Image Filtering to Improve Maize Tassel Detection Accuracy Using Machine Learning Algorithms
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Experimental Design and Image Data Collection
2.2. Procedure for Image Filtration
2.3. The TasselNet Approach
- (a)
- Normalized mean square prediction error (NMSPE).
- (b) Predictive deviance (PD).
2.4. The R-CNN Approach
3. Results
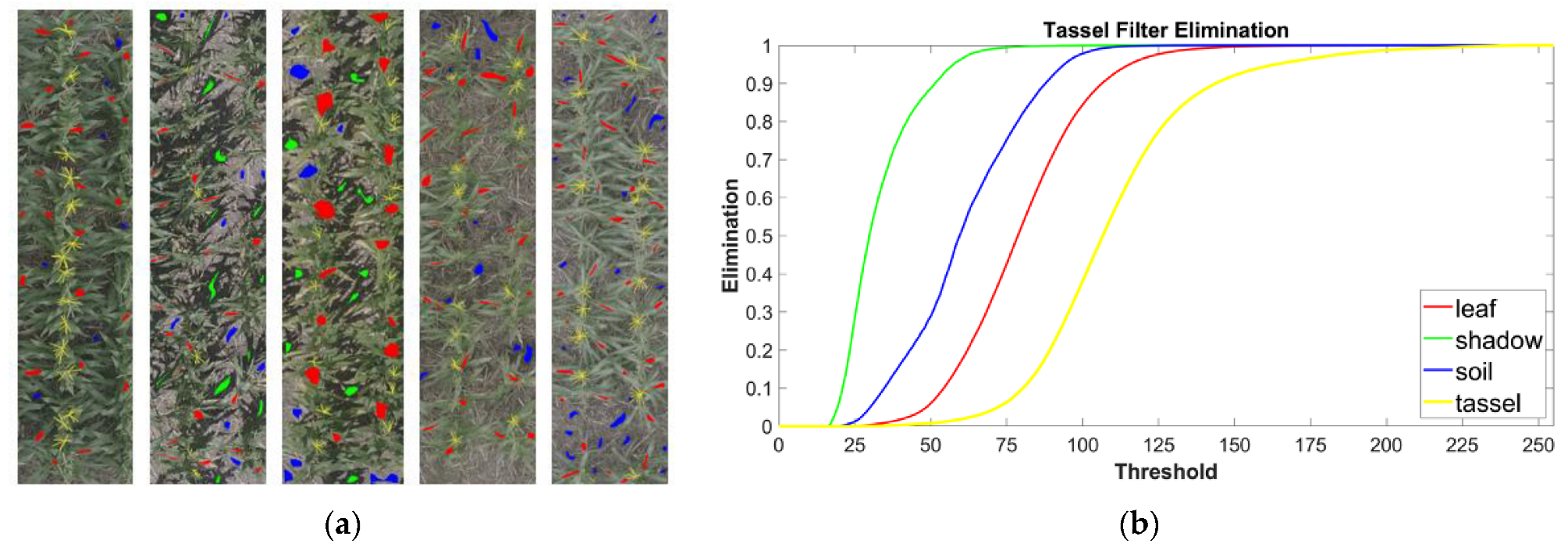
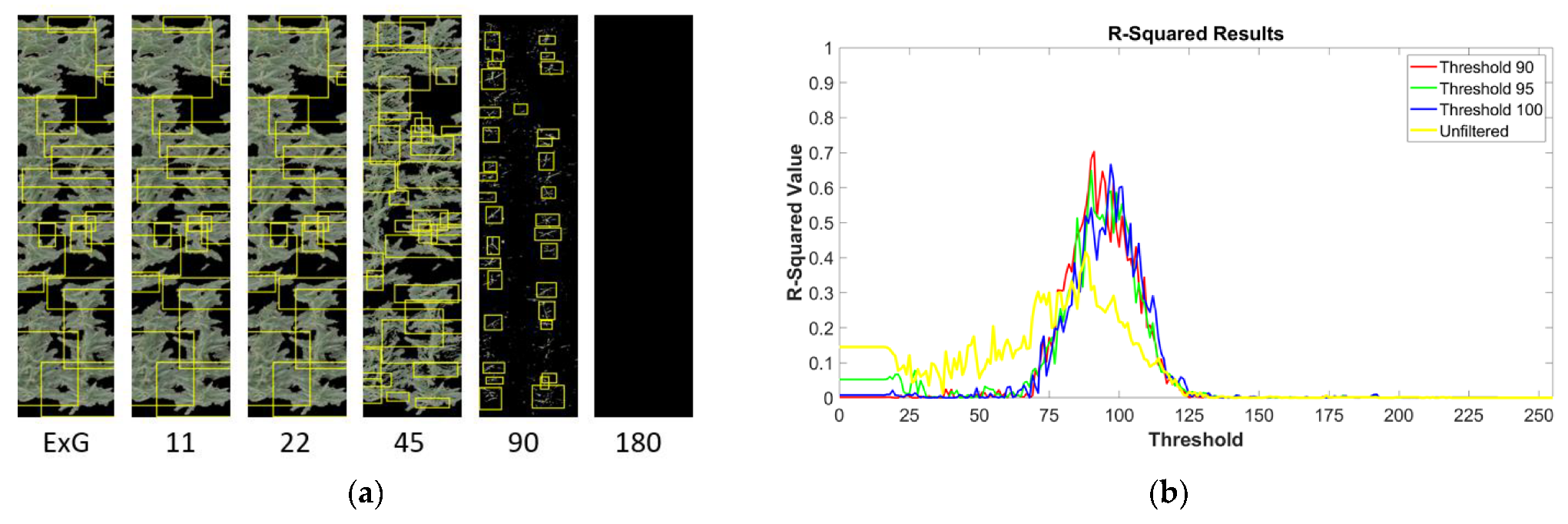
3.1. Image Filtration Removes Non-Tassel Pixels
3.2. Filtered Image Feeds into a CBR Algorithm Implemented in TasselNet
3.3. Filtered Images Improved Tassel Detection for a CBD Algorithm
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.F.; Oliveira, H.R.; Volenec, J.J.; Rainey, K.M.; Brito, L.F. Integrating high-throughput phenotyping and statistical genomic methods to genetically improve longitudinal traits in crops. Front. Plant Sci. 2020, 11, 538244. [Google Scholar] [CrossRef] [PubMed]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.; Ersoz, E.; Flint-Garcia, S.; Garcia, A.; Glaubitz, J.C.; et al. The genetic architecture of maize flowering time. Science 2009, 325, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Cai, E.; Baireddy, S.; Yang, C.; Crawford, M.; Delp, E.J. Panicle counting in UAV images for estimating flowering time in sorghum. arXiv 2021, arXiv:2107.07308v1. [Google Scholar]
- Jiang, Y.; Li, C. Convolutional Neural Networks for Image-Based High-Throughput Plant Phenotyping: A Review. Plant Phenomics 2020, 2020, 4152816. [Google Scholar] [CrossRef] [PubMed]
- Scharr, H.; Bruns, B.; Fischbach, A.; Roussel, J.; Scholtes, L.; Stein, J.V. Germination detection of seedlings in soil: A system, dataset and challenge. In European Conference on Computer Vision; Springer International Publishing: Cham, Switzerland, 2020; pp. 360–374. [Google Scholar]
- Xiong, J.; Liu, Z.; Chen, S.; Liu, B.; Zheng, Z.; Zhong, Z.; Yang, Z.; Peng, H. Visual detection of green mangoes by an unmanned aerial vehicle in orchards based on a deep learning method. Biosyst. Eng. 2020, 194, 261–272. [Google Scholar] [CrossRef]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Okura, F. How convolutional neural networks diagnose plant disease. Plant Phenomics 2019, 2019, 9237136. [Google Scholar] [CrossRef] [PubMed]
- Abdulridha, J.; Ampatzidis, Y.; Kakarla, S.C.; Roberts, P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2020, 21, 955–978. [Google Scholar] [CrossRef]
- Velumani, K.; Lopez-Lozano, R.; Madec, S.; Guo, W.; Gillet, J.; Comar, A.; Baret, F. Estimates of maize plant density from UAV RGB images using faster-RCNN detection model: Impact of the spatial resolution. Plant Phenomics 2021, 2021, 9824843. [Google Scholar] [CrossRef]
- Keller, K.; Kirchgessner, N.; Khanna, R.; Siegwart, R.; Walter, A.; Aasen, H. Soybean leaf coverage estimation with machine learning and thresholding algorithms for field phenotyping. In Proceedings of the British Machine Vision Conference, Newcastle, UK, 3–6 September 2018; BMVA Press: Durham, UK; pp. 3–6.
- Bernotas, G.; Scorza, L.C.; Hansen, M.F.; Hales, I.J.; Halliday, K.J.; Smith, L.N.; Smith, M.L.; McCormick, A.J. A photometric stereo-based 3D imaging system using computer vision and deep learning for tracking plant growth. GigaScience 2019, 8, giz056. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, C.; Han, Q.; Wu, F.; Zou, X. A Performance Analysis of a Litchi Picking Robot System for Actively Removing Obstructions, Using an Artificial Intelligence Algorithm. Agronomy 2023, 13, 2795. [Google Scholar] [CrossRef]
- Ye, L.; Wu, F.; Zou, X.; Li, J. Path planning for mobile robots in unstructured orchard environments: An improved kinematically constrained bi-directional RRT approach. Comput. Electron. Agric. 2023, 215, 108453. [Google Scholar] [CrossRef]
- Meng, F.; Li, J.; Zhang, Y.; Qi, S.; Tang, Y. Transforming unmanned pineapple picking with spatio-temporal convolutional neural networks. Comput. Electron. Agric. 2023, 214, 108298. [Google Scholar] [CrossRef]
- Miao, C.; Hoban, T.P.; Pages, A.; Xu, Z.; Rodene, E.; Ubbens, J.; Stavness, I.; Yang, J.; Schnable, J.C. Simulated plant images improve maize leaf counting accuracy. BioRxiv, 2019; BioRxiv:706994. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Lu, H.; Cao, Z.; Xiao, Y.; Zhuang, B.; Shen, C. TasselNet: Counting maize tassels in the wild via local counts regression network. Plant Methods 2017, 13, 79. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Yang, Y.; Zheng, Y.; Zhu, Q.; Huang, M.; Guo, Y. In-field automatic detection of maize tassels using computer vision. Inf. Process. Agric. 2019, 8, 87–95. [Google Scholar] [CrossRef]
- Liu, Y.; Cen, C.; Che, Y.; Ke, R.; Ma, Y.; Ma, Y. Detection of Maize Tassels from UAV RGB Imagery with Faster R-CNN. Remote Sens. 2020, 12, 338. [Google Scholar] [CrossRef]
- Zou, H.; Lu, H.; Li, Y.; Liu, L.; Cao, Z. Maize tassels detection: A benchmark of the state of the art. Plant Methods 2020, 16, 108. [Google Scholar] [CrossRef] [PubMed]
- Mirnezami, S.V.; Srinivasan, S.; Zhou, Y.; Schnable, P.S.; Ganapathysubramanian, B. Detection of the Progression of Anthesis in Field-Grown Maize Tassels: A Case Study. Plant Phenomics 2021, 2021, 4238701. [Google Scholar] [CrossRef]
- Kumar, A.; Desai, S.V.; Balasubramanian, V.N.; Rajalakshmi, P.; Guo, W.; Naik, B.B.; Balram, M.; Desai, U.B. Efficient maize tassel-detection method using UAV based remote sensing. Remote Sens. Appl. Soc. Environ. 2021, 23, 100549. [Google Scholar] [CrossRef]
- Loy, C.C.; Chen, K.; Gong, S.; Xiang, T. Crowd Counting and Profiling: Methodology and Evaluation. In Modeling, Simulation and Visual Analysis of Crowds: A Multidisciplinary Perspective; Springer: New York, NY, USA, 2013; pp. 347–382. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems 28 (NIPS 2015), Montreal, QC, Canada, 7–12 December 2015; Volume 28, pp. 91–99. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Oñoro-Rubio, D.; López-Sastre, R.J. Towards Perspective-Free Object Counting with Deep Learning. In Proceedings of the Computer Vision—ECCV 2016, Part VII, Amsterdam, The Netherlands, 11–14 October 2016; Volume 9911, pp. 615–629. [Google Scholar]
- Rodene, E.; Xu, G.; Palali Delen, S.; Smith, C.; Ge, Y.; Schnable, J.; Yang, J. A UAV-based high-throughput phenotyping approach to assess time-series nitrogen responses and identify traits associated genetic components in maize. Plant Phenome J. 2022, 5, e20030. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Meyer, G.E.; Hindman, T.W.; Laksmi, K. Machine vision detection parameters for plant species identification. Precis. Agric. Biol. Qual. 1999, 3543, 327–335. [Google Scholar]
- Shao, M.; Nie, C.; Cheng, M.; Yu, X.; Bai, Y.; Ming, B.; Song, H.; Jin, X. Quantifying effect of tassels on near-ground maize canopy RGB images using deep learning segmentation algorithm. Precis. Agric. 2022, 23, 400–418. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Wan, Q.; Pal, R. An ensemble based top performing approach for NCI-DREAM drug sensitivity prediction challenge. PLoS ONE 2014, 9, e101183. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Hinton, G. Learning Multiple Layers of Features from Tiny Images. Master’s Thesis, University of Toronto, Toronto, ON, Canada, 2009. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef]
- Palali Delen, S.; Xu, G.; Velazquez-Perfecto, J.; Yang, J. Estimating the genetic parameters of yield-related traits under different nitrogen conditions in maize. Genetics 2023, 223, iyad012. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, B.; Chapman, S.C.; Laws, K.; George-Jaeggli, B.; Hammer, G.L.; Jordan, D.R.; Potgieter, A.B. Detecting sorghum plant and head features from multispectral UAV imagery. Plant Phenomics 2021, 2021, 9874650. [Google Scholar] [CrossRef]
- Illian, J.; Penttinen, A.; Stoyan, H.; Stoyan, D. Statistical Analysis and Modelling of Spatial Point Patterns; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar]

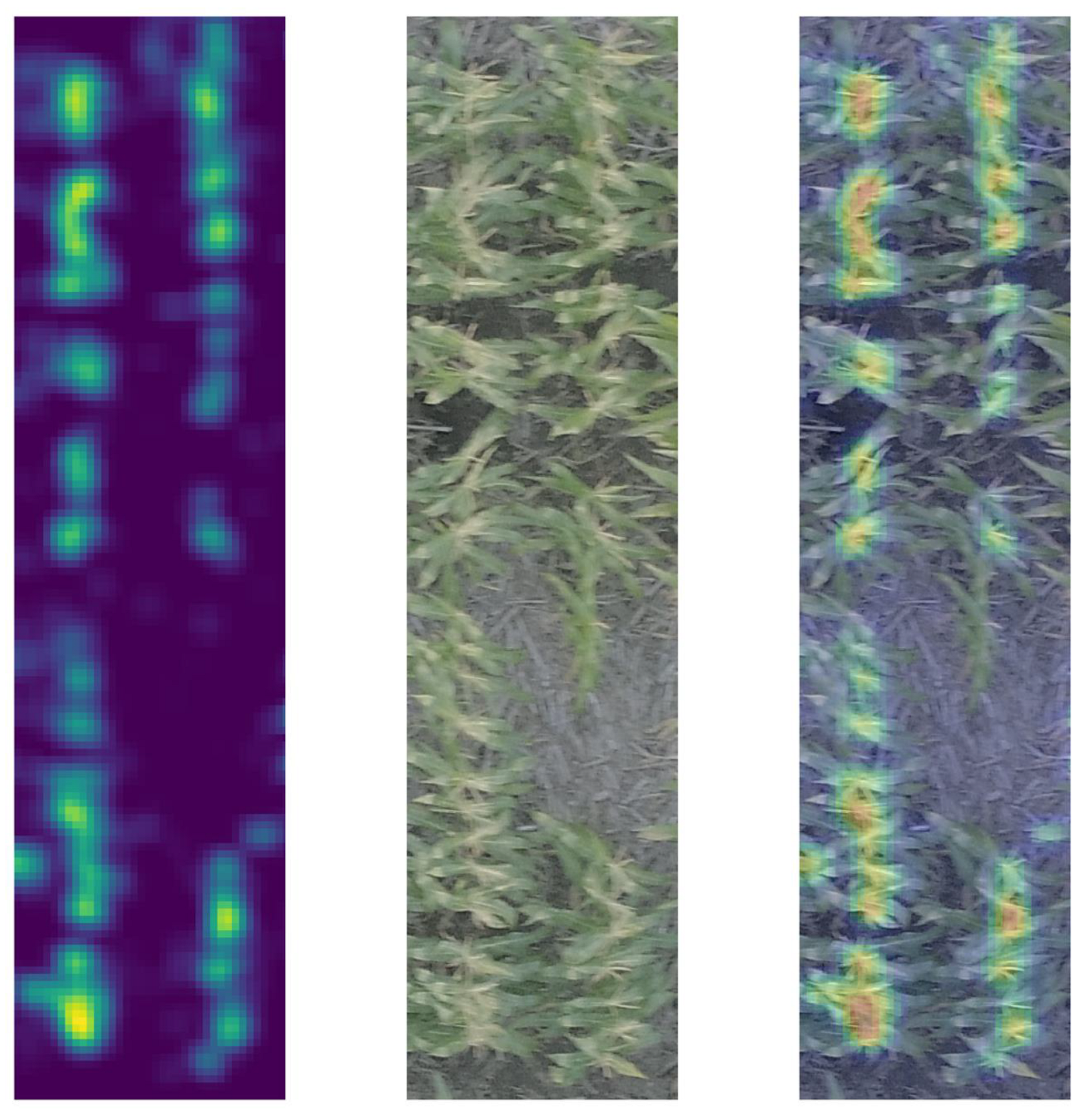
| Image Type | NMSPE | PD |
|---|---|---|
| Unfiltered | 2.7231 | 180.8481 |
| Filtered at threshold 90 | 3.8603 | 227.4901 |
| Filtered at threshold 100 | 5.2032 | 312.1130 |
| Image Type | Spearman’s Correlation |
|---|---|
| Unfiltered | 0.6825 |
| Filtered at threshold 90 | 0.5802 |
| Filtered at threshold 100 | 0.5041 |
| Image Type | Average MAE across Bootstrapped Samples | 95% Uncertainty Intervals |
|---|---|---|
| Unfiltered | 8.90 | (8.73, 9.07) |
| Filtered at threshold 90 | 8.65 | (8.58, 8.72) |
| Filtered at threshold 100 | 9.91 | (9.83, 10.02) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodene, E.; Fernando, G.D.; Piyush, V.; Ge, Y.; Schnable, J.C.; Ghosh, S.; Yang, J. Image Filtering to Improve Maize Tassel Detection Accuracy Using Machine Learning Algorithms. Sensors 2024, 24, 2172. https://doi.org/10.3390/s24072172
Rodene E, Fernando GD, Piyush V, Ge Y, Schnable JC, Ghosh S, Yang J. Image Filtering to Improve Maize Tassel Detection Accuracy Using Machine Learning Algorithms. Sensors. 2024; 24(7):2172. https://doi.org/10.3390/s24072172
Chicago/Turabian StyleRodene, Eric, Gayara Demini Fernando, Ved Piyush, Yufeng Ge, James C. Schnable, Souparno Ghosh, and Jinliang Yang. 2024. "Image Filtering to Improve Maize Tassel Detection Accuracy Using Machine Learning Algorithms" Sensors 24, no. 7: 2172. https://doi.org/10.3390/s24072172
APA StyleRodene, E., Fernando, G. D., Piyush, V., Ge, Y., Schnable, J. C., Ghosh, S., & Yang, J. (2024). Image Filtering to Improve Maize Tassel Detection Accuracy Using Machine Learning Algorithms. Sensors, 24(7), 2172. https://doi.org/10.3390/s24072172










