; Unnikrishnan, A.; Balakrishnan, K. Gray Level Co-Occurrence Matrices: Generalisation and Some New Features. arXiv 2012, arXiv:1205.4831. [Google Scholar]
- Xu, L.; Gao, Q.; Yousefi, N. Brain tumor diagnosis based on discrete wavelet transform, gray-level co-occurrence matrix, and optimal deep belief network. Simulation 2020, 96, 867–879. [Google Scholar] [CrossRef]
- Zhai, Y.; Song, D.; Yang, F.; Wang, Y.; Jia, X.; Wei, S.; Mao, W.; Xue, Y.; Wei, X. Preoperative Prediction of Meningioma Consistency via Machine Learning-Based Radiomics. Front. Oncol. 2021, 11, 657288. [Google Scholar] [CrossRef]
- Alhussainan, N.F.; Youssef, B.B.; Ben Ismail, M.M. A Deep Learning Approach for Brain Tumor Firmness Detection Using YOLOv4. In Proceedings of the 2022 45th International Conference on Telecommunications and Signal Processing (TSP), Prague, Czech Republic, 13–15 July 2022. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.-A. Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Xing, C.; Ma, L.; Yang, X. Stacked Denoise Autoencoder Based Feature Extraction and Classification for Hyperspectral Images. J. Sens. 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Z.; Hu, X. Extracting Semantic Knowledge From GANs with Unsupervised Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 9654–9668. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Zhang, J.; Chen, L.; Zhuo, L.; Liang, X.; Li, J. An Efficient Hyperspectral Image Retrieval Method: Deep Spectral-Spatial Feature Extraction with DCGAN and Dimensionality Reduction Using t-SNE-Based NM Hashing. Remote Sens. 2018, 10, 271. [Google Scholar] [CrossRef]
- Zhang, M.; Gong, M.; Mao, Y.; Li, J.; Wu, Y. Unsupervised Feature Extraction in Hyperspectral Images Based on Wasserstein Generative Adversarial Network. IEEE Trans. Geosci. Remote Sens. 2019, 57, 2669–2688. [Google Scholar] [CrossRef]
- Sharif Razavian, A.; Azizpour, H.; Sullivan, J.; Carlsson, S. CNN features off-the-shelf: An astounding baseline for recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014. [Google Scholar]
- Taleb, A.; Rohrer, C.; Bergner, B.; De Leon, G.; Rodrigues, J.A.; Schwendicke, F.; Lippert, C.; Krois, J. Self-supervised learning methods for label-efficient dental caries classification. Diagnostics 2022, 12, 1237. [Google Scholar] [CrossRef] [PubMed]
- Donahue, J.; Krähenbühl, P.; Darrell, T. Adversarial Feature Learning. In Proceedings of the International Conference on Learning Representations (ICLR), Toulon, France, 2–4 May 2016. [Google Scholar]
- Xu, J.; Li, Z.; Du, B.; Zhang, M.; Liu, J. Reluplex made more practical: Leaky ReLU. In Proceedings of the 2020 IEEE Symposium on Computers and Communications (ISCC), Rennes, France, 8–10 July 2020. [Google Scholar]
- Dubey, S.R.; Singh, S.K.; Chaudhuri, B.B. Activation functions in deep learning: A comprehensive survey and benchmark. Neurocomput 2022, 503, 92–108. [Google Scholar] [CrossRef]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs trained by a two time-scale update rule converge to a local nash equilibrium. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 4–9 December 2017. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Munich, Germany, 18–22 June 2018. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, Hawaii, USA, 21–26 July 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 2015 International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Kong, Q.; Chiang, A.; Aguiar, A.C.; Fernández-Godino, M.G.; Myers, S.C.; Lucas, D.D. Deep convolutional autoencoders as generic feature extractors in seismological applications. Artif. Intell. Geosci. 2021, 2, 96–106. [Google Scholar] [CrossRef]





| Layer Name | Input Shape | Filter Size | Stride | Padding | Output Shape |
|---|---|---|---|---|---|
| conv2d | (56, 56, 3) | (3, 3) | (2, 2) | same | (28, 28, 32) |
| conv_dw_1 | (28, 28, 32) | (3, 3) | (1, 1) | same | (28, 28, 32) |
| conv_pw_1 | (28, 28, 32) | (1, 1) | (1, 1) | same | (28, 28, 64) |
| conv_dw_2 | (29, 29, 64) | (3, 3) | (2, 2) | valid | (14, 14, 64) |
| conv_pw_2 | (14, 14, 64) | (1, 1) | (1, 1) | same | (14, 14, 128) |
| conv_dw_3 | (14, 14, 128) | (3, 3) | (1, 1) | same | (14, 14, 128) |
| conv_pw_3 | (14, 14, 128) | (1, 1) | (1, 1) | same | (14, 14, 128) |
| conv_dw_4 | (15, 15, 128) | (3, 3) | (2, 2) | valid | (7, 7, 128) |
| conv_pw_4 | (7, 7, 128) | (1, 1) | (1, 1) | same | (7, 7, 256) |
| conv_dw_5 | (7, 7, 256) | (3, 3) | (1, 1) | same | (7, 7, 256) |
| conv_pw_5 | (7, 7, 256) | (1, 1) | (1, 1) | same | (7, 7, 256) |
| conv_dw_6 | (8, 8, 256) | (3, 3) | (2, 2) | valid | (3, 3, 256) |
| conv_pw_6 | (3, 3, 256) | (1, 1) | (1, 1) | same | (3, 3, 512) |
| conv_dw_7 | (3, 3, 512) | (3, 3) | (1, 1) | same | (3, 3, 512) |
| conv_pw_7 | (3, 3, 512) | (1, 1) | (1, 1) | same | (3, 3, 512) |
| conv_dw_8 | (3, 3, 512) | (3, 3) | (1, 1) | same | (3, 3, 512) |
| conv_pw_8 | (3, 3, 512) | (1, 1) | (1, 1) | same | (3, 3, 512) |
| conv_dw_9 | (3, 3, 512) | (3, 3) | (1, 1) | same | (3, 3, 512) |
| conv_pw_9 | (3, 3, 512) | (1, 1) | (1, 1) | same | (3, 3, 512) |
| conv_dw_10 | (3, 3, 512) | (3, 3) | (1, 1) | same | (3, 3, 512) |
| conv_pw_10 | (3, 3, 512) | (1, 1) | (1, 1) | same | (3, 3, 512) |
| conv_dw_11 | (3, 3, 512) | (3, 3) | (1, 1) | same | (3, 3, 512) |
| conv_pw_11 | (3, 3, 512) | (1, 1) | (1, 1) | same | (3, 3, 512) |
| conv_dw_12 | (4, 4, 512) | (3, 3) | (2, 2) | valid | (1, 1, 512) |
| conv_pw_12 | (1, 1, 512) | (1, 1) | (1, 1) | same | (1, 1, 1024) |
| conv_dw_13 | (1, 1, 1024) | (3, 3) | (1, 1) | same | (1, 1, 1024) |
| conv_pw_13 | (1, 1, 1024) | (1, 1) | (1, 1) | same | (1, 1, 1024) |
| Approach | Accuracy | Precision | Recall | Weighted F1_Score | Cohen’s Kappa Coefficient |
|---|---|---|---|---|---|
| NASNetLarge [46] | 86.29 (+/−1.12) | 0.72 (+/−0.04) | 0.86 (+/−0.09) | 0.87 (+/−0.01) | 0.68 (+/−0.02) |
| DenseNet201 [47] | 86.72 (+/−3.14) | 0.93 (+/−0.04) | 0.65 (+/−0.10) | 0.86 (+/−0.04) | 0.68 (+/−0.09) |
| ResNet50 [48] | 74.56 (+/−0.93) | 0.43 (+/−0.22) | 0.46 (+/−0.13) | 0.70 (+/−0.05) | 0.19 (+/−0.13) |
| VGG16 [49] | 88.16 (+/−2.17) | 0.85 (+/−0.07) | 0.82 (+/−0.08) | 0.88 (+/−0.02) | 0.74 (+/−0.05) |
| Autoencoder [50] | 84.80 (+/−1.82) | 0.81 (+/−0.06) | 0.79 (+/−0.08) | 0.85 (+/−0.02) | 0.67 (+/−0.04) |
| The approach in [27] | 80.0 (+/−0.01) | 0.77 (+/−0.02) | 0.54 (+/−0.04) | 0.79 (+/−0.01) | 0.50 (+/−0.03) |
| DCGAN [38] | 86.37 (+/−2.49) | 0.87 (+/−0.05) | 0.75 (+/−0.12) | 0.86 (+/−0.03) | 0.70 (+/−0.07) |
| WGAN [39] | 85.34 (+/−2.16) | 0.82 (+/−0.04) | 0.76 (+/−0.05) | 0.85 (+/−0.02) | 0.67 (+/−0.05) |
| Depth-wise separable deep learning-based approach | 72.32 (+/−2.18) | 0.56 (+/−0.03) | 0.80 (+/−0.15) | 0.73 (+/−0.02) | 0.44 (+/−0.05) |
| Basic BiGAN-based approach | 87.52 (+/−1.30) | 0.87 (+/−0.09) | 0.80 (+/−0.12) | 0.87 (+/−0.02) | 0.72 (+/−0.04) |
| Pre-trained BiGAN-based approach | 94.72 (+/−3.34) | 0.94 (+/−0.06) | 0.87 (+/−0.14) | 0.95 (+/−0.04) | 0.86 (+/−0.10) |
| Model | Time (s) |
|---|---|
| NASNetLarge [46] | 33.59 |
| DenseNet201 [47] | 10.77 |
| ResNet50 [48] | 15.94 |
| VGG16 [49] | 168.34 |
| Autoencoder [50] | 18.04 |
| DCGAN [38] | 37.53 |
| WGAN [39] | 35.9 |
| Depth-wise separable deep learning-based approach | 78.87 |
| Basic BiGAN-based approach | 15.38 |
| Pre-trained BiGAN-based approach | 19.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murad, M.; Touir, A.; Ben Ismail, M.M. MRI-Based Meningioma Firmness Classification Using an Adversarial Feature Learning Approach. Sensors 2025, 25, 1397. https://doi.org/10.3390/s25051397
Murad M, Touir A, Ben Ismail MM. MRI-Based Meningioma Firmness Classification Using an Adversarial Feature Learning Approach. Sensors. 2025; 25(5):1397. https://doi.org/10.3390/s25051397
Chicago/Turabian StyleMurad, Miada, Ameur Touir, and Mohamed Maher Ben Ismail. 2025. "MRI-Based Meningioma Firmness Classification Using an Adversarial Feature Learning Approach" Sensors 25, no. 5: 1397. https://doi.org/10.3390/s25051397
APA StyleMurad, M., Touir, A., & Ben Ismail, M. M. (2025). MRI-Based Meningioma Firmness Classification Using an Adversarial Feature Learning Approach. Sensors, 25(5), 1397. https://doi.org/10.3390/s25051397






