Heart Rate Variability and Pulse Rate Variability: Do Anatomical Location and Sampling Rate Matter?
Abstract
1. Introduction
2. Methods
2.1. Ethical Approval
2.2. Study Design and Participants
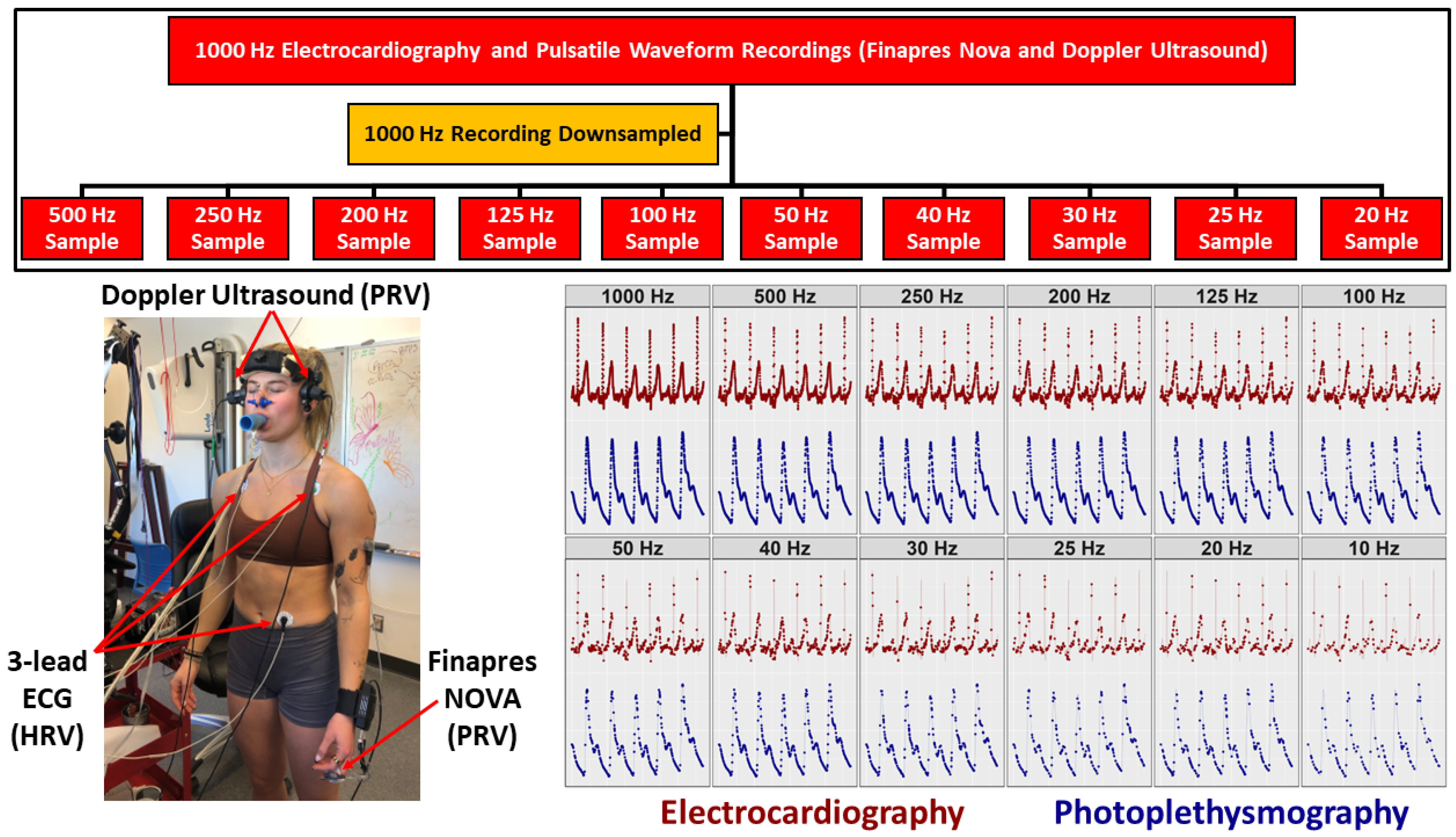
2.3. Instrumentation
2.4. Experimental Protocols
2.5. Data Processing
2.6. Sample Size Calculation
2.7. Statistical Analysis
3. Results
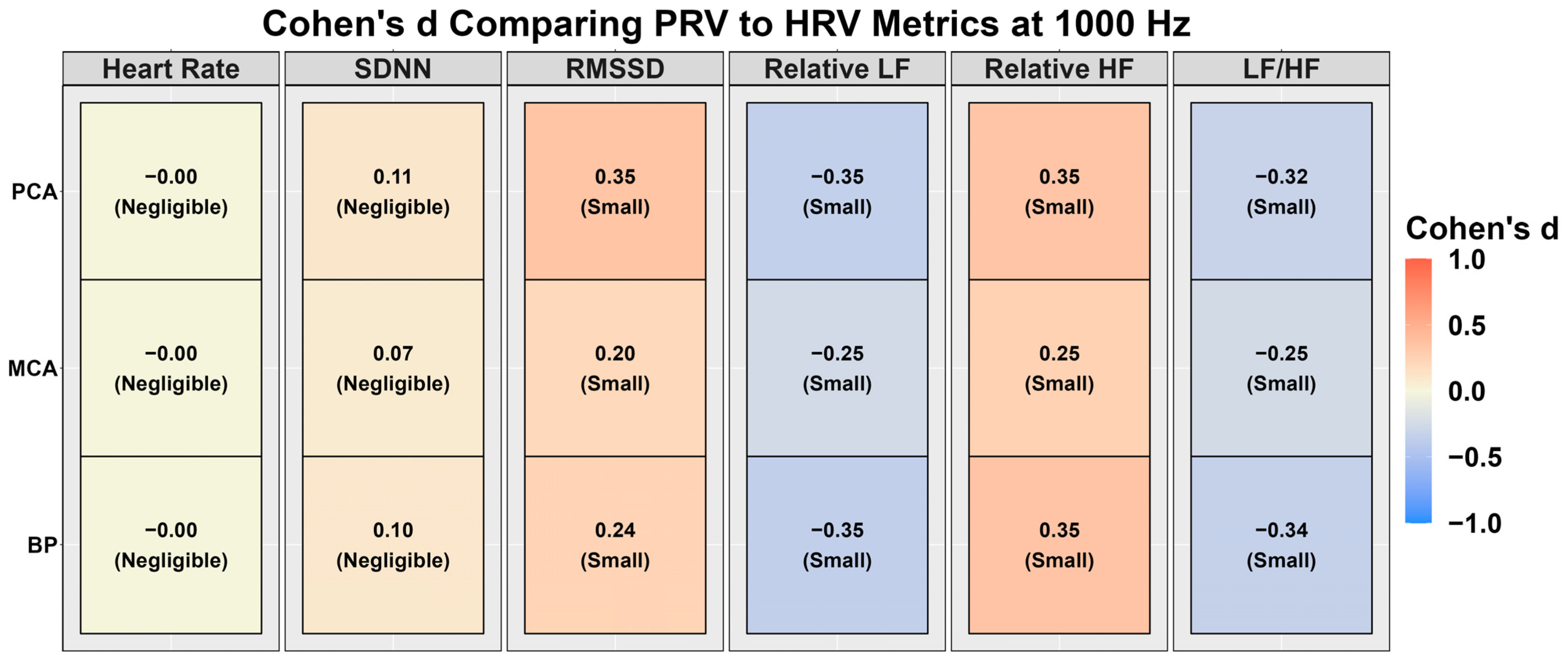
3.1. Heart Rate Variability vs. Pulse Rate Variability at 1000 Hz
3.2. Sampling Frequency Validity for Heart Rate Variability vs. Pulse Rate Variability Metrics
4. Discussion
4.1. Comparison to Previous Research and Physiological Underpinnings
4.2. Future Directions and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaffer, F.; Ginsberg, J.P. An Overview of Heart Rate Variability Metrics and Norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef]
- Shaffer, F.; McCraty, R.; Zerr, C.L. A healthy heart is not a metronome: An integrative review of the heart’s anatomy and heart rate variability. Front. Psychol. 2014, 5, 1040. [Google Scholar] [CrossRef]
- Goldberger, A.L. Is the normal heartbeat chaotic or homeostatic? News Physiol. Sci. 1991, 6, 87–91. [Google Scholar] [CrossRef]
- Rajendra Acharya, U.; Paul Joseph, K.; Kannathal, N.; Lim, C.M.; Suri, J.S. Heart rate variability: A review. Med. Biol. Eng. Comput. 2006, 44, 1031–1051. [Google Scholar] [CrossRef]
- Agorastos, A.; Mansueto, A.C.; Hager, T.; Pappi, E.; Gardikioti, A.; Stiedl, O. Heart Rate Variability as a Translational Dynamic Biomarker of Altered Autonomic Function in Health and Psychiatric Disease. Biomedicines 2023, 11, 1591. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, J.S.; Aldhahir, A.M.; Alghamdi, S.M.; Al Ghamdi, S.S.; AlDraiwiesh, I.A.; Alsulayyim, A.S.; Alqahtani, A.S.; Alobaidi, N.Y.; Al Saikhan, L.; AlRabeeah, S.M.; et al. A systematic review and meta-analysis of heart rate variability in COPD. Front. Cardiovasc. Med. 2023, 10, 1070327. [Google Scholar] [CrossRef] [PubMed]
- Huhn, S.; Axt, M.; Gunga, H.C.; Maggioni, M.A.; Munga, S.; Obor, D.; Sié, A.; Boudo, V.; Bunker, A.; Sauerborn, R.; et al. The Impact of Wearable Technologies in Health Research: Scoping Review. JMIR mHealth uHealth 2022, 10, e34384. [Google Scholar] [CrossRef] [PubMed]
- Petek, B.J.; Al-Alusi, M.A.; Moulson, N.; Grant, A.J.; Besson, C.; Guseh, J.S.; Wasfy, M.M.; Gremeaux, V.; Churchill, T.W.; Baggish, A.L. Consumer Wearable Health and Fitness Technology in Cardiovascular Medicine: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2023, 82, 245–264. [Google Scholar] [CrossRef] [PubMed]
- Žnidarič, M.; Škrinjar, D.; Kapel, A. Electrodermal activity and heart rate variability for detection of peripheral abnormalities in type 2 diabetes: A review. Biomol. Biomed. 2023, 23, 740–751. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, D.; Esparza, A.; Ghamari, M.; Soltanpur, C.; Nazeran, H. A review on wearable photoplethysmography sensors and their potential future applications in health care. Int. J. Biosens. Bioelectron. 2018, 4, 195–202. [Google Scholar] [CrossRef]
- Galli, A.; Montree, R.J.H.; Que, S.; Peri, E.; Vullings, R. An Overview of the Sensors for Heart Rate Monitoring Used in Extramural Applications. Sensors 2022, 22, 4035. [Google Scholar] [CrossRef]
- Kurl, S.; Mäkikallio, T.H.; Rautaharju, P.; Kiviniemi, V.; Laukkanen, J.A. Duration of QRS Complex in Resting Electrocardiogram Is a Predictor of Sudden Cardiac Death in Men. Circulation 2012, 125, 2588–2594. [Google Scholar] [CrossRef]
- Yuda, E.; Shibata, M.; Ogata, Y.; Ueda, N.; Yambe, T.; Yoshizawa, M.; Hayano, J. Pulse rate variability: A new biomarker, not a surrogate for heart rate variability. J. Physiol. Anthropol. 2020, 39, 21. [Google Scholar] [CrossRef]
- Pelaez-Coca, M.D.; Hernando, A.; Lazaro, J.; Gil, E. Impact of the PPG Sampling Rate in the Pulse Rate Variability Indices Evaluating Several Fiducial Points in Different Pulse Waveforms. IEEE J. Biomed. Health Inform. 2022, 26, 539–549. [Google Scholar] [CrossRef]
- Constant, I.; Laude, D.; Murat, I.; Elghozi, J.-L. Pulse rate variability is not a surrogate for heart rate variability. Clin. Sci. 1999, 97, 391–397. [Google Scholar] [CrossRef]
- Wong, J.-S.; Lu, W.-A.; Wu, K.-T.; Liu, M.; Chen, G.-Y.; Kuo, C.-D. A comparative study of pulse rate variability and heart rate variability in healthy subjects. J. Clin. Monit. Comput. 2012, 26, 107–114. [Google Scholar] [CrossRef]
- Yuda, E.; Yamamoto, K.; Yoshida, Y.; Hayano, J. Differences in pulse rate variability with measurement site. J. Physiol. Anthropol. 2020, 39, 4. [Google Scholar] [CrossRef]
- Scholkmann, F.; Kleiser, S.; Metz, A.J.; Zimmermann, R.; Mata Pavia, J.; Wolf, U.; Wolf, M. A review on continuous wave functional near-infrared spectroscopy and imaging instrumentation and methodology. Neuroimage 2014, 85 Pt 1, 6–27. [Google Scholar] [CrossRef]
- Almajidy, R.; Mankodiya, K.; Abtahi, M.; Hofmann, U. A Newcomer’s Guide to Functional Near Infrared Spectroscopy Experiments. IEEE Rev. Biomed. Eng. 2019, 13, 292–308. [Google Scholar] [CrossRef]
- Sung, C.K.; Lee, K.H.; Kim, S.H. Evaluation of factors influencing arterial Doppler waveforms in an in vitro flow phantom. Ultrasonography 2017, 36, 39–52. [Google Scholar] [CrossRef]
- Koep, J.L.; Taylor, C.E.; Coombes, J.S.; Bond, B.; Ainslie, P.N.; Bailey, T.G. Autonomic control of cerebral blood flow: Fundamental comparisons between peripheral and cerebrovascular circulations in humans. J. Physiol. 2022, 600, 15–39. [Google Scholar] [CrossRef]
- Vantanajal, J.S.; Ashmead, J.C.; Anderson, T.J.; Hepple, R.T.; Poulin, M.J. Differential sensitivities of cerebral and brachial blood flow to hypercapnia in humans. J. Appl. Physiol. 2007, 102, 87–93. [Google Scholar] [CrossRef]
- Burma, J.S.; Lapointe, A.P.; Soroush, A.; Oni, I.K.; Smirl, J.D.; Dunn, J.F. Insufficient sampling frequencies skew heart rate variability estimates: Implications for extracting heart rate metrics from neuroimaging and physiological data. J. Biomed. Inform. 2021, 123, 103934. [Google Scholar] [CrossRef]
- Sarhaddi, F.; Kazemi, K.; Azimi, I.; Cao, R.; Niela-Vilén, H.; Axelin, A.; Liljeberg, P.; Rahmani, A.M. A comprehensive accuracy assessment of Samsung smartwatch heart rate and heart rate variability. PLoS ONE 2022, 17, e0268361. [Google Scholar] [CrossRef]
- González-Cañete, F.J.; Casilari, E. A Feasibility Study of the Use of Smartwatches in Wearable Fall Detection Systems. Sensors 2021, 21, 2254. [Google Scholar] [CrossRef]
- Sigcha, L.F.; Pavón, I.; Arezes, P.; Costa, N.; Arcas, G.; Lopez Navarro, J.M. Occupational Risk Prevention through Smartwatches: Precision and Uncertainty Effects of the Built-In Accelerometer. Sensors 2018, 18, 3805. [Google Scholar] [CrossRef]
- Association, W.M. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- Shechter, A.; Boivin, D.B. Sleep, hormones, and circadian rhythms throughout the menstrual cycle in healthy women and women with premenstrual dysphoric disorder. Int. J. Endocrinol. 2010, 2010, 259345. [Google Scholar] [CrossRef]
- Burma, J.S.; Copeland, P.V.; Macaulay, A.; Khatra, O.; Smirl, J.D. Effects of high-intensity intervals and moderate-intensity exercise on baroreceptor sensitivity and heart rate variability during recovery. Appl. Physiol. Nutr. Metab. 2020, 45, 1156–1164. [Google Scholar] [CrossRef]
- Zimmermann-Viehoff, F.; Thayer, J.; Koenig, J.; Herrmann, C.; Weber, C.S.; Deter, H.C. Short-term effects of espresso coffee on heart rate variability and blood pressure in habitual and non-habitual coffee consumers—A randomized crossover study. Nutr. Neurosci. 2016, 19, 169–175. [Google Scholar] [CrossRef]
- Lefrandt, J.D.; Mulder, M.C.; Bosma, E.; Smit, A.J.; Hoogenberg, K. Inverse relationship between blood glucose and autonomic function in healthy subjects. Diabetes Care 2000, 23, 1862–1864. [Google Scholar] [CrossRef] [PubMed]
- van Baak, M.A. Meal-induced activation of the sympathetic nervous system and its cardiovascular and thermogenic effects in man. Physiol. Behav. 2008, 94, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Sammons, E.L.; Samani, N.J.; Smith, S.M.; Rathbone, W.E.; Bentley, S.; Potter, J.F.; Panerai, R.B. Influence of noninvasive peripheral arterial blood pressure measurements on assessment of dynamic cerebral autoregulation. J. Appl. Physiol. 2007, 103, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Omboni, S.; Parati, G.; Frattola, A.; Mutti, E.; Di Rienzo, M.; Castiglioni, P.; Mancia, G. Spectral and sequence analysis of finger blood pressure variability. Comparison with analysis of intra-arterial recordings. Hypertension 1993, 22, 26–33. [Google Scholar] [CrossRef]
- Willie, C.K.; Colino, F.L.; Bailey, D.M.; Tzeng, Y.C.; Binsted, G.; Jones, L.W.; Haykowsky, M.J.; Bellapart, J.; Ogoh, S.; Smith, K.J.; et al. Utility of transcranial Doppler ultrasound for the integrative assessment of cerebrovascular function. J. Neurosci. Methods 2011, 196, 221–237. [Google Scholar] [CrossRef]
- Wieling, W.; Krediet, C.T.; van Dijk, N.; Linzer, M.; Tschakovsky, M.E. Initial orthostatic hypotension: Review of a forgotten condition. Clin. Sci. 2007, 112, 157–165. [Google Scholar] [CrossRef]
- Smith, J.J.; Porth, C.M.; Erickson, M. Hemodynamic response to the upright posture. J. Clin. Pharmacol. 1994, 34, 375–386. [Google Scholar] [CrossRef]
- Burma, J.S.; Graver, S.; Miutz, L.N.; Macaulay, A.; Copeland, P.V.; Smirl, J.D. The validity and reliability of ultra-short-term heart rate variability parameters and the influence of physiological covariates. J. Appl. Physiol. 2021, 130, 1848–1867. [Google Scholar] [CrossRef]
- Malik, M. Heart Rate Variability. Ann. Noninvasive Electrocardiol. 1996, 1, 151–181. [Google Scholar] [CrossRef]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S.H.A. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Compution: Vienna, Austria, 2020. [Google Scholar]
- Pecchia, L.; Castaldo, R.; Montesinos, L.; Melillo, P. Are ultra-short heart rate variability features good surrogates of short-term ones? State-of-the-art review and recommendations. Healthc. Technol. Lett. 2018, 5, 94–100. [Google Scholar] [CrossRef]
- Myles, P.S.; Cui, J. Using the Bland-Altman method to measure agreement with repeated measures. Br. J. Anaesth. 2007, 99, 309–311. [Google Scholar] [CrossRef]
- Giavarina, D. Understanding Bland Altman analysis. Biochem. Medica 2015, 25, 141–151. [Google Scholar] [CrossRef]
- Atkinson, G.; Nevill, A.M. Statistical methods for assessing measurement error (reliability) in variables relevant to sports medicine. Sports Med. 1998, 26, 217–238. [Google Scholar] [CrossRef]
- Burma, J.S.; Van Roessel, R.K.; Oni, I.K.; Dunn, J.F.; Smirl, J.D. Neurovascular coupling on trial: How the number of trials completed impacts the accuracy and precision of temporally derived neurovascular coupling estimates. J. Cereb. Blood Flow. Metab. 2022, 42, 1478–1492. [Google Scholar] [CrossRef]
- Aronhime, S.; Calcagno, C.; Jajamovich, G.H.; Dyvorne, H.A.; Robson, P.; Dieterich, D.; Fiel, M.I.; Martel-Laferriere, V.; Chatterji, M.; Rusinek, H.; et al. DCE-MRI of the liver: Effect of linear and nonlinear conversions on hepatic perfusion quantification and reproducibility. J. Magn. Reson. Imaging 2014, 40, 90–98. [Google Scholar] [CrossRef]
- Lakens, D. Calculating and reporting effect sizes to facilitate cumulative science: A practical primer for t-tests and ANOVAs. Front. Psychol. 2013, 4, 863. [Google Scholar] [CrossRef]
- Halsey, L.G. The reign of the p-value is over: What alternative analyses could we employ to fill the power vacuum? Biol. Lett. 2019, 15, 20190174. [Google Scholar] [CrossRef]
- Panagiotakos, D.B. Value of p-value in biomedical research. Open Cardiovasc. Med. J. 2008, 2, 97–99. [Google Scholar] [CrossRef]
- Amrhein, V.; Greenland, S.; McShane, B. Scientists rise up against statistical significance. Nature 2019, 567, 305–307. [Google Scholar] [CrossRef]
- Maciejewski, M.W.; Qui, H.Z.; Rujan, I.; Mobli, M.; Hoch, J.C. Nonuniform sampling and spectral aliasing. J. Magn. Reson. 2009, 199, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Gregg, R.E.; Zhou, S.H.; Lindauer, J.M.; Helfenbein, E.D.; Giuliano, K.K. What is inside the electrocardiograph? J. Electrocardiol. 2008, 41, 8–14. [Google Scholar] [CrossRef]
- Lefferts, W.K.; DeBlois, J.P.; Augustine, J.A.; Keller, A.P.; Heffernan, K.S. Age, sex, and the vascular contributors to cerebral pulsatility and pulsatile damping. J. Appl. Physiol. 2020, 129, 1092–1101. [Google Scholar] [CrossRef]
- Meinders, J.M.; Hoeks, A.P. Simultaneous assessment of diameter and pressure waveforms in the carotid artery. Ultrasound Med. Biol. 2004, 30, 147–154. [Google Scholar] [CrossRef]
- Bruno, R.M.; Ghiadoni, L.; Seravalle, G.; Dell’Oro, R.; Taddei, S.; Grassi, G. Sympathetic regulation of vascular function in health and disease. Front. Physiol. 2012, 3, 26173. [Google Scholar] [CrossRef]
- Bigger, J.T., Jr.; Albrecht, P.; Steinman, R.C.; Rolnitzky, L.M.; Fleiss, J.L.; Cohen, R.J. Comparison of time- and frequency domain-based measures of cardiac parasympathetic activity in Holter recordings after myocardial infarction. Am. J. Cardiol. 1989, 64, 536–538. [Google Scholar] [CrossRef]
- Clausen, M.; Pendergast, D.R.; Willer, B.; Leddy, J. Cerebral blood flow during treadmill exercise is a marker of physiological postconcussion syndrome in female athletes. J. Head. Trauma Rehabil. 2016, 31, 215–224. [Google Scholar] [CrossRef]
- Thibeault, C.M.; Thorpe, S.; Canac, N.; Wilk, S.J.; Hamilton, R.B. Sex-Based Differences in Transcranial Doppler Ultrasound and Self-Reported Symptoms After Mild Traumatic Brain Injury. Front. Neurol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Wilkinson, I.B.; Mäki-Petäjä, K.M.; Mitchell, G.F. Uses of Arterial Stiffness in Clinical Practice. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1063–1067. [Google Scholar] [CrossRef] [PubMed]



| Variable | Sampling Rate | ECG | BP | MCA | PCA |
|---|---|---|---|---|---|
| Heart rate (beats per minutes) | 1000 | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) |
| 500 | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | |
| 250 | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | |
| 200 | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | |
| 125 | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | |
| 100 | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | |
| 50 | 79.5 (11.1) | 79.8 (10.9) | 79.8 (10.9) | 79.8 (10.9) | |
| 40 | 76.2 (14.0) | 80.0 (10.9) | 80.0 (10.9) | 80.0 (10.9) | |
| 30 | 65.0 (14.2) | 77.4 (9.3) | 78.2 (12.3) | 76.5 (8.6) | |
| 25 | 48.7 (15.2) | 66.0 (9.6) | 67.6 (13.1) | 61.2 (9.7) | |
| 20 | 34.6 (13.1) | 61.9 (11.0) | 56.0 (12.4) | 55.0 (10.6) | |
| SDNN (milliseconds) | 1000 | 61.1 (22.1) | 63.5 (22.6) | 63.4 (22.5) | 62.7 (21.7) |
| 500 | 61.1 (22.1) | 63.5 (22.6) | 63.4 (22.6) | 62.8 (21.7) | |
| 250 | 61.1 (22.1) | 63.5 (22.5) | 63.5 (22.5) | 62.7 (21.6) | |
| 200 | 61.1 (22.2) | 63.6 (22.6) | 63.5 (22.5) | 62.8 (21.6) | |
| 125 | 61.2 (22.0) | 63.6 (22.4) | 63.6 (22.5) | 62.8 (21.6) | |
| 100 | 61.3 (22.2) | 63.6 (22.4) | 63.6 (22.5) | 62.9 (21.6) | |
| 50 | 70.5 (51.9) | 64.4 (22.2) | 64.8 (22.8) | 63.8 (21.6) | |
| 40 | 145.2 (209.4) | 64.3 (22.0) | 64.8 (22.5) | 63.8 (21.3) | |
| 30 | 396.7 (259.4) | 154.7 (106.2) | 141.5 (143.7) | 174.7 (104.6) | |
| 25 | 804.8 (482.3) | 371.2 (228.8) | 352.4 (297.2) | 483.0 (268.2) | |
| 20 | 1441.7 (955.9) | 472.3 (284.1) | 618.9 (358.5) | 633.3 (280.5) | |
| RMSSD (milliseconds) | 1000 | 29.3 (17.6) | 36.6 (23.8) | 33.6 (18.7) | 32.8 (19.2) |
| 500 | 29.3 (17.6) | 36.6 (23.8) | 33.6 (18.7) | 32.9 (19.2) | |
| 250 | 29.4 (17.6) | 36.7 (23.7) | 33.7 (18.6) | 32.9 (18.8) | |
| 200 | 29.5 (17.6) | 36.9 (23.9) | 33.8 (18.6) | 33.0 (19.2) | |
| 125 | 29.9 (17.2) | 37.2 (23.4) | 34.2 (18.5) | 33.4 (18.7) | |
| 100 | 30.4 (17.7) | 37.1 (22.7) | 34.5 (18.4) | 33.9 (19.1) | |
| 50 | 46.1 (73.2) | 40.1 (22.1) | 38.0 (18.2) | 36.8 (18.6) | |
| 40 | 151.2 (288.1) | 41.0 (21.3) | 39.3 (17.6) | 38.2 (17.4) | |
| 30 | 514.5 (345.8) | 194.6 (158.9) | 165.4 (219.1) | 225.4 (159.5) | |
| 25 | 1064.7 (672.2) | 510.3 (328.1) | 480.6 (430.9) | 670.4 (387.9) | |
| 20 | 1872.1 (1159.4) | 661.2 (414.8) | 858.0 (491.2) | 877.3 (398.2) | |
| Relative LF (normalized units) | 1000 | 81.8 (11.0) | 77.4 (13.8) | 77.6 (12.6) | 78.8 (12.4) |
| 500 | 81.8 (11.0) | 77.4 (13.8) | 77.6 (12.5) | 78.8 (12.4) | |
| 250 | 81.8 (10.9) | 77.3 (13.9) | 77.7 (12.5) | 78.8 (12.4) | |
| 200 | 81.6 (11.2) | 77.3 (13.9) | 77.6 (12.6) | 78.7 (12.5) | |
| 125 | 81.5 (11.2) | 77.1 (13.9) | 77.5 (12.5) | 78.7 (12.5) | |
| 100 | 81.4 (11.2) | 77.4 (13.6) | 77.5 (12.7) | 78.5 (12.7) | |
| 50 | 78.5 (12.9) | 76.2 (14.2) | 76.2 (12.6) | 77.5 (13.2) | |
| 40 | 72.8 (14.8) | 76.1 (14.1) | 75.8 (13.3) | 77.2 (13.2) | |
| 30 | 51.7 (14.4) | 51.8 (14.4) | 64.5 (16.7) | 50.7 (14.5) | |
| 25 | 63.5 (12.8) | 51.8 (11.3) | 54.4 (15.1) | 55.1 (12.6) | |
| 20 | 74.8 (16.4) | 53.1 (12.3) | 56.6 (12.7) | 61.2 (12.3) | |
| Relative HF (normalized units) | 1000 | 18.2 (11.0) | 22.6 (13.8) | 22.4 (12.6) | 21.2 (12.4) |
| 500 | 18.2 (11.0) | 22.6 (13.8) | 22.4 (12.5) | 21.2 (12.4) | |
| 250 | 18.2 (10.9) | 22.7 (13.9) | 22.3 (12.5) | 21.2 (12.4) | |
| 200 | 18.4 (11.2) | 22.7 (13.9) | 22.4 (12.6) | 21.3 (12.5) | |
| 125 | 18.5 (11.2) | 22.9 (13.9) | 22.5 (12.5) | 21.3 (12.5) | |
| 100 | 18.6 (11.2) | 22.6 (13.6) | 22.5 (12.7) | 21.5 (12.7) | |
| 50 | 21.5 (12.9) | 23.8 (14.2) | 23.8 (12.6) | 22.5 (13.2) | |
| 40 | 27.2 (14.8) | 23.9 (14.1) | 24.2 (13.3) | 22.8 (13.2) | |
| 30 | 48.3 (14.4) | 48.2 (14.4) | 35.5 (16.7) | 49.3 (14.5) | |
| 25 | 36.5 (12.8) | 48.2 (11.3) | 45.6 (15.1) | 44.9 (12.6) | |
| 20 | 25.2 (16.4) | 46.9 (12.3) | 43.4 (12.7) | 38.8 (12.3) | |
| LF/HF (percent) | 1000 | 6.5 (4.3) | 5.2 (3.7) | 5.1 (3.7) | 5.5 (3.8) |
| 500 | 6.5 (4.3) | 5.2 (3.7) | 5.1 (3.7) | 5.5 (3.8) | |
| 250 | 6.5 (4.3) | 5.2 (3.7) | 5.1 (3.7) | 5.5 (3.7) | |
| 200 | 6.4 (4.3) | 5.2 (3.6) | 5.1 (3.7) | 5.5 (3.7) | |
| 125 | 6.4 (4.2) | 5.1 (3.6) | 5.1 (3.6) | 5.4 (3.7) | |
| 100 | 6.3 (4.1) | 5.2 (3.7) | 5.1 (3.7) | 5.4 (3.6) | |
| 50 | 5.5 (3.9) | 4.8 (3.3) | 4.5 (2.9) | 5.1 (3.4) | |
| 40 | 4.1 (3.3) | 4.7 (3.2) | 4.4 (2.8) | 4.9 (3.2) | |
| 30 | 1.4 (1.2) | 1.4 (1.1) | 2.6 (2.0) | 1.4 (1.4) | |
| 25 | 2.7 (4.0) | 1.2 (0.8) | 1.6 (1.5) | 1.5 (1.2) | |
| 20 | 10.4 (21.7) | 1.4 (1.2) | 1.7 (1.9) | 1.9 (1.1) |
| Variable | Frequency | ECG | BP | MCA | PCA |
|---|---|---|---|---|---|
| Heart rate (beats per minutes) | Intercept | 4.37 (95% CI: 4.31, 4.43); p < 0.001 | 4.37 (95% CI: 4.33, 4.41); p < 0.001 | 4.37 (95% CI: 4.33, 4.41); p < 0.001 | 4.37 (95% CI: 4.34, 4.41); p < 0.001 |
| 500 Hz | −0.00 (95% CI: −0.08, 0.08); p > 0.999 | 0.00 (95% CI: −0.06, 0.06); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | 0.00 (95% CI: −0.05, 0.05); p > 0.999 | |
| 250 Hz | −0.00 (95% CI: −0.08, 0.08); p > 0.999 | −0.00 (95% CI: −0.06, 0.06); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | 0.00 (95% CI: −0.05, 0.05); p > 0.999 | |
| 200 Hz | 0.00 (95% CI: −0.08, 0.08); p = 0.999 | −0.00 (95% CI: −0.06, 0.06); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | 0.00 (95% CI: −0.05, 0.05); p > 0.999 | |
| 125 Hz | 0.00 (95% CI: −0.08, 0.08); p > 0.999 | 0.00 (95% CI: −0.06, 0.06); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | |
| 100 Hz | −0.00 (95% CI: −0.08, 0.08); p > 0.999 | −0.00 (95% CI: −0.06, 0.06); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | |
| 50 Hz | −0.01 (95% CI: −0.09, 0.08); p = 0.875 | −0.00 (95% CI: −0.06, 0.06); p = 0.945 | −0.00 (95% CI: −0.06, 0.05); p = 0.941 | −0.00 (95% CI: −0.06, 0.05); p = 0.941 | |
| 40 Hz | −0.06 (95% CI: −0.14, 0.02); p = 0.167 | −0.00 (95% CI: −0.06, 0.06); p = 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | −0.00 (95% CI: −0.05, 0.05); p > 0.999 | |
| 30 Hz | −0.22 (95% CI: −0.31, −0.14); p < 0.001 | −0.03 (95% CI: −0.08, 0.03); p = 0.404 | −0.04 (95% CI: −0.10, 0.01); p = 0.134 | −0.03 (95% CI: −0.08, 0.02); p = 0.263 | |
| 25 Hz | −0.54 (95% CI: −0.62, −0.46); p < 0.001 | −0.18 (95% CI: −0.24, −0.12); p < 0.001 | −0.27 (95% CI: −0.33, −0.22); p < 0.001 | −0.19 (95% CI: −0.25, −0.14); p < 0.001 | |
| 20 Hz | −0.91 (95% CI: −1.00, −0.83); p < 0.001 | −0.37 (95% CI: −0.43, −0.31); p < 0.001 | −0.38 (95% CI: −0.44, −0.33); p < 0.001 | −0.26 (95% CI: −0.32, −0.21); p < 0.001 | |
| SDNN (milliseconds) | Intercept | 4.04 (95% CI: 3.90, 4.19); p < 0.001 | 4.08 (95% CI: 3.94, 4.23); p < 0.001 | 4.08 (95% CI: 3.95, 4.20); p < 0.001 | 4.08 (95% CI: 3.95, 4.22); p < 0.001 |
| 500 Hz | 0.00 (95% CI: −0.20, 0.21); p = 0.999 | 0.00 (95% CI: −0.21, 0.21); p > 0.999 | 0.00 (95% CI: −0.18, 0.18); p = 0.999 | 0.00 (95% CI: −0.19, 0.19); p = 0.998 | |
| 250 Hz | 0.00 (95% CI: −0.20, 0.21); p = 0.998 | 0.00 (95% CI: −0.21, 0.21); p = 0.997 | 0.00 (95% CI: −0.18, 0.18); p = 0.998 | 0.00 (95% CI: −0.19, 0.19); p = 0.997 | |
| 200 Hz | 0.00 (95% CI: −0.20, 0.21); p = 0.995 | 0.00 (95% CI: −0.21, 0.21); p = 0.994 | 0.00 (95% CI: −0.18, 0.18); p = 0.992 | 0.00 (95% CI: −0.18, 0.19); p = 0.990 | |
| 125 Hz | 0.00 (95% CI: −0.20, 0.21); p = 0.979 | 0.00 (95% CI: −0.20, 0.21); p = 0.983 | 0.00 (95% CI: −0.18, 0.18); p = 0.986 | 0.00 (95% CI: −0.18, 0.19); p = 0.981 | |
| 100 Hz | 0.00 (95% CI: −0.20, 0.21); p = 0.977 | 0.00 (95% CI: −0.20, 0.21); p = 0.976 | 0.00 (95% CI: −0.18, 0.18); p = 0.968 | 0.00 (95% CI: −0.18, 0.19); p = 0.978 | |
| 50 Hz | 0.08 (95% CI: −0.13, 0.29); p = 0.447 | 0.02 (95% CI: −0.18, 0.23); p = 0.827 | 0.02 (95% CI: −0.16, 0.20); p = 0.838 | 0.02 (95% CI: −0.17, 0.21); p = 0.848 | |
| 40 Hz | 0.46 (95% CI: 0.26, 0.67); p < 0.001 | 0.02 (95% CI: −0.18, 0.23); p = 0.814 | 0.02 (95% CI: −0.16, 0.20); p = 0.817 | 0.02 (95% CI: −0.17, 0.20); p = 0.844 | |
| 30 Hz | 1.68 (95% CI: 1.48, 1.89); p < 0.001 | 0.50 (95% CI: 0.29, 0.70); p < 0.001 | 0.91 (95% CI: 0.73, 1.09); p < 0.001 | 0.76 (95% CI: 0.58, 0.95); p < 0.001 | |
| 25 Hz | 2.46 (95% CI: 2.26, 2.67); p < 0.001 | 1.41 (95% CI: 1.20, 1.61); p < 0.001 | 1.89 (95% CI: 1.71, 2.08); p < 0.001 | 1.59 (95% CI: 1.41, 1.78); p < 0.001 | |
| 20 Hz | 3.07 (95% CI: 2.87, 3.28); p < 0.001 | 2.12 (95% CI: 1.91, 2.33); p < 0.001 | 2.21 (95% CI: 2.03, 2.39); p < 0.001 | 1.84 (95% CI: 1.66, 2.03); p < 0.001 | |
| RMSSD (milliseconds) | Intercept | 3.24 (95% CI: 3.06, 3.42); p < 0.001 | 3.40 (95% CI: 3.22, 3.57); p < 0.001 | 3.37 (95% CI: 3.21, 3.52); p < 0.001 | 3.45 (95% CI: 3.29, 3.61); p < 0.001 |
| 500 Hz | 0.00 (95% CI: −0.25, 0.26); p = 0.988 | 0.00 (95% CI: −0.25, 0.25); p = 0.995 | 0.00 (95% CI: −0.22, 0.22); p = 0.989 | 0.00 (95% CI: −0.23, 0.23); p = 0.989 | |
| 250 Hz | 0.01 (95% CI: −0.25, 0.26); p = 0.951 | 0.01 (95% CI: −0.24, 0.26); p = 0.958 | 0.01 (95% CI: −0.21, 0.22); p = 0.949 | 0.01 (95% CI: −0.22, 0.23); p = 0.955 | |
| 200 Hz | 0.02 (95% CI: −0.24, 0.27); p = 0.908 | 0.01 (95% CI: −0.24, 0.26); p = 0.930 | 0.01 (95% CI: −0.21, 0.23); p = 0.926 | 0.01 (95% CI: −0.22, 0.24); p = 0.922 | |
| 125 Hz | 0.04 (95% CI: −0.21, 0.29); p = 0.753 | 0.03 (95% CI: −0.22, 0.28); p = 0.834 | 0.03 (95% CI: −0.19, 0.24); p = 0.804 | 0.03 (95% CI: −0.20, 0.25); p = 0.824 | |
| 100 Hz | 0.06 (95% CI: −0.20, 0.31); p = 0.670 | 0.04 (95% CI: −0.21, 0.29); p = 0.764 | 0.05 (95% CI: −0.17, 0.26); p = 0.677 | 0.04 (95% CI: −0.19, 0.26); p = 0.760 | |
| 50 Hz | 0.30 (95% CI: 0.05, 0.56); p = 0.021 | 0.16 (95% CI: −0.09, 0.41); p = 0.219 | 0.16 (95% CI: −0.06, 0.37); p = 0.162 | 0.14 (95% CI: −0.09, 0.37); p = 0.233 | |
| 40 Hz | 0.88 (95% CI: 0.63, 1.13); p < 0.001 | 0.20 (95% CI: −0.05, 0.46); p = 0.110 | 0.21 (95% CI: −0.01, 0.42); p = 0.060 | 0.17 (95% CI: −0.05, 0.40); p = 0.133 | |
| 30 Hz | 2.65 (95% CI: 2.40, 2.91); p < 0.001 | 1.03 (95% CI: 0.78, 1.28); p < 0.001 | 1.76 (95% CI: 1.54, 1.98); p < 0.001 | 1.51 (95% CI: 1.28, 1.73); p < 0.001 | |
| 25 Hz | 3.52 (95% CI: 3.26, 3.77); p < 0.001 | 2.28 (95% CI: 2.03, 2.53); p < 0.001 | 2.89 (95% CI: 2.67, 3.11); p < 0.001 | 2.50 (95% CI: 2.27, 2.73); p < 0.001 | |
| 20 Hz | 4.14 (95% CI: 3.89, 4.40); p < 0.001 | 3.11 (95% CI: 2.86, 3.36); p < 0.001 | 3.22 (95% CI: 3.01, 3.44); p < 0.001 | 2.78 (95% CI: 2.56, 3.01); p < 0.001 | |
| Relative LF (normalized units) | Intercept | 4.39 (95% CI: 4.34, 4.45); p < 0.001 | 4.34 (95% CI: 4.28, 4.39); p < 0.001 | 4.35 (95% CI: 4.30, 4.41); p < 0.001 | 4.33 (95% CI: 4.27, 4.39); p < 0.001 |
| 500 Hz | −0.00 (95% CI: −0.07, 0.07); p = 0.999 | 0.00 (95% CI: −0.08, 0.08); p = 0.995 | −0.00 (95% CI: −0.08, 0.07); p = 0.989 | −0.00 (95% CI: −0.08, 0.08); p = 0.988 | |
| 250 Hz | 0.00 (95% CI: −0.07, 0.07); p = 0.987 | 0.00 (95% CI: −0.08, 0.08); p = 0.988 | −0.00 (95% CI: −0.07, 0.07); p = 0.994 | −0.00 (95% CI: −0.08, 0.08); p = 0.964 | |
| 200 Hz | −0.00 (95% CI: −0.08, 0.07); p = 0.938 | −0.00 (95% CI: −0.08, 0.08); p = 0.988 | −0.00 (95% CI: −0.08, 0.07); p = 0.973 | −0.00 (95% CI: −0.08, 0.08); p = 0.951 | |
| 125 Hz | −0.00 (95% CI: −0.08, 0.07); p = 0.916 | −0.00 (95% CI: −0.08, 0.08); p = 0.962 | −0.00 (95% CI: −0.08, 0.07); p = 0.964 | −0.00 (95% CI: −0.09, 0.08); p = 0.909 | |
| 100 Hz | −0.01 (95% CI: −0.08, 0.07); p = 0.884 | −0.00 (95% CI: −0.08, 0.08); p = 0.946 | −0.01 (95% CI: −0.08, 0.07); p = 0.880 | 0.00 (95% CI: −0.08, 0.08); p = 0.995 | |
| 50 Hz | −0.05 (95% CI: −0.12, 0.03); p = 0.218 | −0.02 (95% CI: −0.10, 0.06); p = 0.625 | −0.02 (95% CI: −0.09, 0.06); p = 0.620 | −0.02 (95% CI: −0.10, 0.06); p = 0.656 | |
| 40 Hz | −0.13 (95% CI: −0.20, −0.06); p = 0.001 | −0.03 (95% CI: −0.11, 0.05); p = 0.497 | −0.02 (95% CI: −0.10, 0.05); p = 0.523 | −0.02 (95% CI: −0.10, 0.06); p = 0.637 | |
| 30 Hz | −0.49 (95% CI: −0.56, −0.42); p < 0.001 | −0.21 (95% CI: −0.29, −0.13); p < 0.001 | −0.46 (95% CI: −0.54, −0.39); p < 0.001 | −0.42 (95% CI: −0.51, −0.34); p < 0.001 | |
| 25 Hz | −0.26 (95% CI: −0.34, −0.19); p < 0.001 | −0.38 (95% CI: −0.46, −0.30); p < 0.001 | −0.37 (95% CI: −0.44, −0.29); p < 0.001 | −0.41 (95% CI: −0.49, −0.32); p < 0.001 | |
| 20 Hz | −0.11 (95% CI: −0.18, −0.03); p = 0.005 | −0.32 (95% CI: −0.40, −0.25); p < 0.001 | −0.26 (95% CI: −0.34, −0.19); p < 0.001 | −0.38 (95% CI: −0.47, −0.30); p < 0.001 | |
| Relative HF (normalized units) | Intercept | 2.74 (95% CI: 2.58, 2.90); p < 0.001 | 2.95 (95% CI: 2.81, 3.10); p < 0.001 | 2.89 (95% CI: 2.75, 3.03); p < 0.001 | 2.94 (95% CI: 2.80, 3.09); p < 0.001 |
| 500 Hz | −0.00 (95% CI: −0.23, 0.23); p = 0.994 | −0.00 (95% CI: −0.20, 0.20); p = 0.998 | 0.00 (95% CI: −0.20, 0.20); p = 0.988 | 0.00 (95% CI: −0.20, 0.20); p = 0.996 | |
| 250 Hz | −0.00 (95% CI: −0.23, 0.23); p = 0.996 | 0.00 (95% CI: −0.20, 0.20); p > 0.999 | 0.00 (95% CI: −0.20, 0.20); p = 0.985 | 0.00 (95% CI: −0.20, 0.20); p = 0.972 | |
| 200 Hz | 0.01 (95% CI: −0.22, 0.24); p = 0.936 | 0.00 (95% CI: −0.20, 0.20); p = 0.979 | 0.00 (95% CI: −0.19, 0.20); p = 0.962 | 0.01 (95% CI: −0.20, 0.21); p = 0.956 | |
| 125 Hz | 0.01 (95% CI: −0.21, 0.24); p = 0.898 | 0.01 (95% CI: −0.19, 0.21); p = 0.927 | 0.01 (95% CI: −0.19, 0.20); p = 0.948 | 0.02 (95% CI: −0.18, 0.22); p = 0.874 | |
| 100 Hz | 0.02 (95% CI: −0.21, 0.25); p = 0.850 | 0.01 (95% CI: −0.19, 0.21); p = 0.933 | 0.02 (95% CI: −0.18, 0.22); p = 0.860 | 0.00 (95% CI: −0.20, 0.21); p = 0.964 | |
| 50 Hz | 0.16 (95% CI: −0.07, 0.39); p = 0.172 | 0.08 (95% CI: −0.12, 0.29); p = 0.415 | 0.06 (95% CI: −0.14, 0.26); p = 0.554 | 0.06 (95% CI: −0.14, 0.27); p = 0.535 | |
| 40 Hz | 0.41 (95% CI: 0.18, 0.64); p < 0.001 | 0.10 (95% CI: −0.11, 0.30); p = 0.350 | 0.09 (95% CI: −0.11, 0.28); p = 0.398 | 0.07 (95% CI: −0.13, 0.27); p = 0.490 | |
| 30 Hz | 1.08 (95% CI: 0.85, 1.31); p < 0.001 | 0.50 (95% CI: 0.29, 0.70); p < 0.001 | 0.94 (95% CI: 0.74, 1.14); p < 0.001 | 0.88 (95% CI: 0.68, 1.08); p < 0.001 | |
| 25 Hz | 0.90 (95% CI: 0.70, 1.10); p < 0.001 | 0.86 (95% CI: 0.66, 1.06); p < 0.001 | 0.76 (95% CI: 0.53, 0.99); p < 0.001 | 0.90 (95% CI: 0.70, 1.10); p < 0.001 | |
| 20 Hz | 0.86 (95% CI: 0.66, 1.06); p < 0.001 | 0.71 (95% CI: 0.52, 0.91); p < 0.001 | 0.82 (95% CI: 0.62, 1.02); p < 0.001 | 0.86 (95% CI: 0.66, 1.06); p < 0.001 | |
| LF/HF (percent) | Intercept | 1.39 (95% CI: 1.19, 1.58); p < 0.001 | 1.46 (95% CI: 1.27, 1.65); p < 0.001 | 1.65 (95% CI: 1.44, 1.86); p < 0.001 | 1.39 (95% CI: 1.19, 1.58); p < 0.001 |
| 500 Hz | −0.00 (95% CI: −0.28, 0.28); p = 0.994 | −0.00 (95% CI: −0.27, 0.26); p = 0.988 | 0.00 (95% CI: −0.29, 0.29); p = 0.996 | −0.00 (95% CI: −0.28, 0.28); p = 0.994 | |
| 250 Hz | −0.01 (95% CI: −0.28, 0.27); p = 0.969 | −0.00 (95% CI: −0.27, 0.26); p = 0.987 | 0.00 (95% CI: −0.29, 0.29); p = 0.994 | −0.01 (95% CI: −0.28, 0.27); p = 0.969 | |
| 200 Hz | −0.01 (95% CI: −0.28, 0.27); p = 0.954 | −0.01 (95% CI: −0.27, 0.26); p = 0.964 | −0.01 (95% CI: −0.30, 0.28); p = 0.934 | −0.01 (95% CI: −0.28, 0.27); p = 0.954 | |
| 125 Hz | −0.02 (95% CI: −0.30, 0.26); p = 0.881 | −0.01 (95% CI: −0.27, 0.26); p = 0.951 | −0.02 (95% CI: −0.31, 0.27); p = 0.899 | −0.02 (95% CI: −0.30, 0.26); p = 0.881 | |
| 100 Hz | −0.00 (95% CI: −0.28, 0.27); p = 0.975 | −0.02 (95% CI: −0.29, 0.24); p = 0.862 | −0.03 (95% CI: −0.32, 0.27); p = 0.855 | −0.00 (95% CI: −0.28, 0.27); p = 0.975 | |
| 50 Hz | −0.08 (95% CI: −0.36, 0.20); p = 0.559 | −0.08 (95% CI: −0.35, 0.19); p = 0.563 | −0.21 (95% CI: −0.50, 0.09); p = 0.170 | −0.08 (95% CI: −0.36, 0.20); p = 0.559 | |
| 40 Hz | −0.09 (95% CI: −0.37, 0.19); p = 0.521 | −0.11 (95% CI: −0.38, 0.16); p = 0.420 | −0.54 (95% CI: −0.83, −0.25); p < 0.001 | −0.09 (95% CI: −0.37, 0.19); p = 0.521 | |
| 30 Hz | −1.30 (95% CI: −1.58, −1.02); p < 0.001 | −1.40 (95% CI: −1.67, −1.13); p < 0.001 | −1.57 (95% CI: −1.86, −1.27); p < 0.001 | −1.30 (95% CI: −1.58, −1.02); p < 0.001 | |
| 25 Hz | −1.30 (95% CI: −1.58, −1.03); p < 0.001 | −1.23 (95% CI: −1.49, −0.96); p < 0.001 | −1.02 (95% CI: −1.31, −0.73); p < 0.001 | −1.30 (95% CI: −1.58, −1.03); p < 0.001 | |
| 20 Hz | −1.24 (95% CI: −1.52, −0.96); p < 0.001 | −0.97 (95% CI: −1.24, −0.71); p < 0.001 | −1.25 (95% CI: −1.55, −0.94); p < 0.001 | −1.24 (95% CI: −1.52, −0.96); p < 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burma, J.S.; Griffiths, J.K.; Lapointe, A.P.; Oni, I.K.; Soroush, A.; Carere, J.; Smirl, J.D.; Dunn, J.F. Heart Rate Variability and Pulse Rate Variability: Do Anatomical Location and Sampling Rate Matter? Sensors 2024, 24, 2048. https://doi.org/10.3390/s24072048
Burma JS, Griffiths JK, Lapointe AP, Oni IK, Soroush A, Carere J, Smirl JD, Dunn JF. Heart Rate Variability and Pulse Rate Variability: Do Anatomical Location and Sampling Rate Matter? Sensors. 2024; 24(7):2048. https://doi.org/10.3390/s24072048
Chicago/Turabian StyleBurma, Joel S., James K. Griffiths, Andrew P. Lapointe, Ibukunoluwa K. Oni, Ateyeh Soroush, Joseph Carere, Jonathan D. Smirl, and Jeff F. Dunn. 2024. "Heart Rate Variability and Pulse Rate Variability: Do Anatomical Location and Sampling Rate Matter?" Sensors 24, no. 7: 2048. https://doi.org/10.3390/s24072048
APA StyleBurma, J. S., Griffiths, J. K., Lapointe, A. P., Oni, I. K., Soroush, A., Carere, J., Smirl, J. D., & Dunn, J. F. (2024). Heart Rate Variability and Pulse Rate Variability: Do Anatomical Location and Sampling Rate Matter? Sensors, 24(7), 2048. https://doi.org/10.3390/s24072048





