Estimation of Muscle Forces of Lower Limbs Based on CNN–LSTM Neural Network and Wearable Sensor System
Abstract
1. Introduction
2. Materials and Methods
2.1. Calculation of Muscle Force
2.2. Neural Network for Muscle Forces Estimation
2.2.1. Structure of Standard CNN
2.2.2. Structure of Standard LSTM
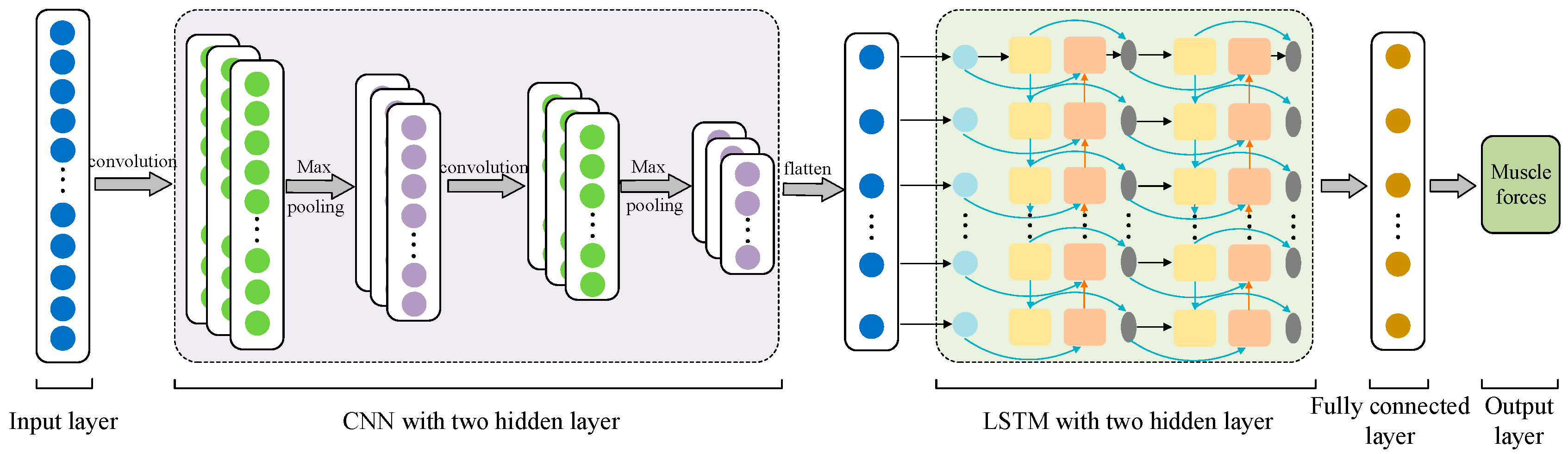
2.2.3. Structure of CNN–LSTM
2.2.4. Evaluation Method
3. Experiment
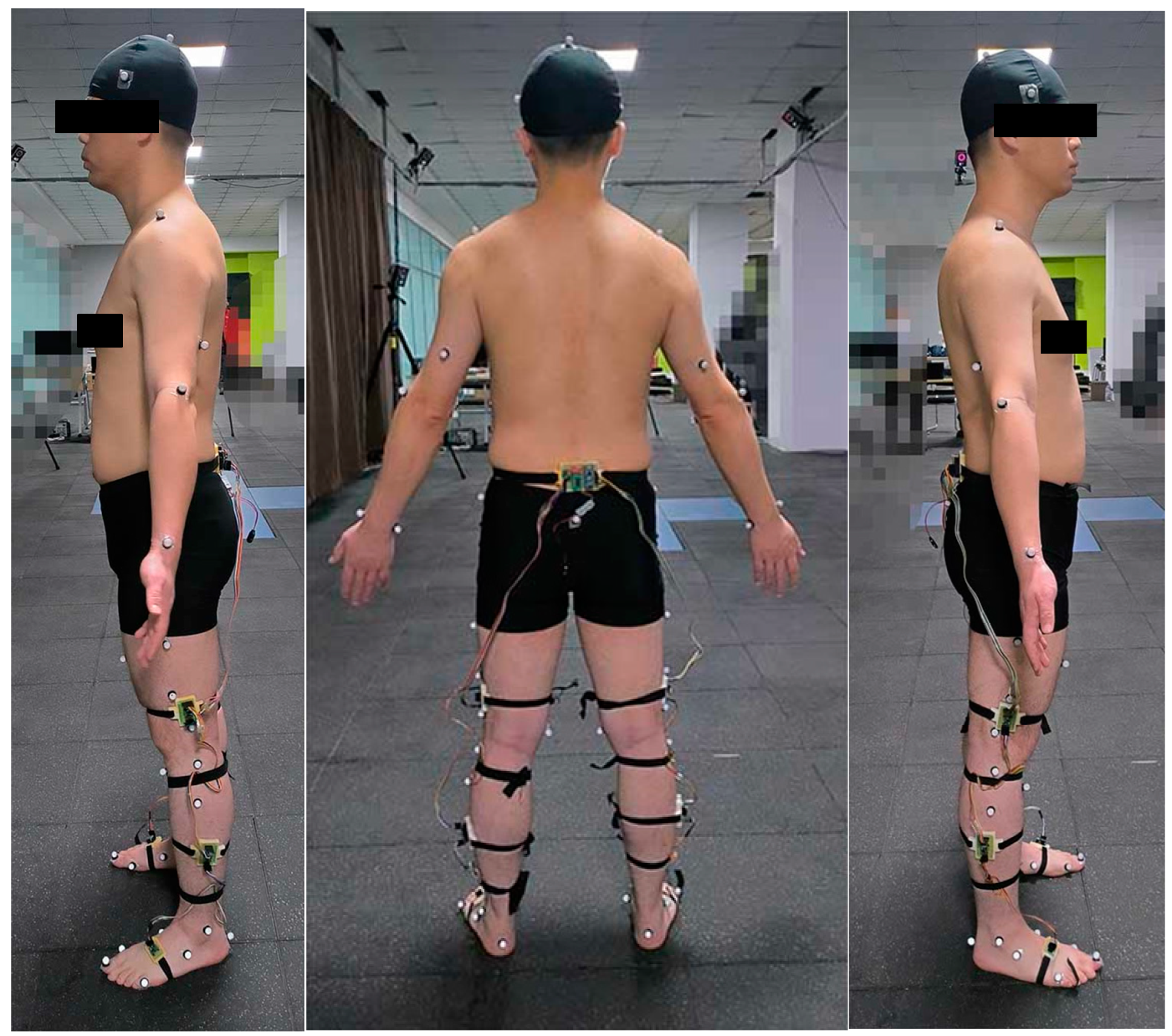
3.1. Data Collection and Preprocessing
3.2. Participants
3.2.1. Training Validation Experiment
3.2.2. External Testing Experiment
4. Results and Discussion
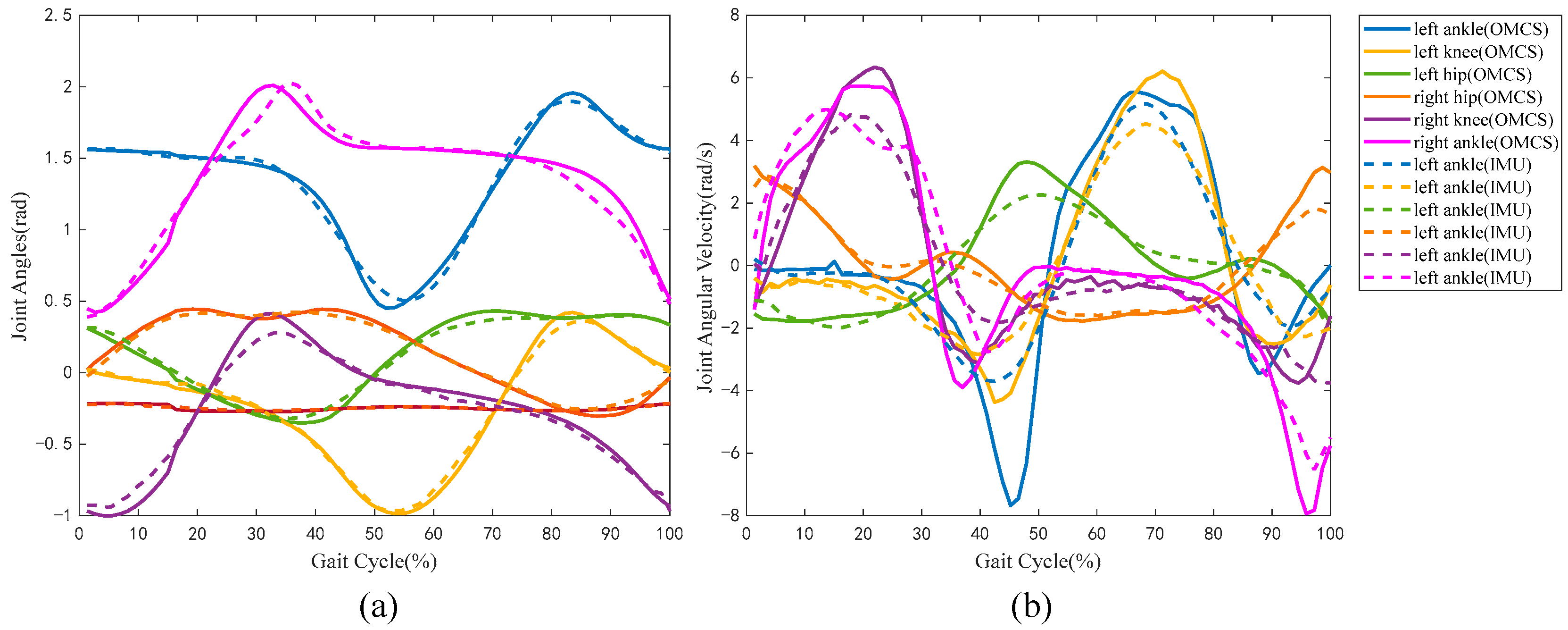
4.1. Accuracy Verification of Sensor System
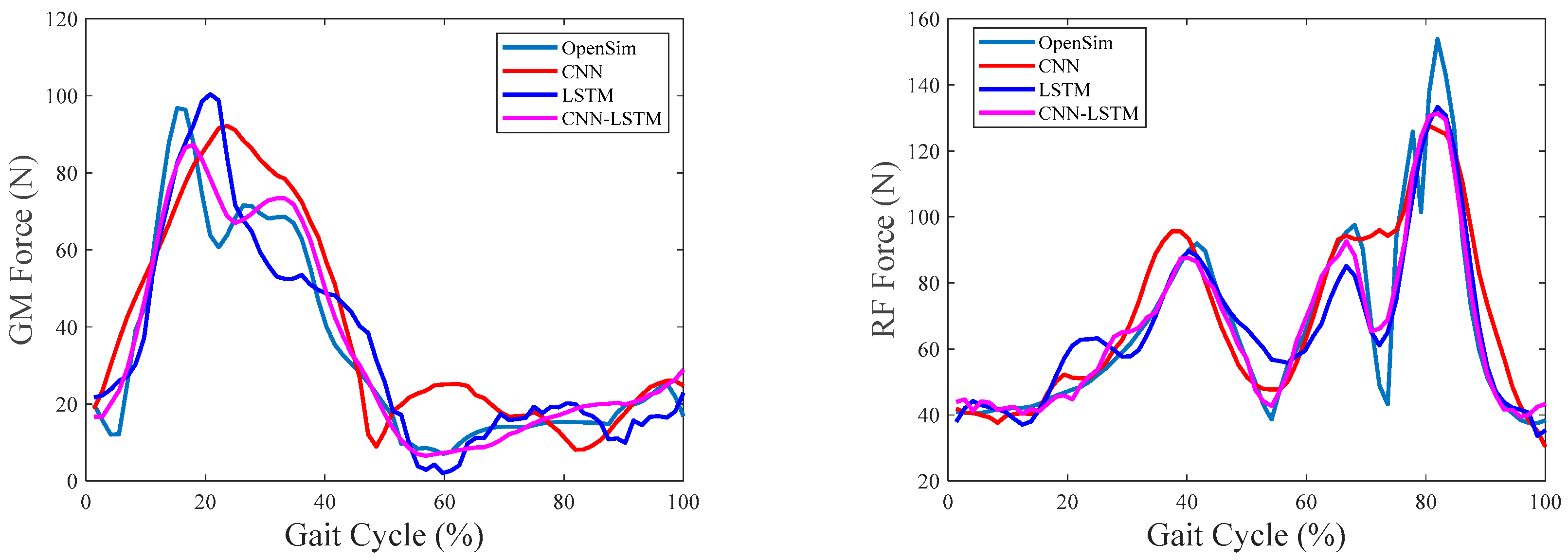
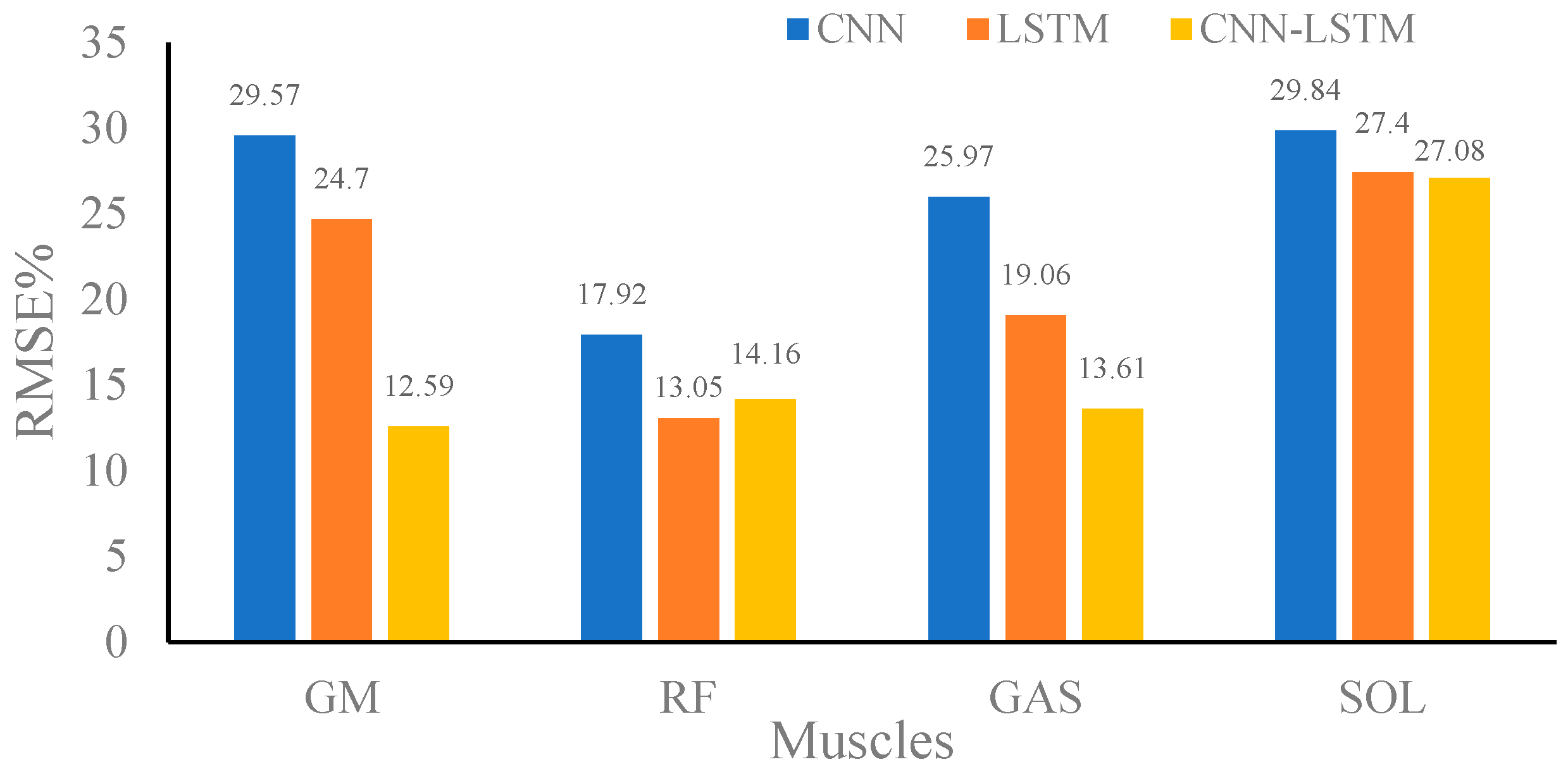
4.2. Overall Comparisons
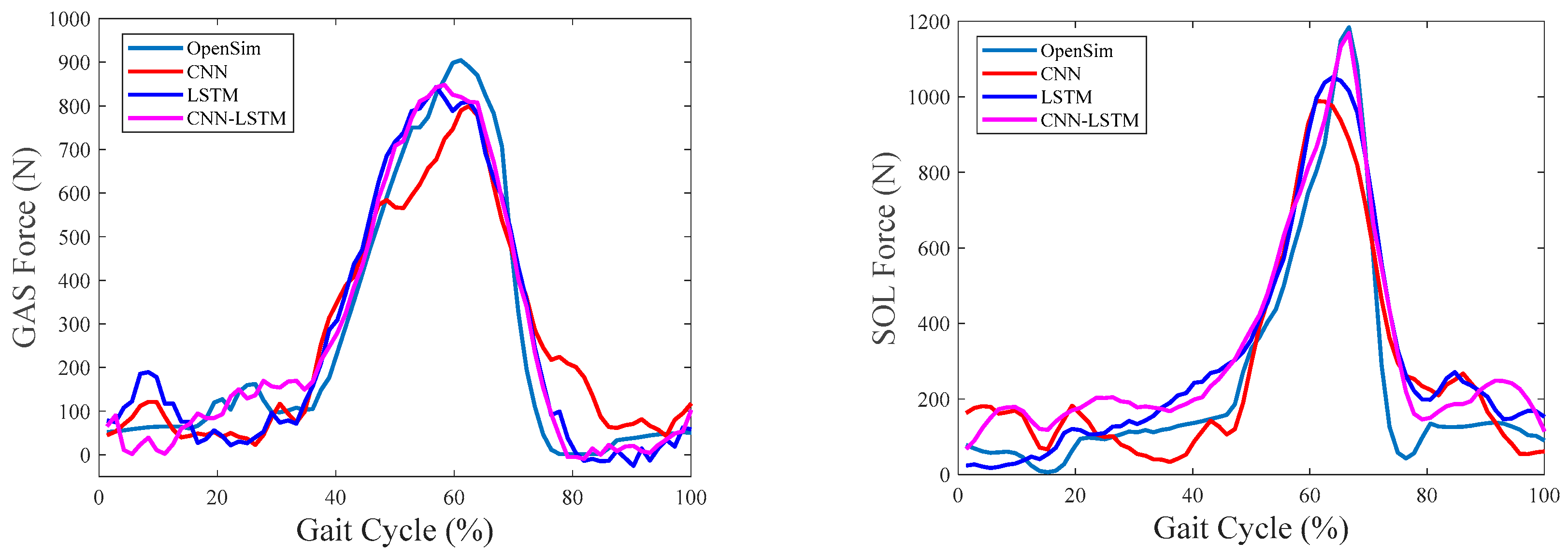
4.3. Evaluation of Intrasession Scenario
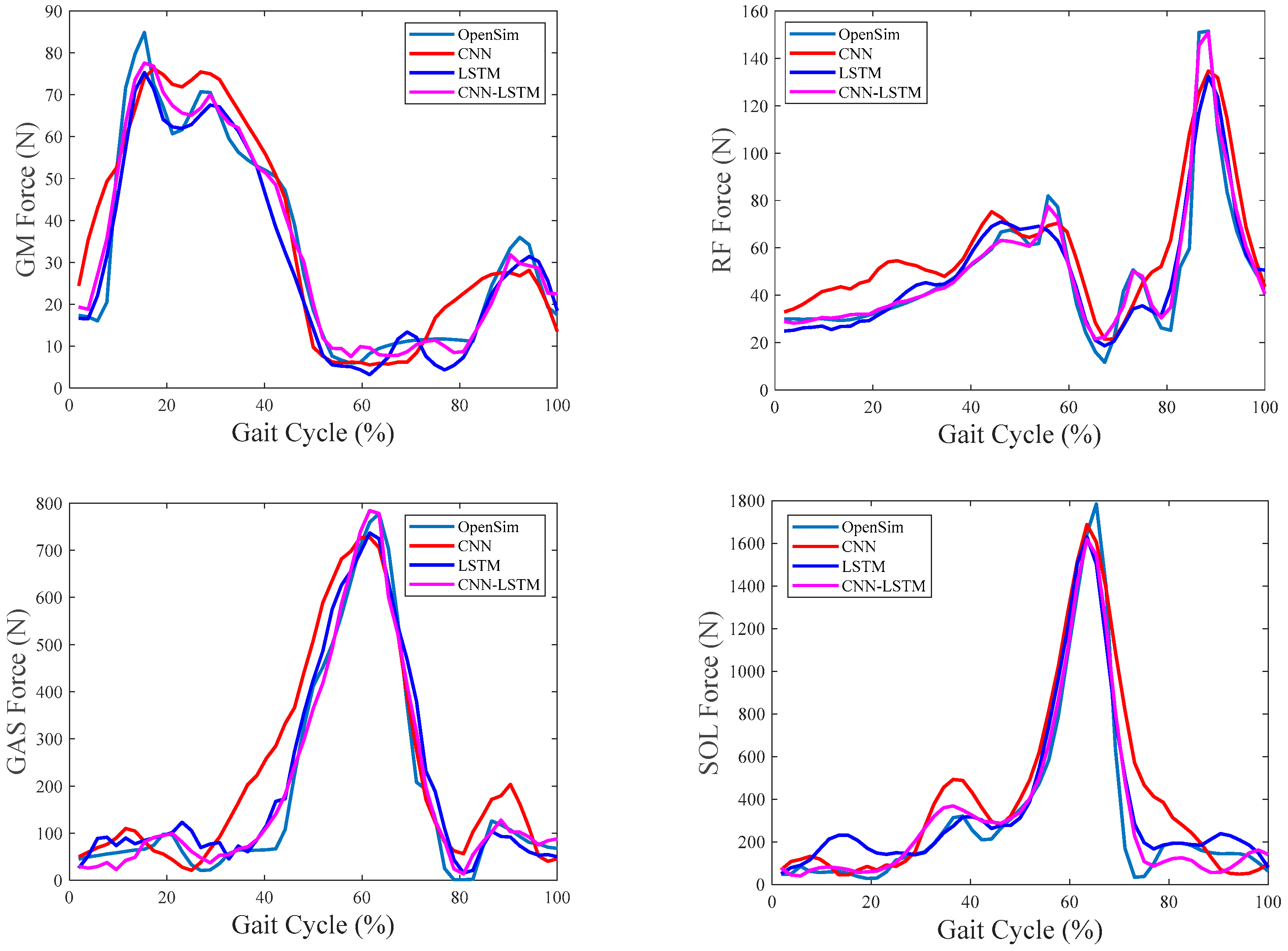
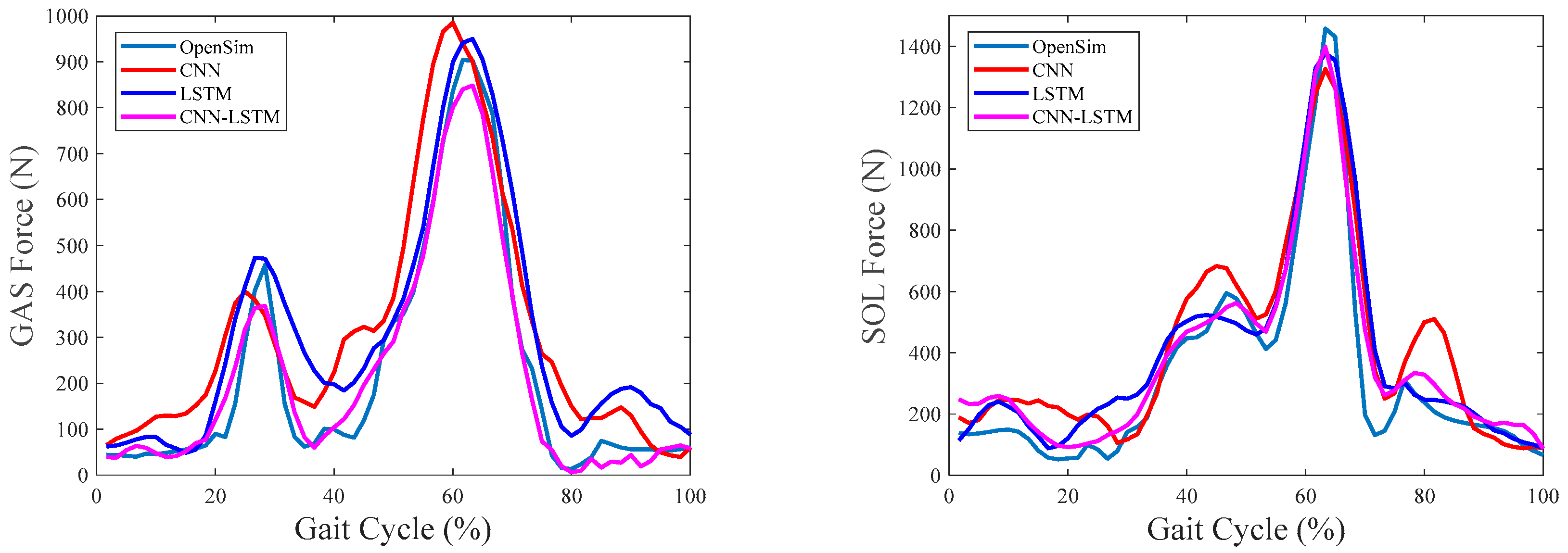
4.4. Evaluation of Intersession Scenario
4.5. Advantages and Limitations of the CNN–LSTM
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Harrington, M.S.; Burkhart, T.A. Validation of a musculoskeletal model to investigate hip joint mechanics in response to dynamic multiplanar tasks. J. Biomech. 2023, 158, 111767. [Google Scholar] [CrossRef]
- An, Q.; Yamakawa, H.; Yamashita, A.H. Temporal Structure of Muscle Synergy of Human Stepping Leg During Sit-to-Walk Motion. Intell. Auton. Syst. 2017, 531, 91–102. [Google Scholar]
- Teasell, R. Stroke recovery and rehabilitation. Stroke 2003, 34, 365–366. [Google Scholar] [CrossRef]
- Sicherer, S.T.; Venkatarama, R.S.; Grasman, J.M. Recent Trends in Injury Models to Study Skeletal Muscle Regeneration and Repair. Bioengineering 2020, 7, 76. [Google Scholar] [CrossRef]
- Laschowski, B.; McPhee, J. Energy-Efficient Actuator Design Principles for Robotic Leg Prostheses and Exoskeletons: A Review of Series Elasticity and Backdrivability. J. Comput. Nonlinear Dyn. 2023, 18, 6. [Google Scholar] [CrossRef]
- Chen, Q.; Guo, S.J.; Wang, J.; Wang, J.X.; Zhang, D.; Jin, S.H. Biomechanical and Physiological Evaluation of Biologically-inspired Hip Assistance with Belt-type Soft Exosuits. IEEE Trans. Neural. Syst. Rehabil. Eng. 2022, 30, 2802–2814. [Google Scholar] [CrossRef]
- Wang, D.J.; Gu, X.P.; Li, W.Z.; Jin, Y.X.; Yang, M.S.; Yu, H.L. Evaluation of safety-related performance of wearable lower limb exoskeleton robot (WLLER): A systematic review. Rob. Auton. Syst. 2023, 160, 104308. [Google Scholar] [CrossRef]
- Sung, Y.S.; Kristen, H.; Matt, G.; Louis, N.; Conor, J.; Arun, J. Soft robotic exosuit augmented high intensity gait training on stroke survivors: A pilot study. J. Neuroeng. Rehabil. 2022, 19, 1–12. [Google Scholar]
- Dao, T.T.; Pouletaut, P.; Charleux, F.; Lazáry, Á.; Eltes, P.; Varga, P.P.; Ho, B.; Tho, M.C. Multimodal medical imaging (CT and dynamic MRI) data and computer-graphics multi-physical model for the estimation of patient specific lumbar spine muscle forces. Data Knowl. Eng. 2015, 96, 3–18. [Google Scholar] [CrossRef]
- Yang, K.W.; Tang, W.T.; Liu, S.H. Muscle Contributions to Take-Off Velocity in the Long Jump. Med. Sci. Sports Exerc. 2023, 55, 1434–1444. [Google Scholar] [CrossRef]
- Eskinazi, I.; Fregly, B.J. A computational framework for simultaneous estimation of muscle and joint contact forces and body motion using optimization and surrogate modeling. Med. Eng. Phys. 2018, 54, 56–64. [Google Scholar] [CrossRef]
- Son, J.; Hwang, S.; Kim, Y. A hybrid static optimisation method to estimate muscle forces during muscle co-activation. Comput. Methods Biomech. Biomed. Eng. 2012, 15, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.; Artemiadis, P. A model-based analysis of supraspinal mechanisms of inter-leg coordination in human gait: Toward modelinformed robot-assisted rehabilitation. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Q.; Anderson, F.C.; Schwartz, M.H.; Delp, S.L. Muscle contributions to support and progression over a range of walking speeds. J. Biomech. 2008, 41, 3243–3252. [Google Scholar] [CrossRef] [PubMed]
- Delp, S.L.; Anderson, F.C.; Arnold, A.S. OpenSim: Opensource software to create and analyze dynamic simulations of movement. IEEE. Trans. Biomed. Eng. 2007, 54, 1940–1950. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.D.; Henley, B.J.; Sleap, S.B.; Sloan, S.W. Kinetic model selection and the Hill model in geochemistry. Int. J. Environ. Sci. Technol. 2015, 12, 2545–2558. [Google Scholar] [CrossRef]
- Clancy, E.A.; Hogan, N. Relating agonist-antagonist electromyograms to joint torque during isometric, quasi-isotonic, nonfatiguing contractions. IEEE Trans. Biomed. Eng. 1997, 44, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Johns, G.; Morin, E.; Hashtrudi-Zaad, K. Force Modelling of Upper Limb Biomechanics Using Ensemble Fast Orthogonal Search on High-Density Electromyography. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 24, 1041–1050. [Google Scholar] [CrossRef]
- Hashemi, J.; Morin, E.; Mousavi, P.; Hashtrudi-Zaad, K. Enhanced Dynamic EMG-Force Estimation Through Calibration and PCI Modeling. IEEE Trans. Neural Syst. Rehabil. Eng. 2015, 23, 41–50. [Google Scholar] [CrossRef]
- Hua, S.Y.; Wang, C.Q.; Wu, X.V. A novel sEMG-based force estimation method using deep-learning algorithm. Complex Intell. Syst. 2021, 8, 1949–1961. [Google Scholar] [CrossRef]
- Xu, L.F.; Chen, X.; Cao, S.; Zhang, X.; Chen, X. Feasibility Study of Advanced Neural Networks Applied to sEMG-Based Force Estimation. Sensors 2018, 18, 3226. [Google Scholar] [CrossRef]
- Choi, C.; Kwon, S.; Park, W.; Lee, H.D.; Kim, J. Real-time pinch force estimation by surface electromyography using an artificial neural network. Med. Eng. Phys. 2010, 32, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Hou, Z.-G.; Peng, L.; Hu, J.; Wang, W. An sEMG-driven musculoskeletal model of shoulder and elbow based on neural networks. In Proceedings of the 7th International Conference on Advanced Computational Intelligence, Wuyi, China, 27–29 March 2015. [Google Scholar]
- Zhang, J.; Zhao, Y.H.; Shone, F.; Li, Z.H.; Frang, A.F. Physics-Informed Deep Learning for Musculoskeletal Modeling: Predicting Muscle Forces and Joint Kinematics From Surface EMG. IEEE Trans. Neural Syst. Rehabil. Eng. 2023, 31, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Chen, X.; Zhang, H.; Zhang, X.; Chen, X. A novel myoelectric control scheme supporting synchronous gesture recognition and muscle force estimation. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Tobias, S.; Sandro, W.; Ingrid, B.; Travieso, C.M. Analyzing gait symmetry with automatically synchronized wearable sensors in daily life. Microprocess. Microsyst. 2020, 77, 103118. [Google Scholar]
- Huang, D.B.; Wu, C.; Wang, Y.W.; Zhang, Z.Y. Episode-level prediction of freezing of gait based on wearable inertial signals using a deep neural network model. Biomed. Signal Process. Control. 2024, 88, 105613. [Google Scholar] [CrossRef]
- Brognara, L.; Sempere-Bigorra, M.; Mazzotti, A.; Artioli, E.; Julián-Rochina, I.; Cauli, O. Wearable sensors-based postural analysis and fall risk assessment among patients with diabetic foot neuropathy. J. Tissue Viability 2023, 32, 516–526. [Google Scholar]
- Lu, Y.; Cao, Y.D.; Chen, Y.; Li, H.; Li, W.H.; Du, H.P. Investigation of a wearable piezoelectric-IMU multi-modal sensing system for real-time muscle force estimation. Smart Mater. Struct. 2023, 32, 065013. [Google Scholar] [CrossRef]
- Dao, T.T. From deep learning to transfer learning for the prediction of skeletal muscle forces. Med. Biol. Eng. Comput. 2019, 57, 1049–1058. [Google Scholar] [CrossRef]
- Suin, K.; Ro, K.K.; Bae, J. Estimation of Individual Muscular Forces of the Lower Limb during Walking Using a Wearable Sensor System. J. Sens. 2017, 2017, 6747921. [Google Scholar]
- Liu, K.; Liu, Y.; Ji, S.; Gao, C.; Zhang, S.Z. A Novel Gait Phase Recognition Method Based on DPF-LSTM-CNN using Wearable inertial sensors. Sensors 2023, 23, 5905. [Google Scholar] [CrossRef]
- Liu, K.; Ji, S.; Liu, Y.; Gao, C.; Zhang, S.Z.; Fu, J.; Dai, L. Analysis of Ankle Muscle Dynamics during the STS Process Based on Wearable Sensors. Sensors 2023, 23, 6607. [Google Scholar] [CrossRef] [PubMed]
- Thelen, D.G.; Anderson, F.C.; Delp, S.L. Generating dynamic simulations of movement using computed muscle control. J. Biomech. 2003, 36, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Qureshi, A.N.; Li, J.Q.; Mahmood, T. On the Analyses of Medical Images Using Traditional Machine Learning Techniques and Convolutional Neural Networks. Arch. Comput. Methods Eng. 2023, 30, 3173–3233. [Google Scholar] [CrossRef] [PubMed]
- Aysa, Z.; Ablimit, M.; Hamdulla, A. Multi-Scale Feature Learning for Language Identification of Overlapped Speech. Appl. Sci. 2023, 13, 4235. [Google Scholar] [CrossRef]
- Alzahrani, N.; Al-Baity, H.H. Object Recognition System for the Visually Impaired: A Deep Learning Approach using Arabic Annotation. Electronics 2023, 12, 541. [Google Scholar] [CrossRef]
- Han, Z.Y.; Li, Z.Y.; Yamanashi, Y.; Yoshikawa, N. Design of Max Pooling Operation Circuit for Binarized Neural Networks Using Single-Flux-Quantum Circuit. IEEE Trans. Appl. Supercond. 2023, 33, 1–5. [Google Scholar] [CrossRef]
- Su, S.S.W.; Kek, S.L. An Improvement of Stochastic Gradient Descent Approach for Mean-Variance Portfolio Optimization Problem. J. Math. 2021, 2021, 8892636. [Google Scholar] [CrossRef]


| Kinematics Parameters | RMSE% | r |
|---|---|---|
| Left ankle joint angle (rad) | 13.03 | 0.969 |
| Left knee joint angle (rad) | 10.36 | 0.981 |
| Left hip joint angle (rad) | 9.51 | 0.979 |
| Right hip joint angle (rad) | 10.84 | 0.982 |
| Right knee joint angle (rad) | 11.06 | 0.969 |
| Right ankle joint angle (rad) | 14.31 | 0.971 |
| Left ankle joint angular velocity (rad/s) | 18.72 | 0.932 |
| Left knee joint angular velocity (rad/s) | 13.87 | 0.924 |
| Left hip joint angular velocity (rad/s) | 19.31 | 0.926 |
| Right hip joint angular velocity (rad/s) | 16.42 | 0.947 |
| Right knee joint angular velocity (rad/s) | 17.14 | 0.953 |
| Right ankle joint angular velocity (rad/s) | 16.77 | 0.921 |
| Gait Speed | Muscle | CNN | LSTM | CNN–LSTM |
|---|---|---|---|---|
| Slow speed | GM | 0.9135 | 0.9608 | 0.9798 |
| RF | 0.8943 | 0.9514 | 0.9749 | |
| GAS | 0.9077 | 0.9681 | 0.9841 | |
| SOL | 0.8986 | 0.9591 | 0.9816 | |
| Average | 0.9035 | 0.9599 | 0.9801 | |
| Normal speed | GM | 0.9388 | 0.9768 | 0.9838 |
| RF | 0.9186 | 0.9336 | 0.9814 | |
| GAS | 0.9399 | 0.9813 | 0.9809 | |
| SOL | 0.9434 | 0.9704 | 0.9776 | |
| Average | 0.9352 | 0.9655 | 0.9829 | |
| Fast speed | GM | 0.9282 | 0.9665 | 0.9765 |
| RF | 0.8855 | 0.9488 | 0.9762 | |
| GAS | 0.9055 | 0.9720 | 0.9877 | |
| SOL | 0.9199 | 0.9555 | 0.9832 | |
| Average | 0.9098 | 0.9607 | 0.9809 |
| Muscle | CNN | LSTM | CNN–LSTM | |
|---|---|---|---|---|
| Pearson’s correlation coefficient (r) | GM | 0.9174 | 0.9364 | 0.9776 |
| RF | 0.9017 | 0.9488 | 0.9798 | |
| GAS | 0.9217 | 0.9674 | 0.9848 | |
| SOL | 0.9329 | 0.9604 | 0.9808 | |
| Average | 0.9184 | 0.9532 | 0.9807 | |
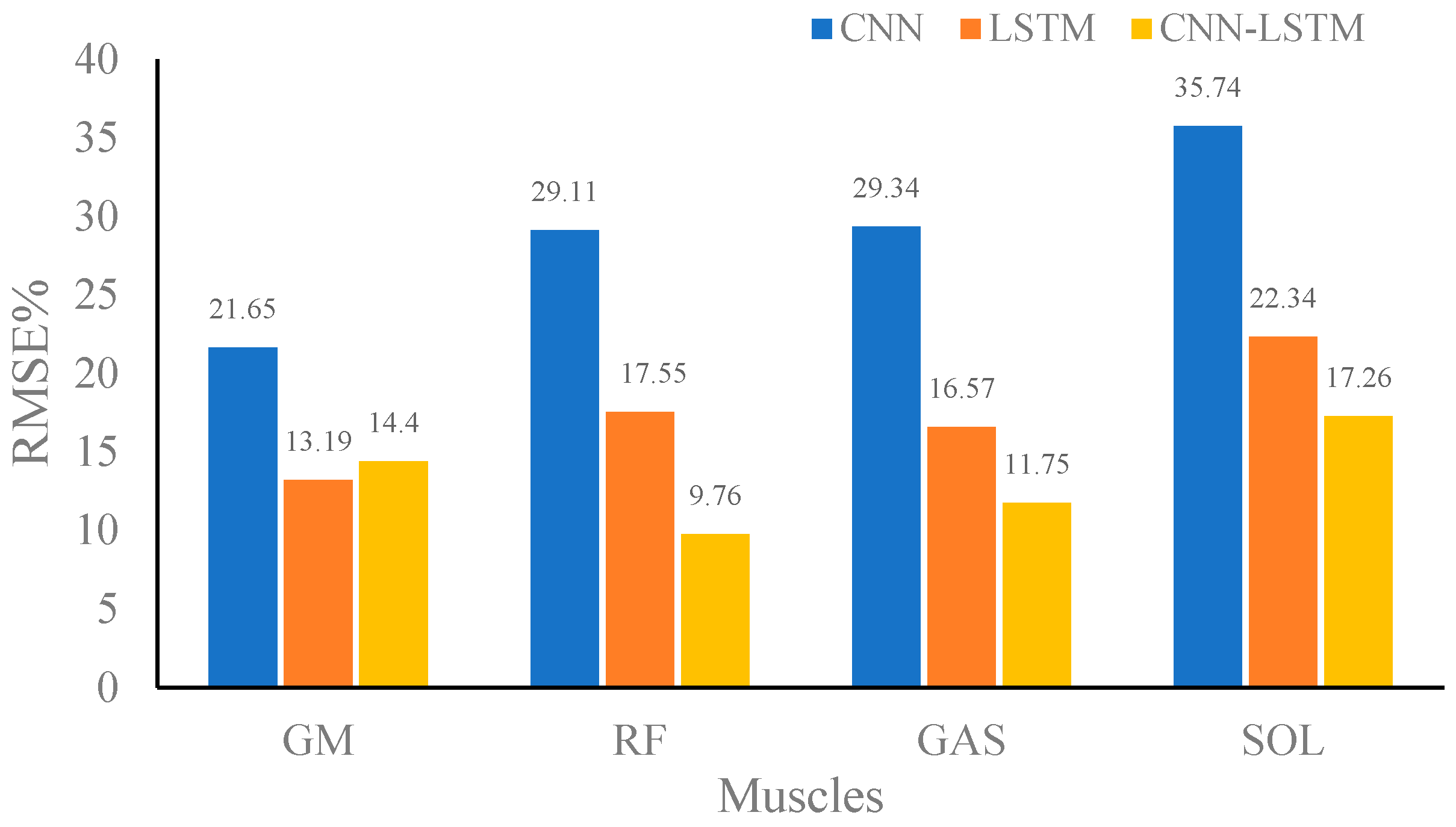
| Percentage root mean square error (RMSE%) | GM | 27.37 | 20.19 | 12.54 |
| RF | 32.64 | 14.83 | 15.01 | |
| GAS | 29.87 | 22.31 | 14.23 | |
| SOL | 30.69 | 21.22 | 18.78 | |
| Average | 30.15 | 19.64 | 15.14 |
| Gait Speed | GM | RF | GAS | SOL | |
|---|---|---|---|---|---|
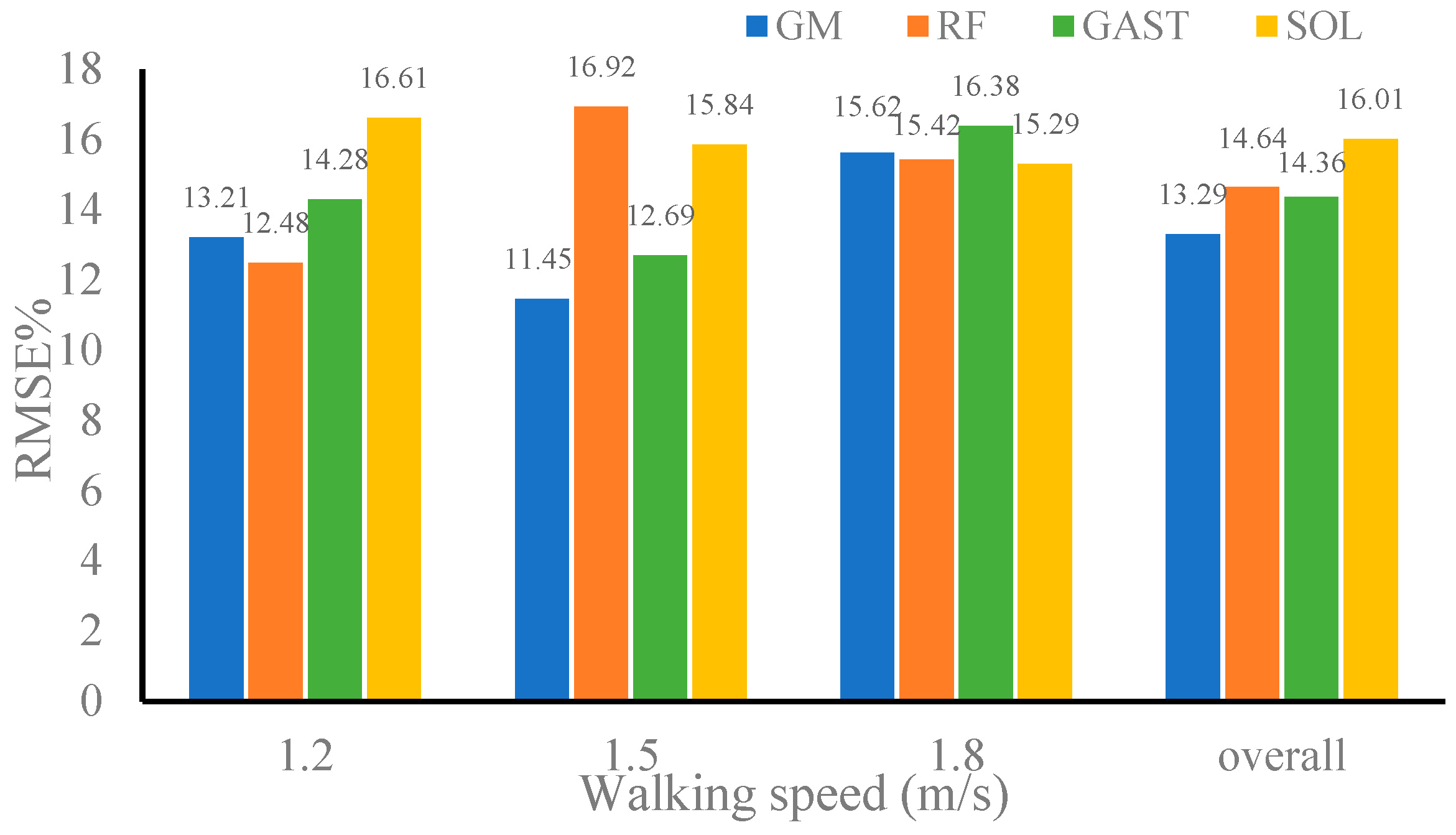
| Dataset 1 | 1.2 m/s | 0.9765 | 0.9823 | 0.9689 | 0.9758 |
| 1.5 m/s | 0.9828 | 0.9874 | 0.9808 | 0.9783 | |
| 1.8 m/s | 0.9804 | 0.9768 | 0.9728 | 0.9806 | |
| Overall | 0.9793 | 0.9846 | 0.9719 | 0.9802 | |
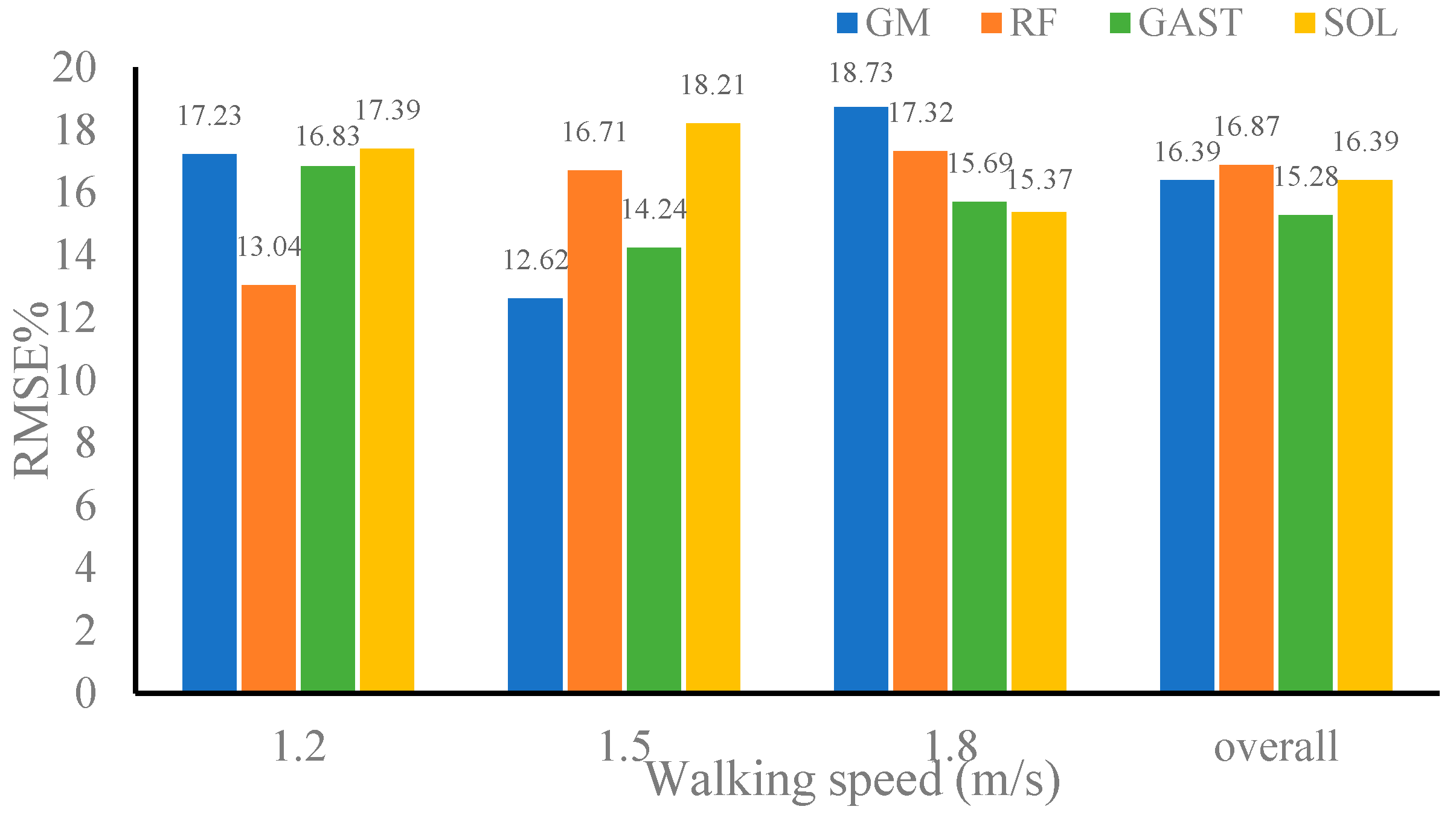
| Dataset 2 | 1.1 m/s | 0.9737 | 0.9721 | 0.9763 | 0.9803 |
| 1.6 m/s | 0.9698 | 0.9687 | 0.9742 | 0.9763 | |
| Overall | 0.9728 | 0.9669 | 0.9738 | 0.9733 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Liu, Y.; Ji, S.; Gao, C.; Fu, J. Estimation of Muscle Forces of Lower Limbs Based on CNN–LSTM Neural Network and Wearable Sensor System. Sensors 2024, 24, 1032. https://doi.org/10.3390/s24031032
Liu K, Liu Y, Ji S, Gao C, Fu J. Estimation of Muscle Forces of Lower Limbs Based on CNN–LSTM Neural Network and Wearable Sensor System. Sensors. 2024; 24(3):1032. https://doi.org/10.3390/s24031032
Chicago/Turabian StyleLiu, Kun, Yong Liu, Shuo Ji, Chi Gao, and Jun Fu. 2024. "Estimation of Muscle Forces of Lower Limbs Based on CNN–LSTM Neural Network and Wearable Sensor System" Sensors 24, no. 3: 1032. https://doi.org/10.3390/s24031032
APA StyleLiu, K., Liu, Y., Ji, S., Gao, C., & Fu, J. (2024). Estimation of Muscle Forces of Lower Limbs Based on CNN–LSTM Neural Network and Wearable Sensor System. Sensors, 24(3), 1032. https://doi.org/10.3390/s24031032






