Med-Diffusion: Diffusion Model-Based Imputation of Multimodal Sensor Data for Surgical Patients
Abstract
1. Introduction
- We propose a novel diffusion model-based multimodal medical data imputation method, Med-Diffusion, and implement it for the tabular imputation of static medical sequences.
- We propose a comprehensive pipeline for effectively training Med-Diffusion on categorical features, utilizing advanced encoding and embedding techniques.
- Our experimental results show that Med-Diffusion reduces error rates and the RMSE (Root Mean Square Error) by 4.4% and 5.1%, respectively, compared to existing probabilistic methods for healthcare and environmental data.
2. Related Works
2.1. Disease Risk Prediction Based on Multimodal Medical Data
2.2. Multimodal Medical Data Imputation
2.3. Deep Learning-Based Data Imputation
2.4. Generative Artificial Intelligence and Diffusion Methods
3. Methodology
3.1. Overview
- Handling Categorical Variables: Categorical variables are processed using one-hot encoding, simulated bit encoding, and embedding encoding methods. Among these, the embedding method (feature tokenization/FT) is found to perform better. Thus, categorical variables are processed using FT in the experimental implementation, and this is reflected in Table 2.
- Forward Diffusion Process: Both the complete dataset and the incomplete dataset are gradually transformed into a Gaussian distribution by adding noise, which generates a series of progressively noisy states.
- Reverse Generation Process: A neural network is trained on the complete dataset to model the reverse process. This process enables the model to gradually recover the original data from noisy states, essentially reversing the effects of the forward diffusion process.
- Imputing Missing Data: For the incomplete dataset, the reverse process, which is trained on the complete dataset, is used to gradually recover the missing data points from the Gaussian noise state. This process combines the reverse diffusion model with the conditional observed values.
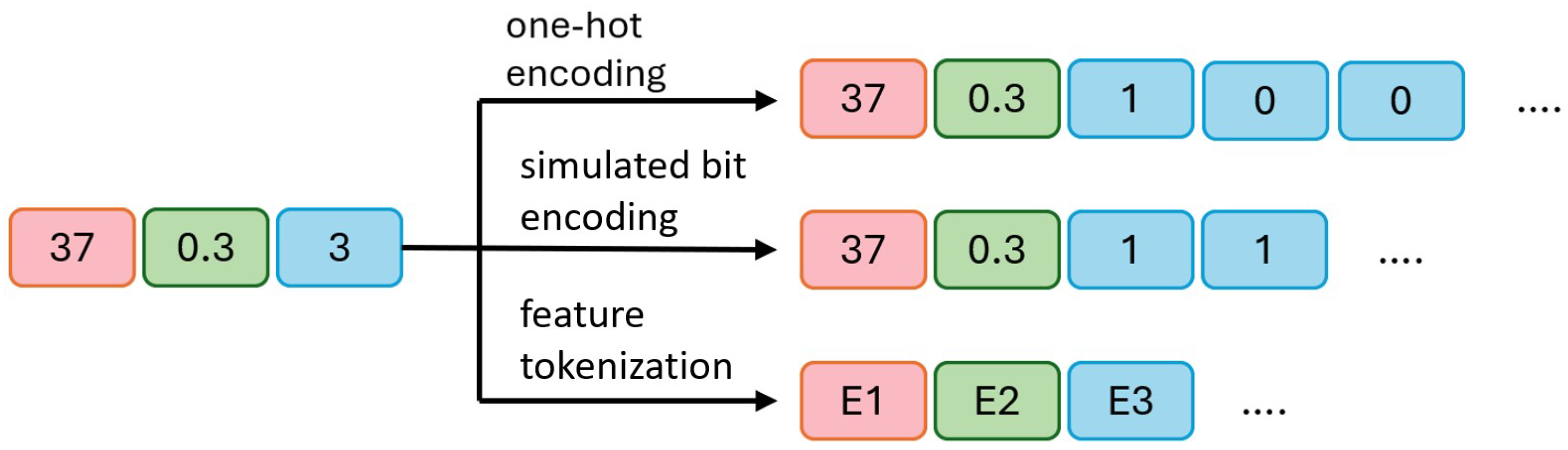
3.2. Categorical Variable Processing
- a.
- In one-hot encoding, 45 and 0.7 remain unchanged, while the categorical variable 3 is converted into three binary bits, [1, 0, 0], indicating the third category is selected (1) and the other two are unselected (0).
- b.
- In simulated bit encoding, 45 and 0.7 remain unchanged, with each category assigned a unique fixed-length binary code. The categorical variable 3 is encoded as two binary bits, [1, 1], where a more compact representation requires fewer bits.
- c.
- In processing categorical variables, embedding encoding is used to convert all variables (both numerical and categorical) into embeddings of the same length. E1, E2, and E3 represent the embedded representations of the three original variables. This approach maps all variables to a unified dimensional space. During output processing, the embedding vector is converted back to the original numerical or categorical values. For numerical variables, the final value is obtained by averaging the elements of the embedding vector divided by the output of the diffusion model. For categorical variables, the predicted category is determined by calculating the distance between the embedding vector and the diffusion model output, selecting the nearest embedding vector.
3.3. Diffusion Probabilistic Model for Numerical Imputation
3.3.1. Objective and Evaluation Metric
- (1)
- Defining the Conditional Diffusion Model: Med-Diffusion introduces the Conditional Diffusion Model to impute the missing values conditioned on observed data. This includes the following. Conditioning Setup: For each sample , define the observed part (non-missing values) and the missing part y. The goal is to learn the conditional distribution . In the diffusion model, the conditional information is incorporated into the reverse process. Forward Process: The forward process is the same as in the standard diffusion model, where noise is gradually added to the true data , generating . For numerical variables, Gaussian noise is directly added; for categorical variables, one-hot encoding is applied first, and then noise is added. Reverse Process: The reverse process is conditioned on the observed values and progressively denoised to generate imputed values. Med-Diffusion modifies the reverse process to focus on imputing the missing part while keeping the observed part unchanged.
- (2)
- Processing Numerical and Categorical Variables: Med-Diffusion employs distinct modeling strategies for numerical and categorical variables:
- a.
- Numerical Variables (e.g., heart rate and blood pressure). Modeling: Numerical variables are treated as continuous values, fitting the Gaussian noise assumption of the diffusion model. Imputation: During the reverse diffusion process, missing numerical values are progressively denoised to reasonable real values. For example, heart rate might recover from a noisy value to 80 beats per minute, and blood pressure may return to 120/80 mmHg. Loss Function: Mean squared error (MSE) is used to optimize the imputation results, ensuring the imputed values are close to the true distribution.
- b.
- Categorical Variables (e.g., alcohol consumption level and heart rhythm status). Modeling: Categorical variables are transformed into continuous vectors through one-hot encoding. However, the Gaussian noise of the diffusion model may disrupt the discrete nature. Therefore, Med-Diffusion introduces a Discrete Diffusion Model or a post-processing step. Imputation: During the reverse diffusion process, the one-hot (embedding) vector for categorical variables is denoised, generating a probability distribution (e.g., alcohol consumption level might produce [0.7, 0.2, 0.1]). Post-processing: The probability distribution is converted into discrete categories by selecting the category with the highest probability (e.g., [0.7, 0.2, 0.1] becomes “mild”). Loss Function: Cross-entropy loss is used to optimize the imputation results for categorical variables, ensuring the imputed category aligns with the true distribution.
- (3)
- Training the Imputation Function f: Neural Network Design: Med-Diffusion employs a neural network based on a diffusion architecture to parameterize the mean of the reverse diffusion process. Input: Current time step t, noisy data , and conditional information . Output: Denoised data , which corresponds to the imputed data. Training Goal: The goal is to optimize the neural network parameters so that the predicted noise approaches the true noise, thus making the denoising process as realistic as possible. The objective function iswhere is the noise predicted by the neural network, and represents the conditional information. : A time step t is randomly selected from the diffusion process. : An original data point is sampled from the training set, which may contain missing or non-missing values. : Noise sampled from a standard normal distribution. : Given an original sample and a time step t, the noisy version is obtained using this formula. is a time-dependent noise decay factor, which is predetermined in this experiment (usually provided by a pre-defined linear noise schedule). : The model predicts the most likely original noise given the noisy data , the current time step t, and the conditional information (i.e., the observed non-missing part). : The standard mean squared error (MSE) between the model’s predicted noise and the true noise. Minimizing this error is equivalent to instructing the model to “reverse the noise I added.” Training Process. a. Sample from the training data . b. Randomly select a time step t and generate the noisy version . c. Use the observed values as conditioning information to predict the noise . d. Optimize the loss function and update the network parameters .
- (4)
- Inference Process (Generation Process):
- a.
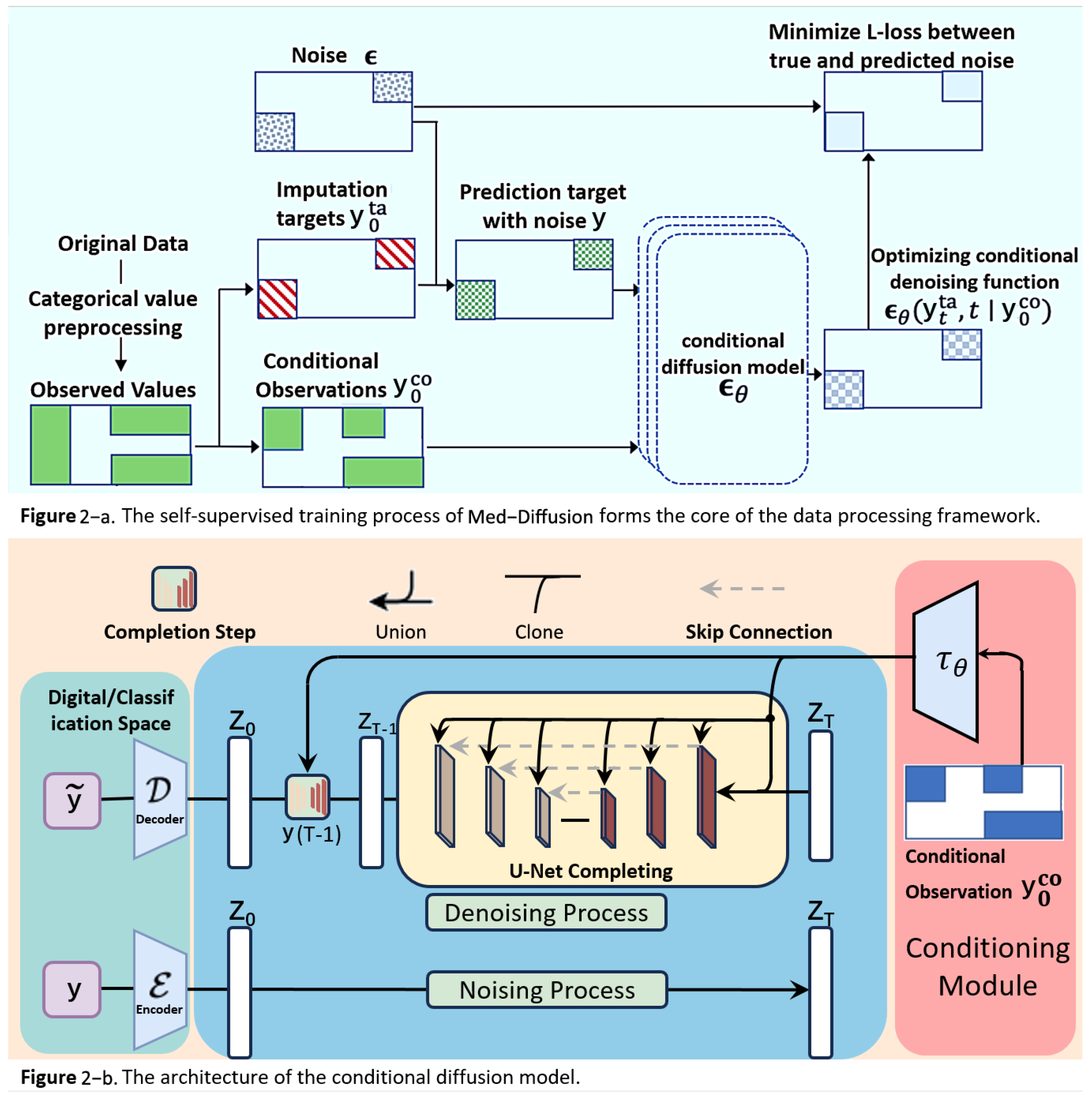
- Input Sample. The input sample , which contains missing values, is represented as . This vector has several missing dimensions to be predicted (denoted by a mask M, where indicates the j-th dimension is missing and indicates the j-th dimension is observed). We divided the data into two parts—the conditional observed values as the observable part and the target missing values y as the part to be imputed. The observed and missing sections are represented by the blue and red areas in Figure 2.
- b.
- Initialization of Missing Value Region, Forming . To initiate the reverse diffusion process, we need an initial “fully noisy” vector. The observed region retains the original ; the missing region y is initialized with random noise (Gaussian distribution). We constructed an initial statewhere , introducing noise in only the missing region, while the observed region retained the true values.
- c.
- Conditional Control Mechanism Input—Feeding into Multi-layer Denoising U-Net for T Steps. Next, we feed the current noisy data , the current time step t, and the conditional observed values into a neural network function (in this paper, U-Net). represents the network parameters, and the network predicts the noise term . In other words, the network asks the following: “How should I remove the noise to make the missing region more realistic?”The “conditioning mechanism” (highlighted in red in the figure) is crucial as it ensures that the network knows which areas are the observed values and which are missing, allowing for the imputation of the missing values according to the completion process of the U-Net.
- d.
- Step-by-Step Reverse Diffusion, from to . Using the noise output from the forward process, we update according to the reverse formulaNote that the observed region is “condition-locked” and does not participate in sampling or noise updating. The reverse diffusion mechanism generates the predicted missing values based on the conditional observed data. Starting from , we update step-by-step to , gradually denoising the data from a fully noisy state to the true data space, generating the imputed values for the missing regions.
- e.
- Obtaining the Imputed Result . Finally, when , the network outputs the predicted values for the features , which are decoded to obtain . At this point, the missing regions have been filled through multiple steps of denoising, and the denoised output will be as close as possible to the true data distribution, with the observed regions remaining unchanged. This output can directly serve as the imputed result for further analysis.Through these two steps—the training phase and the inference phase—the Med-Diffusion model effectively learns to recover the missing parts of samples with missing values, providing an accurate and efficient imputation function f.
- f.
- Post-processing. The processed input is then used to train the model. After obtaining the raw output, different processing schemes require different recovery processes.
3.3.2. Evaluation Process
4. Experiments
4.1. Datasets
4.2. Experimental Details
5. Results and Discussion
Discussion
- (1)
- Mean/Mode Baseline Method:
- (2)
- MICE (Multiple Imputation by Chained Equations) Linear Regression Version [44]:
- (3)
- MissForest Random Forest Imputation [11]:
- (4)
- GAINs (Generative Adversarial Imputation Networks) [12]:
- (5)
- CSDI [45]:
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moon, K.S.; Lee, S.Q. A wearable multimodal wireless sensing system for respiratory monitoring and analysis. Sensors 2023, 23, 6790. [Google Scholar] [CrossRef]
- Choi, S.H.; Yoon, H.; Baek, H.J.; Long, X. Biomedical Signal Processing and Health Monitoring Based on Sensors. Sensors 2025, 25, 641. [Google Scholar] [CrossRef] [PubMed]
- Yashudas, A.; Gupta, D.; Prashant, G.C.; Dua, A.; AlQahtani, D.; Reddy, A.S.K. Deep-cardio: Recommendation system for cardiovascular disease prediction using IoT network. IEEE Sens. J. 2024, 24, 14539–14547. [Google Scholar] [CrossRef]
- Shieh, A.; Tran, B.; He, G.; Kumar, M.; Freed, J.A.; Majety, P. Assessing ChatGPT 4.0’s test performance and clinical diagnostic accuracy on USMLE STEP 2 CK and clinical case reports. Sci. Rep. 2024, 14, 9330. [Google Scholar] [CrossRef]
- Abdulhussain, S.H.; Mahmmod, B.M.; Alwhelat, A.; Shehada, D.; Shihab, Z.I.; Mohammed, H.J.; Abdulameer, T.H.; Alsabah, M.; Fadel, M.H.; Ali, S.K.; et al. A Comprehensive Review of Sensor Technologies in IoT: Technical Aspects, Challenges, and Future Directions. Computers 2025, 14, 342. [Google Scholar] [CrossRef]
- Campello, V.M.; Gkontra, P.; Izquierdo, C.; Martín-Isla, C.; Sojoudi, A.; Full, P.M. Multi-centre, multi-vendor and multi-disease cardiac segmentation: The M&Ms challenge. IEEE Trans. Med. Imag. 2021, 40, 3543–3554. [Google Scholar]
- Vaduganathan, M.; Mensah, G.A.; Turco, J.V.; Fuster, V.; Roth, G.A. The global burden of cardiovascular diseases and risk: A compass for future health. J. Am. Coll. Cardiol. 2022, 80, 2361–2371. [Google Scholar] [CrossRef]
- Behrad, F.; Abadeh, M.S. An overview of deep learning methods for multimodal medical data mining. Expert Syst. Appl. 2022, 200, 117006. [Google Scholar] [CrossRef]
- Soenksen, L.R.; Ma, Y.; Zeng, C.; Boussioux, L.; Carballo, K.V.; Na, L.; Wiberg, H.M.; Li, M.L.; Fuentes, I.; Bertsimas, D. Integrated multimodal artificial intelligence framework for healthcare applications. NPJ Digit. Med. 2022, 5, 149. [Google Scholar] [CrossRef]
- Bardak, B.; Tan, M. Improving clinical outcome predictions using convolution over medical entities with multimodal learning. Artif. Intell. Med. 2021, 117, 102112. [Google Scholar] [CrossRef]
- Hong, S.; Lynn, H.S. Accuracy of random-forest-based imputation of missing data in the presence of non-normality, non-linearity, and interaction. BMC Med. Res. Methodol. 2020, 20, 199. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Jordon, J.; Schaar, M. Gain: Missing data imputation using generative adversarial nets. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 5689–5698. [Google Scholar]
- Li, Y.; Daho, M.E.H.; Conze, P.; Zeghlache, R.; Boité, H.L.; Tadayoni, R.; Cochener, B.; Lamard, M.; Quellec, G. A review of deep learning-based information fusion techniques for multimodal medical image classification. Comput. Biol. Med. 2024, 108635. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liao, I.Y.; Zhong, N.; Toshihiro, F.; Wang, Y.; Wang, S. Generative AI enables the detection of autism using EEG signals. In Proceedings of the Chinese Conference on Biometric Recognition, Xuzhou, China, 1–3 December 2023; pp. 375–384. [Google Scholar]
- Ho, J.; Jain, A.; Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 2020, 33, 6840–6851. [Google Scholar]
- Zheng, S.; Charoenphakdee, N. Diffusion models for missing value imputation in tabular data. arXiv 2022, arXiv:2210.17128. [Google Scholar]
- Mahmmod, B.M.; Abdulhussain, S.H.; Naser, M.A.; Alsabah, M.; Hussain, A.; Al-Jumeily, D. 3D object recognition using fast overlapped block processing technique. Sensors 2022, 22, 9209. [Google Scholar] [CrossRef]
- Alsabah, M.; Naser, M.A.; Albahri, A.S.; Albahri, O.S.; Alamoodi, A.H.; Abdulhussain, S.H.; Alzubaidi, L. A comprehensive review on key technologies toward smart healthcare systems based IoT: Technical aspects, challenges and future directions. Artif. Intell. Rev. 2025, 58, 343. [Google Scholar] [CrossRef]
- Hameed, I.M.; Abdulhussain, S.H.; Mahmmod, B.M.; Hussain, A. Content based image retrieval based on feature fusion and support vector machine. In Proceedings of the 14th International Conference on Developments in eSystems Engineering (DeSE), Sharjah, United Arab Emirates, 7–10 December 2021; pp. 552–558. [Google Scholar]
- Chen, P.-F.; Chen, L.; Lin, Y.-K.; Li, G.-H.; Lai, F.; Lu, C.-W.; Yang, C.-Y.; Chen, K.-C.; Lin, T.-Y. Predicting postoperative mortality with deep neural networks and natural language processing: Model development and validation. JMIR Med. Inform. 2022, 10, e38241. [Google Scholar] [CrossRef]
- Hui, V.; Litton, E.; Edibam, C.; Geldenhuys, A.; Hahn, R.; Larbalestier, R.; Wright, B.; Pavey, W. Using machine learning to predict bleeding after cardiac surgery. Eur. J. Cardio-Thorac. Surg. 2023, 64, ezad297. [Google Scholar] [CrossRef]
- McLean, K.A.; Sgrò, A.; Brown, L.R.; Buijs, L.F.; Mountain, K.E.; Shaw, C.A.; Drake, T.M.; Pius, R.; Knight, S.R.; Fairfield, C.J.; et al. Multimodal machine learning to predict surgical site infection with healthcare workload impact assessment. NPJ Digit. Med. 2025, 8, 121. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, Z.; Fan, G.; Li, N.; Xu, C.; Li, B.; Zhao, G.; Zhou, S. FocusMorph: A novel multi-scale fusion network for 3D brain MR image registration. Pattern Recognit. 2025, 167, 111761. [Google Scholar] [CrossRef]
- Rao, P.K.; Chatterjee, S.; Nagaraju, K.; Khan, S.B.; Almusharraf, A.; Alharbi, A.I. Fusion of graph and tabular deep learning models for predicting chronic kidney disease. Diagnostics 2023, 13, 1981. [Google Scholar] [CrossRef]
- Psychogyios, K.; Ilias, L.; Ntanos, C.; Askounis, D. Missing value imputation methods for electronic health records. IEEE Access 2023, 11, 21562–21574. [Google Scholar] [CrossRef]
- Fan, C.-C.; Peng, L.; Wang, T.; Yang, H.; Zhou, X.-H.; Ni, Z.-L.; Wang, G.; Chen, S.; Zhou, Y.-J.; Hou, Z.-G. TR-Gan: Multi-session future MRI prediction with temporal recurrent generative adversarial Network. IEEE Trans. Med. Imag. 2022, 41, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Noh, Y.-K.; Hu, M.; Shen, X.; Shah, V.H.; Kamath, P.S. Use of Artificial Intelligence–Generated Synthetic Data to Augment and Enhance the Performance of Clinical Prediction Models in Patients with Alcohol-Associated Hepatitis and Acute Cholangitis. Gastro Hep Adv. 2025, 4, 100643. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Yuan, H.; Ning, Y.; Ong, M.E.H.; Feng, M.; Hsu, W.; Chakraborty, B.; Liu, N. Deep learning for temporal data representation in electronic health records: A systematic review of challenges and methodologies. J. Biomed. Inform. 2022, 126, 103980. [Google Scholar] [CrossRef]
- Liaghat, B.; Ussing, A.; Petersen, B.H.; Andersen, H.K.; Barfod, K.W.; Jensen, M.B.; Hoegh, M.; Tarp, S.; Juul-Kristensen, B.; Brorson, S. Supervised training compared with no training or self-training in patients with subacromial pain syndrome: A systematic review and meta-analysis. Arch. Phys. Med. Rehabil. 2021, 102, 2428–2441. [Google Scholar] [CrossRef]
- Fortuin, V.; Baranchuk, D.; Rätsch, G.; Mandt, S. Gp-vae: Deep probabilistic time series imputation. In Proceedings of the International Conference on Artificial Intelligence and Statistics, Online, 26–28 August 2020; pp. 1651–1661. [Google Scholar]
- Waikel, R.L.; Othman, A.A.; Patel, T.; Hanchard, S.L.; Hu, P.; Tekendo-Ngongang, C.; Duong, D.; Solomon, B.D. Recognition of genetic conditions after learning with images created using generative artificial intelligence. JAMA Netw. Open 2024, 7, e242609. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. MissForest—Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, Z.; Fan, G.; Li, B.; Zhou, S.; Xu, C. MambaVesselNet: A Novel Approach to Blood Vessel Segmentation Based on State-Space Models. IEEE J. Biomed. Health Inform. 2024, 29, 2034–2047. [Google Scholar] [CrossRef]
- Yang, D.; Tian, J.; Tan, X.; Huang, R.; Liu, S.; Guo, H.; Chang, X.; Shi, J.; Zhao, S.; Bian, J.; et al. Uniaudio: Towards universal audio generation with large language models. In Proceedings of the International Conference on Machine Learning, Vienna, Austria, 21–27 July 2024. [Google Scholar]
- Zhang, Z.; Zhang, J.; Zhang, X.; Mai, W. A comprehensive overview of Generative AI (GAI): Technologies, applications, and challenges. Neurocomputing 2025, 632, 129645. [Google Scholar] [CrossRef]
- Guo, X.; Zhao, L. A systematic survey on deep generative models for graph generation. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 5370–5390. [Google Scholar] [CrossRef]
- Kollovieh, M.; Ansari, A.F.; Bohlke-Schneider, M.; Zschiegner, J.; Wang, H.; Wang, Y. Predict, refine, synthesize: Self-guiding diffusion models for probabilistic time series forecasting. Adv. Neural Inf. Process. Syst. 2023, 36, 28341–28364. [Google Scholar]
- Alcaraz, J.M.L.; Strodthoff, N. Diffusion-based time series imputation and forecasting with structured state space models. arXiv 2022, arXiv:2208.09399. [Google Scholar]
- Silva, I.; Moody, G.; Mark, R.; Celi, L.A. Predicting Mortality of ICU Patients: The PhysioNet/Computing in Cardiology Challenge 2012. Available online: https://physionet.org/content/challenge-2012/1.0.0/ (accessed on 1 August 2025).
- Dua, D.; Graff, C. UCI Machine Learning Repository. Available online: https://archive.ics.uci.edu/dataset/17/breast+cancer+wisconsin+diagnostic (accessed on 1 August 2025).
- Pytlak, K. Personal Key Indicators of Heart Disease dataset. Available online: https://www.kaggle.com/datasets/kamilpytlak/personal-key-indicators-of-heart-disease (accessed on 1 August 2025).
- Teboul, A. Diabetes Health Indicators Dataset. Available online: https://www.kaggle.com/datasets/alexteboul/diabetes-health-indicators-dataset (accessed on 1 August 2025).
- Tanmoy, X. COVID-19 Patient Pre-Condition Dataset. Available online: https://www.kaggle.com/datasets/tanmoyx/covid19-patient-precondition-dataset (accessed on 1 August 2025).
- Royston, P.; White, I.R. Multiple imputation by chained equations (MICE): Implementation in Stata. J. Stat. Softw. 2011, 45, 1–20. [Google Scholar] [CrossRef]
- Tashiro, Y.; Song, J.; Song, Y.; Ermon, S. Csdi: Conditional score-based diffusion models for probabilistic time series imputation. Adv. Neural Inf. Process. Syst. 2021, 34, 24804–24816. [Google Scholar]


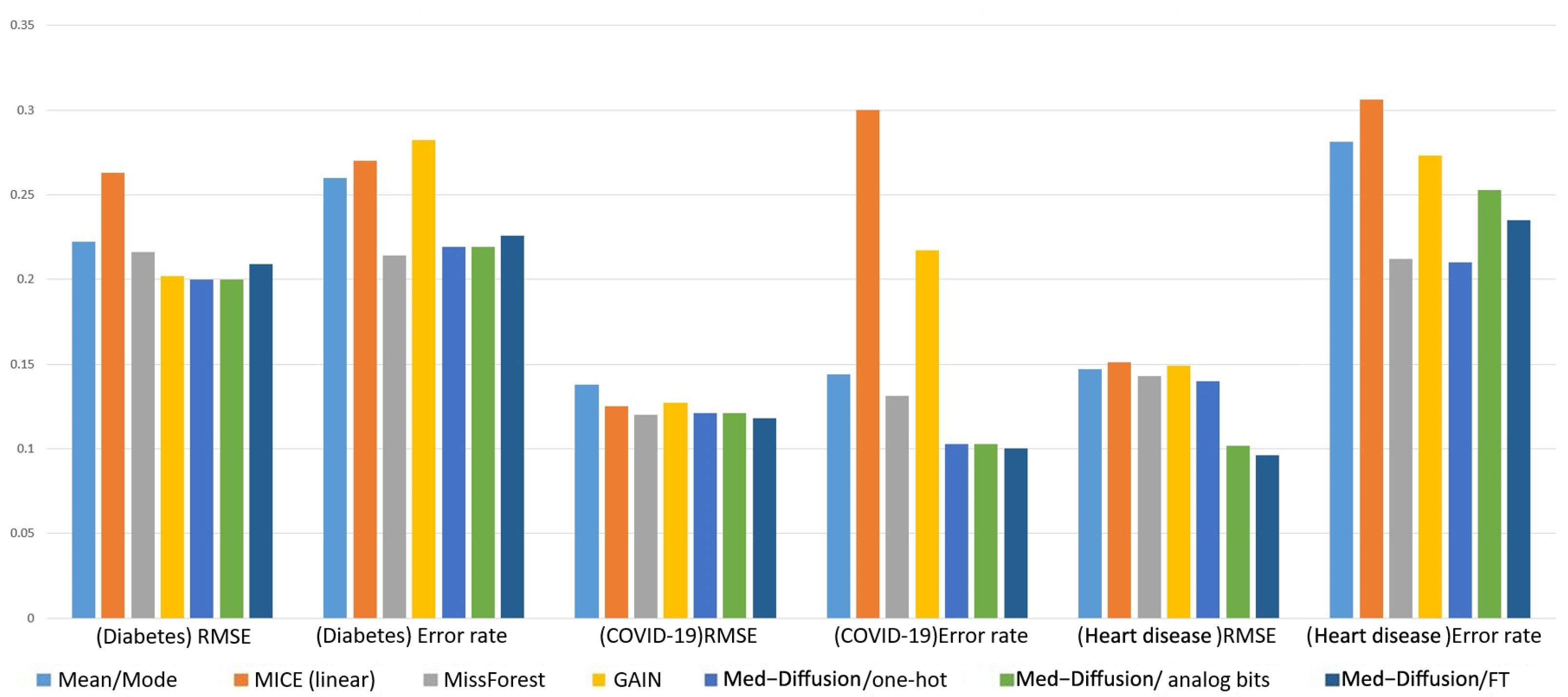
| Diabetes | COVID-19 | Heart Disease | ||||
|---|---|---|---|---|---|---|
| RMSE | Error Rate | RMSE | Error Rate | RMSE | Error Rate | |
| Mean/Mode | 0.222 (0.003) | 0.260 (0.004) | 0.138 (0.002) | 0.144 (0.002) | 0.147 (0.004) | 0.281 (0.005) |
| MICE (linear) | 0.263 (0.002) | 0.270 (0.004) | 0.125 (0.003) | 0.300 (0.038) | 0.151 (0.005) | 0.306 (0.013) |
| MissForest | 0.216 (0.003) | 0.214 (0.001) | 0.120 (0.002) | 0.131 (0.002) | 0.143 (0.002) | 0.212 (0.017) |
| GAINs | 0.202 (0.003) | 0.282 (0.005) | 0.127 (0.002) | 0.217 (0.011) | 0.149 (0.045) | 0.273 (0.006) |
| Med-Diffusion/one-hot | 0.200 (0.001) | 0.219 (0.004) | 0.121 (0.005) | 0.103 (0.010) | 0.140 (0.001) | 0.210 (0.009) |
| Med-Diffusion/simulated bits | 0.200 (0.001) | 0.219 (0.004) | 0.121 (0.005) | 0.103 (0.010) | 0.102 (0.004) | 0.253 (0.009) |
| Med-Diffusion/FT | 0.209 (0.002) | 0.226 (0.003) | 0.118 (0.003) | 0.100 (0.002) | 0.096 (0.001) | 0.235 (0.008) |
| Mean | MICE (Linear) | MissForest | GAINs | CSDI | Med-Diffusion | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | |
| PhysioNet | 0.251 (0.013) | 0.2002 | 0.214 (0.013) | 0.1707 | 0.170 (0.012) | 0.1356 | 0.168 (0.010) | 0.1340 | 0.177 (0.027) | 0.1412 | 0.150 (0.008) | 0.1197 |
| Breast Cancer | 0.263 (0.009) | 0.2097 | 0.154 (0.011) | 0.1229 | 0.163 (0.014) | 0.1301 | 0.165 (0.006) | 0.1317 | 0.195 (0.012) | 0.1556 | 0.143 (0.005) | 0.1141 |
| Heart Disease | 0.237 (0.012) | 0.1891 | 0.167 (0.016) | 0.1332 | 0.175 (0.010) | 0.1396 | 0.183 (0.008) | 0.146 | 0.169 (0.019) | 0.1348 | 0.153 (0.006) | 0.1221 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Z.; Zhang, B.; Hu, Y.; Du, Y.; Liu, T.; Zhang, Z.; Lu, C.; Zhou, S.; Cui, Z. Med-Diffusion: Diffusion Model-Based Imputation of Multimodal Sensor Data for Surgical Patients. Sensors 2025, 25, 6175. https://doi.org/10.3390/s25196175
Cheng Z, Zhang B, Hu Y, Du Y, Liu T, Zhang Z, Lu C, Zhou S, Cui Z. Med-Diffusion: Diffusion Model-Based Imputation of Multimodal Sensor Data for Surgical Patients. Sensors. 2025; 25(19):6175. https://doi.org/10.3390/s25196175
Chicago/Turabian StyleCheng, Zhenyu, Boyuan Zhang, Yanbo Hu, Yue Du, Tianyong Liu, Zhenxi Zhang, Chang Lu, Shoujun Zhou, and Zhuoxu Cui. 2025. "Med-Diffusion: Diffusion Model-Based Imputation of Multimodal Sensor Data for Surgical Patients" Sensors 25, no. 19: 6175. https://doi.org/10.3390/s25196175
APA StyleCheng, Z., Zhang, B., Hu, Y., Du, Y., Liu, T., Zhang, Z., Lu, C., Zhou, S., & Cui, Z. (2025). Med-Diffusion: Diffusion Model-Based Imputation of Multimodal Sensor Data for Surgical Patients. Sensors, 25(19), 6175. https://doi.org/10.3390/s25196175





