Figure 1.
Block diagram showing the main steps of the study’s workflow: input data (a) undergo manual tumour segmentation (b), that is the starting point for both ZoT and tRIM detection (c); then, features are generated from both ZoT and tRIM regions (d) and used for predictive purposes in HCC-CT and LARC-MRI studies (e).
Figure 1.
Block diagram showing the main steps of the study’s workflow: input data (a) undergo manual tumour segmentation (b), that is the starting point for both ZoT and tRIM detection (c); then, features are generated from both ZoT and tRIM regions (d) and used for predictive purposes in HCC-CT and LARC-MRI studies (e).
Figure 2.
Four-step procedure developed for ZoT detection. The process begins with the segmented tumour ROIs, which stand for the input, shown in (a), for LARC and (b) HCC. The first step is the morphological edge detection (c,d), which is followed by the gradient profile analysis (e,f). In this way, more details are depicted in (g–j) for a representative edge pixel, analysed along its gradient direction with an assumed rectangular kernel, for instance of 15 × 2 pixels in size. Then, this steps yield binary masks of pixel-based gradient transition zones, which are thickened in the fourth step (k,l). Finally, the last step is the ZoT detection, arising from the computation of the density maps on the previous masks and their automatic segmentation, which ultimately provides the ZoT’s outer and inner borders (m,n).
Figure 2.
Four-step procedure developed for ZoT detection. The process begins with the segmented tumour ROIs, which stand for the input, shown in (a), for LARC and (b) HCC. The first step is the morphological edge detection (c,d), which is followed by the gradient profile analysis (e,f). In this way, more details are depicted in (g–j) for a representative edge pixel, analysed along its gradient direction with an assumed rectangular kernel, for instance of 15 × 2 pixels in size. Then, this steps yield binary masks of pixel-based gradient transition zones, which are thickened in the fourth step (k,l). Finally, the last step is the ZoT detection, arising from the computation of the density maps on the previous masks and their automatic segmentation, which ultimately provides the ZoT’s outer and inner borders (m,n).
Figure 3.
Summary of features considered for the and studies. Each grey box indicates the ROI considered for feature generation (tRIM, ZoT, or tumour) and image type; that is, the gradient magnitude (GM) or grey level (GL). Two more grey boxes refer to volume features and RCS measurements. Each coloured path highlights the pool of imaging features analysed by each study.
Figure 3.
Summary of features considered for the and studies. Each grey box indicates the ROI considered for feature generation (tRIM, ZoT, or tumour) and image type; that is, the gradient magnitude (GM) or grey level (GL). Two more grey boxes refer to volume features and RCS measurements. Each coloured path highlights the pool of imaging features analysed by each study.
Figure 4.
Examples of the ZoT (in red), tRIM (in green), and tumour (the shaded area bounded by the blue line) ROIs. For each image, the patient identification code () and slice number () are indicated, along with the dataset the samples belong to (i.e., HCC or LARC) and their membership class (MVI+ or MVI− for the HCC dataset; R or NR for the LARC dataset). In addition, the pink arrows highlight the key differences between the tRIM and ZoT ROIs.
Figure 4.
Examples of the ZoT (in red), tRIM (in green), and tumour (the shaded area bounded by the blue line) ROIs. For each image, the patient identification code () and slice number () are indicated, along with the dataset the samples belong to (i.e., HCC or LARC) and their membership class (MVI+ or MVI− for the HCC dataset; R or NR for the LARC dataset). In addition, the pink arrows highlight the key differences between the tRIM and ZoT ROIs.
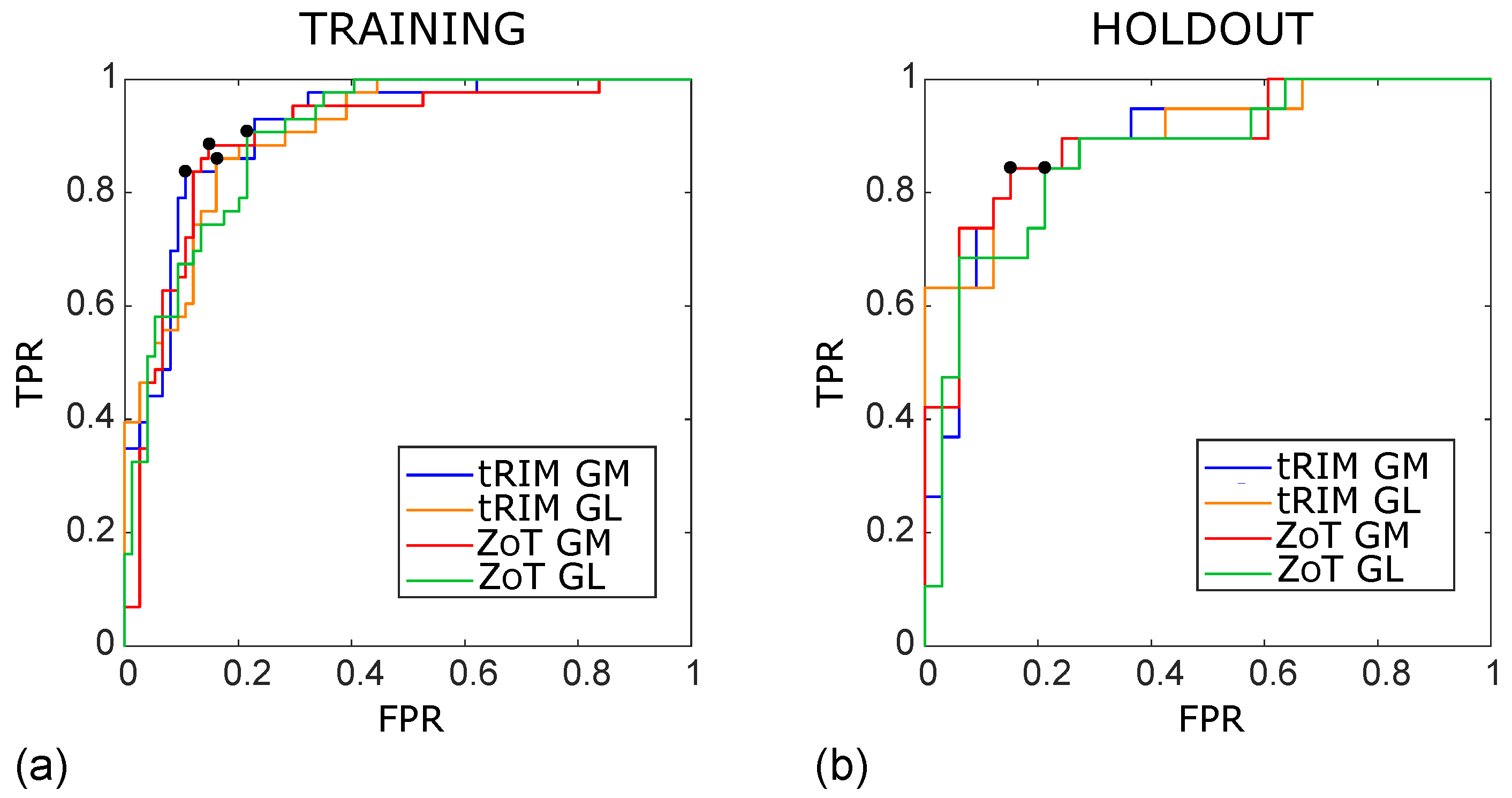
Figure 5.
ROC curves of the SVM classifiers arising from the studies achieved for the training (a) and holdout (b) sets, referring to the tRIM or ZoT analysis performed on the gradient magnitude (GM) or original grey level (GL) images. The outcomes for the training subset were as follows: tRIM GM: AUC ROC = 0.91 (95% CI 0.85–0.96), tRIM GL: AUC ROC = 0.91 (95% CI 0.84–0.95), ZoT GM: AUC ROC = 0.90 (95% CI 0.84–0.94), and ZoT GL: AUC ROC = 0.90 (95% CI 0.83–0.94). The outcomes for the holdout were as follows: tRIM GM: AUC ROC = 0.89 (95% CI 0.77–0.96), tRIM GL: AUC ROC = 0.87 (95% CI 0.74–0.96), ZoT GM: AUC ROC = 0.89 (95% CI 0.75–0.95), and ZoT GL: AUC ROC = 0.87 (95% CI 0.75–0.95).
Figure 5.
ROC curves of the SVM classifiers arising from the studies achieved for the training (a) and holdout (b) sets, referring to the tRIM or ZoT analysis performed on the gradient magnitude (GM) or original grey level (GL) images. The outcomes for the training subset were as follows: tRIM GM: AUC ROC = 0.91 (95% CI 0.85–0.96), tRIM GL: AUC ROC = 0.91 (95% CI 0.84–0.95), ZoT GM: AUC ROC = 0.90 (95% CI 0.84–0.94), and ZoT GL: AUC ROC = 0.90 (95% CI 0.83–0.94). The outcomes for the holdout were as follows: tRIM GM: AUC ROC = 0.89 (95% CI 0.77–0.96), tRIM GL: AUC ROC = 0.87 (95% CI 0.74–0.96), ZoT GM: AUC ROC = 0.89 (95% CI 0.75–0.95), and ZoT GL: AUC ROC = 0.87 (95% CI 0.75–0.95).
Figure 6.
Box plot of the classification achieved by the SVM models in the training sets for the tRIM (a,b) and ZoT (c,d) analyses on gradient magnitude (GM) or rgw original grey-level (GL) series. The p-values for the Wilcoxon rank-sum test are p∼ (a), p∼ (b,d), and p∼ (c).
Figure 6.
Box plot of the classification achieved by the SVM models in the training sets for the tRIM (a,b) and ZoT (c,d) analyses on gradient magnitude (GM) or rgw original grey-level (GL) series. The p-values for the Wilcoxon rank-sum test are p∼ (a), p∼ (b,d), and p∼ (c).
Figure 7.
Box plot of the classification achieved by SVM models in the holdout sets for tRIM (a,b) and ZoT (c,d) analyses on gradient magnitude (GM) or original grey-level (GL) series. The p-values of Wilcoxon rank-sum test are p∼ (a,c) and p∼ (b,d).
Figure 7.
Box plot of the classification achieved by SVM models in the holdout sets for tRIM (a,b) and ZoT (c,d) analyses on gradient magnitude (GM) or original grey-level (GL) series. The p-values of Wilcoxon rank-sum test are p∼ (a,c) and p∼ (b,d).
Figure 8.
Box plot of S−E (a), S (b), and K (c), referring the t tumour, tRIM, and ZoT analyses for the GM images ( studies) for discriminating the R and NR groups.
Figure 8.
Box plot of S−E (a), S (b), and K (c), referring the t tumour, tRIM, and ZoT analyses for the GM images ( studies) for discriminating the R and NR groups.
Figure 9.
ROC curves of the most discriminant features between the R and NR groups arising from the analysis of tumour (blue line), tRIM (orange line), and ZoT (red line) on gradient magnitude (GM) images, with the Youden cutoff which defines the I value highlighted by black circles. The tumour’s ROC yielded an AUC = 0.86 (95% CI, 0.63–0.92) and an I = 0.62, tRIM’s ROC an AUC = 0.81 (95% CI, 0.65–0.91) and an I = 0.58, and ZoT’s ROC an AUC = 0.82 (95% CI, 0.63–0.93) and an I = 0.68.
Figure 9.
ROC curves of the most discriminant features between the R and NR groups arising from the analysis of tumour (blue line), tRIM (orange line), and ZoT (red line) on gradient magnitude (GM) images, with the Youden cutoff which defines the I value highlighted by black circles. The tumour’s ROC yielded an AUC = 0.86 (95% CI, 0.63–0.92) and an I = 0.62, tRIM’s ROC an AUC = 0.81 (95% CI, 0.65–0.91) and an I = 0.58, and ZoT’s ROC an AUC = 0.82 (95% CI, 0.63–0.93) and an I = 0.68.
Table 1.
HCC dataset description.
Table 1.
HCC dataset description.
| Study Population | Number of Samples |
|---|
| True-positive class | MVI+ |
| True-negative class | MVI− |
| Initial dataset | 89 (32 MVI+, 57 MVI−) |
| Oversampled dataset (OD) | 169 (62 MVI+, 107 MVI−) |
| OD training set | 117 (43 MVI+, 74 MVI−) |
| OD test set | 52 (19 MVI+, 33 MVI−) |
Table 2.
LARC dataset description.
Table 2.
LARC dataset description.
| Study Population | Number of Samples |
|---|
| True-positive class | Responders (R) |
| True-negative class | Non responder (NR) |
| Study population | 46 (18 R, 28 NR) |
Table 3.
Summary of the feature selection performed on all the studies . Columns report from left to right the study’s identifier, the number of features initially selected by LASSO (), all quadruples (), the uncorrelated quadruples (), the significant quadruples in the Wilcoxon rank-sum test with Holm-Bonferroni correction (), the finally selected quadruples (through the single feature identifiers), and their corresponding I.
Table 3.
Summary of the feature selection performed on all the studies . Columns report from left to right the study’s identifier, the number of features initially selected by LASSO (), all quadruples (), the uncorrelated quadruples (), the significant quadruples in the Wilcoxon rank-sum test with Holm-Bonferroni correction (), the finally selected quadruples (through the single feature identifiers), and their corresponding I.
| | | | | | Quadruple | I |
|---|
| : tRIM GM | 19 | 3876 | 1951 | 26 | [F136, F256, F592, F754] | 0.67 |
| : tRIM GL | 18 | 3060 | 1893 | 21 | [F136, F466, F592, F754] | 0.67 |
| : ZoT GM | 16 | 1820 | 767 | 18 | [F72, F136, F465, F754] | 0.74 |
| : ZoT GL | 13 | 715 | 268 | 40 | [F136, F465, F688, F754] | 0.70 |
Table 4.
Association between the identifier (ID) and the feature name. In particular, A or V indicate whether the feature refers to the arterial or venous phases, respectively. If the abbreviation “T” is reported as the feature that is computed on tumour ROIs; otherwise it originates from tRIM or ZoT ROIs depending on the study () from which it has been selected.
Table 4.
Association between the identifier (ID) and the feature name. In particular, A or V indicate whether the feature refers to the arterial or venous phases, respectively. If the abbreviation “T” is reported as the feature that is computed on tumour ROIs; otherwise it originates from tRIM or ZoT ROIs depending on the study () from which it has been selected.
| ID | Feature Name |
|---|
| F72 | S - E (A) |
| F136 | E - K (T, A) |
| F256 | E - K (V) |
| F465 | K - MAD (T, V) |
| F466 | K - IQR (T, V) |
| F592 | RCS of K − IQR |
| F688 | RCS of IQR (T) |
| F754 | RCS of MAD − K (T) |
Table 5.
ROC-related metrics achieved by the SVM classifiers in the training sets for the prediction of MVI+.
Table 5.
ROC-related metrics achieved by the SVM classifiers in the training sets for the prediction of MVI+.
| | AUC | I | SN | SP | TP | TN | FP | FN | ACC | NPV | PPV | DOR |
|---|
| HCC1 | 0.91 | 0.73 | 84% | 89% | 36 | 66 | 8 | 7 | 87% | 90% | 82% | 42 |
| HCC2 | 0.91 | 0.70 | 86% | 84% | 37 | 62 | 12 | 6 | 85% | 91% | 76% | 31 |
| HCC3 | 0.90 | 0.74 | 88% | 85% | 38 | 63 | 11 | 5 | 86% | 93% | 78% | 43 |
| HCC4 | 0.90 | 0.69 | 91% | 78% | 39 | 58 | 16 | 4 | 83% | 94% | 71% | 35 |
Table 6.
ROC-related metrics achieved by the SVM classifiers in the test sets for the prediction of MVI+.
Table 6.
ROC-related metrics achieved by the SVM classifiers in the test sets for the prediction of MVI+.
| | AUC | I | SN | SP | TP | TN | FP | FN | ACC | NPV | PPV | DOR |
|---|
| HCC1 | 0.89 | 0.69 | 84% | 85% | 16 | 28 | 5 | 3 | 88% | 90% | 76% | 30 |
| HCC2 | 0.87 | 0.69 | 84% | 85% | 16 | 28 | 5 | 3 | 88% | 90% | 76% | 30 |
| HCC3 | 0.89 | 0.69 | 84% | 85% | 16 | 28 | 5 | 3 | 88% | 90% | 76% | 30 |
| HCC4 | 0.87 | 0.63 | 84% | 79% | 16 | 26 | 7 | 3 | 81% | 90% | 70% | 18 |
Table 7.
ROC-related metrics achieved by S−E, S, and K referring to the tumour, tRIM, and ZoT analyses ( studies).
Table 7.
ROC-related metrics achieved by S−E, S, and K referring to the tumour, tRIM, and ZoT analyses ( studies).
| | AUC | I | SN | SP | TP | TN | FP | FN | ACC |
|---|
| 0.86 | 0.62 | 89% | 72% | 13 | 25 | 3 | 5 | 83% |
| 0.81 | 0.58 | 83% | 75% | 15 | 21 | 7 | 3 | 78% |
| 0.82 | 0.67 | 89% | 79% | 16 | 22 | 6 | 2 | 83% |