1. Introduction
Parkinson’s disease (PD) is the second most common dementia in the world, affecting a significant proportion of the elderly population [
1]. According to [
2], approximately 9 million people in the ten most populous countries will suffer from this disease by 2030. PD is characterized by a severe loss of dopamine in the forebrain, resulting in motor symptoms such as tremors, muscle stiffness, bradykinesia, postural instability, as well as non-motor symptoms including hyposmia, sleep disturbances, and autonomic dysfunction [
3]. The disability rate among PD patients is notably high [
4]. Furthermore, there is currently no cure for PD. All drug treatments can only relieve symptoms, reduce complications, and prolong life. Although Parkinson’s disease is incurable, a quantitative assessment of PD symptoms is necessary because it can help doctors use appropriate targeted interventions. To effectively assess these motor symptoms, rating scales have been widely adopted, such as the MDS-Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) [
5]. However, these assessments typically occur in clinical settings with infrequent annual visits, and the MDS-UPDRS evaluation is time-consuming, requiring at least 30 min and specialized training [
6]. These factors all contribute to the difficulty in effectively monitoring PD. As a result, convenient and objective PD assessment tools are required to better assist patients.
With the widespread adoption of wearable devices and the advancements in machine learning technology [
7,
8], there has been a significant body of research dedicated to the objective assessment of PD symptom severity using wearable inertial sensors. For instance, ref. [
9] used a wearable device on the hand to detect the number of finger-taps to assess the severity of bradykinesia, ref. [
10] developed a CNN-LSTM network to detect PD gait freezing using pressure sensors in insoles, and [
11] used two IMU sensors on the wrist and then asked subjects to perform 11 tasks to detect the early stages of PD. These works demonstrate the great potential of using wearable devices to assess PD.
Despite the significant promise demonstrated by wearable technology in monitoring PD symptoms, the development of a system and algorithm for assessing the disease stage of PD patients remains a challenging endeavor. Previous studies have predominantly concentrated on the detection of individual symptoms, such as tremor [
12], bradykinesia [
13,
14], and gait freezing [
15]. While these studies have effectively gauged the severity of specific PD symptoms, they fall short in providing a comprehensive evaluation of the overall disease stage of PD patients.
The clinical scale MDS-UPDRS evaluates the severity of PD based on symptoms manifested during various activities. Previous research has been limited to a single symptom and activity. To achieve a more comprehensive assessment of PD severity, it is imperative to consider the severity of multiple symptoms. However, real-world scenarios present the following two crucial challenges:
Limited annotation: In practical scenarios, obtaining detailed symptom annotations is a time-consuming endeavor [
16]. Especially in a free-living environment, the workload of obtaining accurate annotations of when PD symptoms begin and end is huge. PD symptoms are intermittent, which means that PD symptoms may be sparsely distributed and their time of appearance is unpredictable. However, we can only obtain coarse-grained annotations of PD stages, which makes training a supervised classifier to evaluate PD stages difficult. In fact, most current approaches are to cut the signal into shorter time segments, and then assign the overall disease severity label to each segment. However, this approach introduces a lot of noise segments, especially when symptoms are sparsely distributed.
Figure 1 shows the weak-label problem. This situation is commonly recognized as a weakly supervised problem, which has motivated us to develop a recognition framework within the context of weak supervision.
Class imbalance: The majority of patients tend to fall into the category of mild PD, with a relatively small proportion classified as severe (in our collected dataset, there was only about a quarter as many severe patients as mild patients). This results in a class imbalance issue. Additionally, there is often substantial variability in motor performance among PD patients at the same disease stage. Hence, we must explore effective strategies for leveraging our data to address this challenge.
The factors mentioned above present formidable challenges when it comes to assessing the stage of PD patients. Leveraging these challenges as a foundation, we introduce an innovative framework for PD diagnosis under weakly supervised conditions, with the goal of addressing the PD diagnostic problem in natural and daily environments characterized by limited annotation and data imbalance.
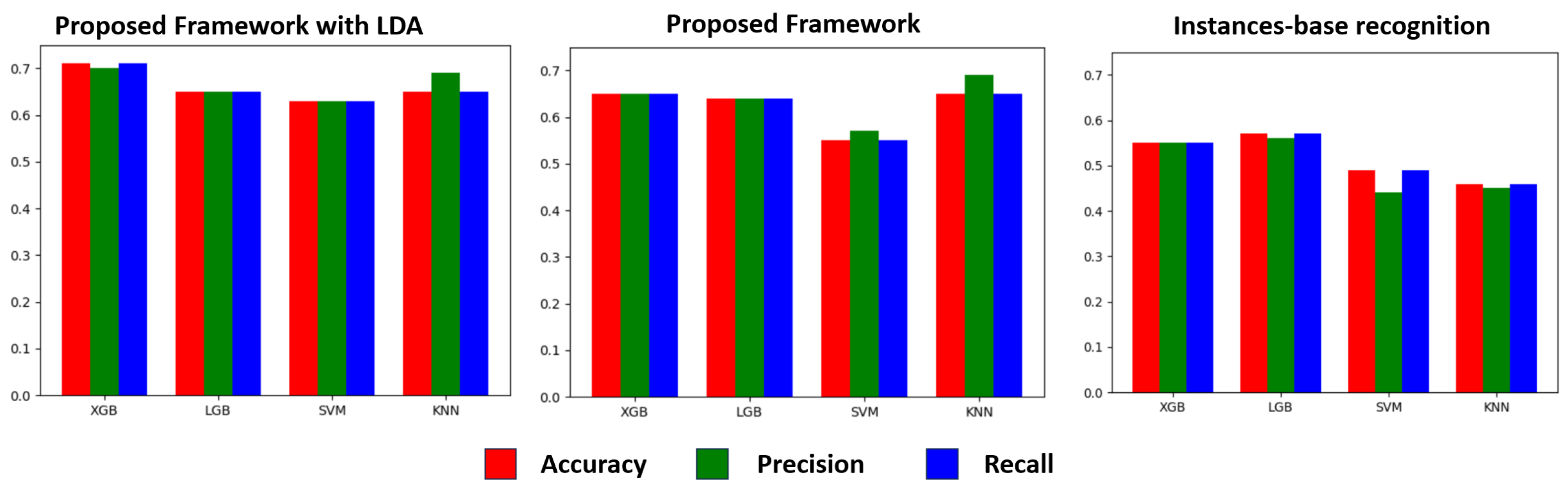
Our approach comprises two key components. First, we establish a PD learning framework under weak supervision. Initially, we employ fixed-window segments to extract features from the sensor signal data generated during various activities by PD patients. Subsequently, we apply k-means clustering to group together segments from the same activity across all patients. To uncover latent associations among different activities of PD patients, we utilize latent Dirichlet allocation (LDA) topic models to generate global features from these clustering labels. These two sets of features are then fused to create a refined representation of the PD patient features. Additionally, we introduce a data augmentation technique that identifies similar patient pairs through similarity comparisons and mixes them. Simultaneously, we disrupt the sequence of different segments within the data to reduce the reliance on segment position and time, thereby generating more diverse samples. These generated pseudo-data samples are used for training. Our approach is grounded in the belief that fine-grained features extracted from short-term fixed windows may not adequately capture the overall disease stage of a patient. Thus, we aim to unveil implicit associations between various activities of patients through unsupervised topic modeling. Furthermore, our data augmentation method enriches feature expressions while maintaining semantic consistency. Finally, we conduct a PD-stage classification test using a real-world dataset comprising 83 individuals in a free-living environment. Our approach achieves an accuracy rate of 73.48% in classifying PD stages (normal, mild, moderate, severe), surpassing segment-based PD-stage classification by 17%. This outcome serves as compelling evidence of the effectiveness of our method.
In summary, the main contributions of this work can be outlined as follows:
We propose a framework that utilizes the idea of multi-instance learning to combine symptom representations from multiple different activities to assess PD severity.
We propose a novel framework to accurately assess the status of PD patients within a weakly annotated context. We first combine local features from multiple segments with global topic features from various activities to perform classification. To address the problem of class imbalance, we present a straightforward yet highly effective data augmentation technique designed to generate additional data, enriching the original dataset, and enhancing the classification performance, particularly for minority classes.
To fully demonstrate the efficacy of our proposed framework, we collected a dataset containing wearable-sensor signals from 83 individuals in real-life, free-living conditions. Not only that, but this dataset also contains comprehensive and diverse wearable-sensor signals of a total of 12 human activities from each individual, which has never been provided in any previous works (to the best of our knowledge).
The detailed experimental results shows that our framework achieves an astounding 73.48% accuracy in the fine-grained (normal, mild, moderate, severe) classification of PD severity based on hand movements, which verifies the feasibility of accurately identifying PD patients based on machine learning and wearable-sensor data.
The remainder of this paper is structured as follows:
Section 2 delves into the relevant literature, while
Section 3 covers data collection and data preprocessing. In
Section 4, we introduce our framework for PD-stage assessment, followed by an evaluation and presentation of the results in
Section 5. Finally,
Section 6 provides the concluding remarks for this paper.
3. Data Collection
The data utilized in this research study were gathered by our team at the hospital over the period spanning from 15 January 2021 to 30 July 2022. The study involved 70 individuals diagnosed with PD and 15 healthy volunteers who willingly participated. Every participant provided informed consent. The total of 85 individuals can be divided into four categories, namely, healthy people, mild-PD patients (mild), moderate-PD patients (moderate), and severe-PD patients (severe). Please refer to
Table 1 for detailed demographics of this dataset.
Throughout the study, participants were equipped with Shimmer3 inertial measurement units (Dublin) (IMUs) on various body parts, including the left wrist, right wrist, left ankle, right ankle, and waist, for the collection of acceleration and gyroscope signal data. In subsequent experiments, data solely from the right wrist sensor were utilized to ease the burden on patients. The Shimmer3 IMU communicated wirelessly with a computer via Bluetooth. The ConsensysPRO software (Ver1.5.0) on the computer was employed to collect signal data at a high sampling frequency of 200 Hz. Each participant engaged in 12 distinct activities, with a 1-min rest interval between each activity. Prior to commencing the experiments, researchers provided instructions to the PD patients regarding the activity requirements. Once the experiments were underway, no further guidance or interference from the investigators was provided. Video recordings were made during the data collection process, and a neurologist subsequently scored all the data according to the Hoehn–Yahr (H –Y) scale, considering the participant’s performance across multiple activities. Individual-level labels were assigned based on this assessment.
Table 2 and
Figure 2 present a comprehensive list of the activities performed during the experimental setup. After collecting the data, we excluded patients whose activity lasted less than 20 s, and finally 83 subjects met the requirements.
4. Methodology
4.1. Framework Overview
The comprehensive framework is visually represented in
Figure 3. This framework is organized into five distinct components. The initial part encompasses data preprocessing and sliding window segmentation. The second component extracts the features from the original signal. In the third part, these fragment features are used for k-means clustering and aggregation. We then fit the distribution of these cluster labels through the LDA model to generate new global features. In the fourth part, we use data augmentation methods to generate more data and alleviate the negative effects of data imbalance. The final segment pertains to the training and testing phase, where machine learning models are trained and assessed using these features. This approach aims to develop a machine learning model which is capable of evaluating the disease severity of PD patients. Detailed descriptions of each step follow in the subsequent sections.
4.2. Problem Statement
We formulate the research problem as a four-class classification task. Given that the input features x represent the activities of participants, the objective of the model is to predict the PD stage y, which can fall into one of four categories: healthy, mild, moderate, or severe PD. This problem, distinct from binary classification between PD patients and non-PD patients, presents a much greater challenge due to the potential similarity in features exhibited by PD patients at various stages during specific time intervals.
To solve the problem of inaccurate supervision, we adopt the multiple-instance learning (MIL) method. In MIL, each learned sample is defined as a bag containing multiple instances. Different from traditional single-instance learning, each package contains the feature space of different instances. We define the bag collection ; for each bag has m instances . Here, we adopt MIL’s bag assumption: each instance is independent of the label, and the bag is related to the label. The classification tasks occur at the bag level.
4.3. Data Preprocessing and Segmentation
Tremor in PD patients can be classified into three types: rest tremor, 3–6 Hz; postural tremor, 4–12 Hz; and kinetic tremor, 2–7 Hz [
30]. To smooth the signal and remove the gravity component, we use a 4th-order Butterworth filter with a bandpass range of 0.3–20 Hz. After applying Z-score normalization to the signal, the data are sliced at 300 data points (1.5 s) with a 50% overlap. Finally, for each window signal, we compute the time- and frequency-domain-related features (standard deviation, variance, skewness, kurtosis, root mean square, energy, median, range, correlation). The preprocessing method is depicted in
Figure 4.
4.4. Feature Extraction and Fusion
Clustering for Document Creation: As depicted in
Figure 3, the activity signals performed by the patient are segmented into segments represented as
where
m denotes the
m-th activity,
n represents the
n-th window segment of the activity, and
i signifies the feature set of the segment. Subsequently, the feature sets of the same activity segment from different patients are clustered using k-means, and the resulting clustered labels are employed as words to construct documents:
Here, represents the k-means clustering label, b denotes the b-th activity, and p indicates the p-th subject. This method involves aggregating words from multiple patient activities to generate a document, followed by utilizing a topic model to derive global features.
LDA Topic Model for Global Feature Generation: In our framework, latent Dirichlet allocation (LDA) [
31] is utilized to discover global topic features across various activities. The document–word matrix serves as input to LDA, which subsequently outputs the document–topic distribution as global features. The document–topic distribution is defined as
The variables
and
represent Dirichlet distributions,
signifies the topic distribution, and
represents the word distribution. In the LDA model, a topic
z is selected from the topic distribution
, and a word
w is chosen from the word distribution
. A document comprises a collection of
N words, while a corpus
D consists of
M documents, and
K signifies the total number of topics in the corpus. LDA generates documents based on input parameters
,
, and
K, governing the creation of topics and words [
32]. As depicted in
Figure 3, we input a document–word matrix into LDA, which produces a topic distribution. The probabilities associated with each topic are utilized as features. Ultimately, these feature vectors are combined horizontally to form the comprehensive set of features for PD recognition. This approach provides an advantage as each patient’s various activities generate a topic distribution feature, derived from the global information across multiple activities, resulting in richer information and enhanced feature expression.
Figure 5 illustrates the final feature vector. Algorithm 1 demonstrates the entire process.
| Algorithm 1 Bag Generation for Multiple Activity Instances |
- 1:
function Bag generation for multiple activity instances - 2:
Input: Instance of different activities - 3:
Output: Bag vector - 4:
- 5:
for to do - 6:
for instance in do - 7:
= - 8:
end for - 9:
Create Document Feature - 10:
Create LDA Feature - 11:
Horizontal binding vector - 12:
end for - 13:
- 14:
Return - 15:
end function
|
4.5. Data Augmentation
As mentioned in
Section 1, data augmentation serves two primary purposes. Firstly, it aims to enhance the prediction accuracy of minority classes, addressing the issue of class imbalance. Secondly, it seeks to reduce variability between patients, thus improving the model’s robustness.
Figure 6 illustrates the data augmentation method. Initially, we organize the instance clustering labels into vectors A and B according to their original chronological order. Subsequently, we compute the distances between different patients using the following (
3) and select pairs with close distances:
We employ two methods for mixing the samples: ① We randomly shuffle similar sample pairs to generate new samples. These bag pairs originate from patients at the same PD stage, so the labels remain unchanged. This process increases sample diversity and mitigates intra-class differences. ② We shuffle the order of instances within each bag to address the uncertainty surrounding when patients exhibit symptoms. Through shuffling, we generate samples that are independent of time.
6. Conclusions
In this study, we aimed to assess PD stages using a single wearable sensor attached to the right hand. We conducted our research on a real dataset comprising 85 individuals with PD. During our investigation, we identified two key challenges: weak labeling and data imbalance. To address these issues, we introduced a framework for PD-stage evaluation represented by symbols. We employed topic modeling to enhance feature representation within this framework. Additionally, we incorporated a data augmentation component to diversify our dataset, exploring various sample generation techniques such as SMOTE, ADASYN, and SMOTETomek, among others. Furthermore, we introduced a novel similarity-based pattern mixing method. As a result, our final model achieved an impressive accuracy rate of 73.48%. This demonstrates the framework’s ability to mitigate the impact of weak annotations and to enhance data diversity. In summary, our research contributes to more accurate self-diagnosis of PD in real-world settings, offering the potential for remote guidance on medical interventions by healthcare professionals.
In future work, we will continue to dig deeper into the performance of using wearable sensors to accurately assess the severity of PD conditions. In this work, we only independently analyze and utilize the sensor data corresponding to a single activity. Considering the inevitable connection between multiple activities performed by each patient, a more natural and promising approach is to use multi-task learning [
40,
41] to fully mining the complex relationship between multiple activities, so as to utilize the inherent shared information among multiple activities, further improving the predictive performance and robustness of the model.