Explainable Machine Learning Techniques to Predict Muscle Injuries in Professional Soccer Players through Biomechanical Analysis
Abstract
:1. Introduction
2. State of the Art
3. Soccer Player Injury Classification Architecture
3.1. Dataset for Biomechanical Tests
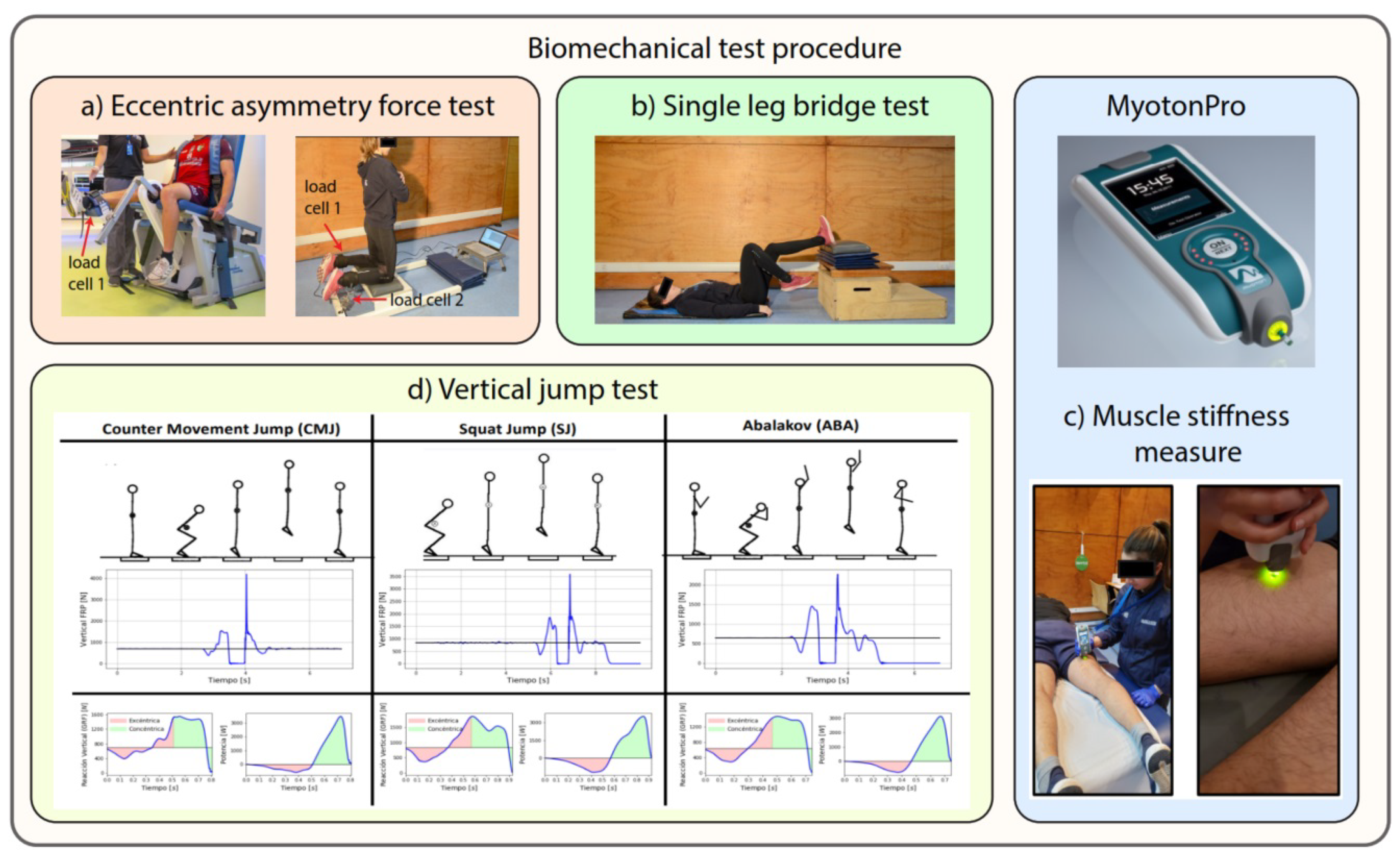
Biomechanical Tests
- Eccentric asymmetry force test (Nordic Hamstring): The participants assume a kneeling position with aligned hips and trunk support (see Figure 2a). An assistant or, in this case, load cells, is responsible for securing the heels, ensuring continual contact with the ground during the exercise. Load cells are utilized to measure the eccentric activation of the hamstring muscles. This test yields two parameters: the maximum right hamstring eccentric force (N) and the maximum left hamstring eccentric force (N).
- Eccentric force test (Maximum Force Quadriceps): The participants use a quadriceps chair (Figure 2a) connected to load cells to measure the eccentric activation of the quadriceps muscles. This test delivers the parameters of the right quadriceps muscle’s maximal eccentric force (N) and the left quadriceps muscle’s maximal force (N).
- Single-leg bridge test: This clinical test assesses susceptibility to hamstring injury. The participant is instructed to lie on the floor supine, with the heel of the designated leg placed inside a 60 cm high box. With hands crossed over the chest, the subject must push with the heel to elevate the glutes off the ground. Each repetition requires the participant to touch the ground before raising the glutes again without resting (see Figure 2b). This test yields the number of repetitions for the right leg and the number of repetitions for the left leg.
- Muscle stiffness measure (Myotonometry): This technique involves an objective and non-invasive digital palpation method for superficial skeletal muscles. The measurement explicitly targets the hamstring muscles (see Figure 2c) and is conducted using a MyotonPRO device. The parameter obtained for both extremities includes the S–stiffness (N/m), which reflects the resistance to force or contraction that induces structural or tissue deformation.
- Vertical jump test (Bosco test): This series of vertical jumps serves to evaluate various aspects, including morphophysiological characteristics (muscle fiber type), functional attributes (height and mechanical jump power), and neuromuscular features (utilization of elastic energy and myotatic reflex, fatigue resistance) of the lower limb extensor muscles based on the attained jump height and mechanical power in different types of vertical jumps. The Bosco test employs three jumps on a force platform. The execution of these jumps can be observed in Figure 2d, encompassing data from the countermovement jump (both two-legged and one-legged), squat jump (both two-legged and one-legged), and Abalakov (both bipodal and unipodal) jump.
3.2. Pre-Processing
3.3. Classification
3.4. Most Important Features
3.5. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baroni, B.M.; Ruas, C.V.; Ribeiro-Alvares, J.B.; Pinto, R.S. Hamstring-to-quadriceps torque ratios of professional male soccer players: A systematic review. J. Strength Cond. Res. 2020, 34, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Nho, K.; Kang, B.; Sohn, K.A.; Kim, D. Predicting Alzheimer’s disease progression using multi-modal deep learning approach. Sci. Rep. 2019, 9, 1952. [Google Scholar] [CrossRef] [PubMed]
- Cumps, E.; Verhagen, E.; Annemans, L.; Meeusen, R. Injury rate and socioeconomic costs resulting from sports injuries in Flanders: Data derived from sports insurance statistics 2003. Br. J. Sport. Med. 2008, 42, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Díaz, M.; Ulloa-Jiménez, R.; Saavedra, C.; Salas, R. Wavelet-based semblance analysis to determine muscle synergy for different handstand postures of Chilean circus athletes. Comput. Methods Biomech. Biomed. Eng. 2021, 24, 1053–1063. [Google Scholar] [CrossRef]
- Rosado-Portillo, A.; Chamorro-Moriana, G.; Gonzalez-Medina, G.; Perez-Cabezas, V. Acute hamstring injury prevention programs in eleven-a-side football players based on physical exercises: Systematic review. J. Clin. Med. 2021, 10, 2029. [Google Scholar] [CrossRef]
- Grazioli, R.; Lopez, P.; Andersen, L.L.; Machado, C.L.F.; Pinto, M.D.; Cadore, E.L.; Pinto, R.S. Hamstring rate of torque development is more affected than maximal voluntary contraction after a professional soccer match. Eur. J. Sport Sci. 2019, 19, 1336–1341. [Google Scholar] [CrossRef]
- Crema, M.D.; Guermazi, A.; Tol, J.L.; Niu, J.; Hamilton, B.; Roemer, F.W. Acute hamstring injury in football players: Association between anatomical location and extent of injury—a large single-center MRI report. J. Sci. Med. Sport 2016, 19, 317–322. [Google Scholar] [CrossRef]
- Ekstrand, J.; Hägglund, M.; Kristenson, K.; Magnusson, H.; Waldén, M. Fewer ligament injuries but no preventive effect on muscle injuries and severe injuries: An 11-year follow-up of the UEFA Champions League injury study. Br. J. Sport. Med. 2013, 47, 732–737. [Google Scholar] [CrossRef]
- Szymski, D.; Krutsch, V.; Achenbach, L.; Gerling, S.; Pfeifer, C.; Alt, V.; Krutsch, W.; Loose, O. Epidemiological analysis of injury occurrence and current prevention strategies on international amateur football level during the UEFA Regions Cup 2019. Arch. Orthop. Trauma Surg. 2022, 142, 271–280. [Google Scholar] [CrossRef]
- Biz, C.; Nicoletti, P.; Baldin, G.; Bragazzi, N.L.; Crimì, A.; Ruggieri, P. Hamstring strain injury (HSI) prevention in professional and semi-professional football teams: A systematic review and meta-analysis. Int. J. Environ. Res. Public Health 2021, 18, 8272. [Google Scholar] [CrossRef]
- Musahl, V.; Karlsson, J.; Krutsch, W.; Mandelbaum, B.R.; Espregueira-Mendes, J.; d’Hooghe, P. Return to Play in Football: An Evidence-Based Approach; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Cos, F.; Cos, M.Á.; Buenaventura, L.; Pruna, R.; Ekstrand, J. Modelos de análisis para la prevención de lesiones en el deporte. Estudio epidemiológico de lesiones: El modelo Union of European Football Associations en el fútbol. Apunts. Med. De L’Esport 2010, 45, 95–102. [Google Scholar] [CrossRef]
- Halilaj, E.; Rajagopal, A.; Fiterau, M.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Machine learning in human movement biomechanics: Best practices, common pitfalls, and new opportunities. J. Biomech. 2018, 81, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Handelman, G.; Kok, H.; Chandra, R.; Razavi, A.; Lee, M.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Tseng, P.Y.; Chen, Y.T.; Wang, C.H.; Chiu, K.M.; Peng, Y.S.; Hsu, S.P.; Chen, K.L.; Yang, C.Y.; Lee, O.K.S. Prediction of the development of acute kidney injury following cardiac surgery by machine learning. Crit. Care 2020, 24, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.L.; Zhang, S. From fighting COVID-19 pandemic to tackling sustainable development goals: An opportunity for responsible information systems research. Int. J. Inf. Manag. 2020, 55, 102196. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, P.N.; Karnuta, J.M.; Haeberle, H.S.; Owusu-Akyaw, K.A.; Warner, T.S.; Rodeo, S.A.; Nwachukwu, B.U.; Williams III, R.J. Association between preoperative mental health and clinically meaningful outcomes after osteochondral allograft for cartilage defects of the knee: A machine learning analysis. Am. J. Sport. Med. 2021, 49, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Jamaludin, A.; Lootus, M.; Kadir, T.; Zisserman, A.; Urban, J.; Battié, M.C.; Fairbank, J.; McCall, I. Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur. Spine J. 2017, 26, 1374–1383. [Google Scholar] [CrossRef]
- Mak, W.K.; Bin Abd Razak, H.R.; Tan, H.C.A. Which Patients Require a Contralateral Total Knee Arthroplasty Within 5 Years of Index Surgery? J. Knee Surg. 2019, 33, 1029–1033. [Google Scholar] [CrossRef]
- Masís, S. Interpretable Machine Learning with Python: Learn to Build Interpretable High-Performance Models with Hands-On Real-World Examples; Packt Publishing Ltd.: Birmingham, UK, 2021. [Google Scholar]
- Chen, H.; Michalopoulos, G.; Subendran, S.; Yang, R.; Quinn, R.; Oliver, M.; Butt, Z.; Wong, A. Interpretability of ml models for health data-a case study. In Proceedings of the First Annual International Workshop on Interpretability: Methodologies and Algorithms—IMA 2019, Adelaide, Australia, 2 December 2019. [Google Scholar]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable ai: A review of machine learning interpretability methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef]
- Brownlee, J. XGBoost With Python: Gradient Boosted Trees with XGBoost and Scikit-Learn; Machine Learning Mastery: Victoria, AU, USA, 2016. [Google Scholar]
- Van Beijsterveldt, A.; van de Port, I.G.; Vereijken, A.; Backx, F. Risk factors for hamstring injuries in male soccer players: A systematic review of prospective studies. Scand. J. Med. Sci. Sport. 2013, 23, 253–262. [Google Scholar] [CrossRef]
- Hägglund, M.; Waldén, M.; Ekstrand, J. Previous injury as a risk factor for injury in elite football: A prospective study over two consecutive seasons. Br. J. Sport. Med. 2006, 40, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Arnason, A.; Sigurdsson, S.B.; Gudmundsson, A.; Holme, I.; Engebretsen, L.; Bahr, R. Risk factors for injuries in football. Am. J. Sport. Med. 2004, 32, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Namazi, P.; Zarei, M.; Hovanloo, F.; Abbasi, H. The association between the isokinetic muscle strength and lower extremity injuries in young male football players. Phys. Ther. Sport 2019, 39, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Mok, K.M.; Chan, H.C.; Yung, P.S.; Chan, K.M. Eccentric hamstring strength deficit and poor hamstring-to-quadriceps ratio are risk factors for hamstring strain injury in football: A prospective study of 146 professional players. J. Sci. Med. Sport 2018, 21, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Green, B.; Bourne, M.N.; van Dyk, N.; Pizzari, T. Recalibrating the risk of hamstring strain injury (HSI): A 2020 systematic review and meta-analysis of risk factors for index and recurrent hamstring strain injury in sport. Br. J. Sport. Med. 2020, 54, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Freckleton, G.; Pizzari, T. Risk factors for hamstring muscle strain injury in sport: A systematic review and meta-analysis. Br. J. Sport. Med. 2013, 47, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Fousekis, K.; Tsepis, E.; Poulmedis, P.; Athanasopoulos, S.; Vagenas, G. Intrinsic risk factors of non-contact quadriceps and hamstring strains in soccer: A prospective study of 100 professional players. Br. J. Sport. Med. 2011, 45, 709–714. [Google Scholar] [CrossRef]
- Henderson, G.; Barnes, C.A.; Portas, M.D. Factors associated with increased propensity for hamstring injury in English Premier League soccer players. J. Sci. Med. Sport 2010, 13, 397–402. [Google Scholar] [CrossRef]
- Fyfe, J.J.; Opar, D.A.; Williams, M.D.; Shield, A.J. The role of neuromuscular inhibition in hamstring strain injury recurrence. J. Electromyogr. Kinesiol. 2013, 23, 523–530. [Google Scholar] [CrossRef]
- Warren, P.; Gabbe, B.J.; Schneider-Kolsky, M.; Bennell, K.L. Clinical predictors of time to return to competition and of recurrence following hamstring strain in elite Australian footballers. Br. J. Sport. Med. 2010, 44, 415–419. [Google Scholar] [CrossRef]
- Blackburn, J.T.; Norcross, M.F. The effects of isometric and isotonic training on hamstring stiffness and anterior cruciate ligament loading mechanisms. J. Electromyogr. Kinesiol. 2014, 24, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Watsford, M.L.; Murphy, A.J.; McLachlan, K.A.; Bryant, A.L.; Cameron, M.L.; Crossley, K.M.; Makdissi, M. A prospective study of the relationship between lower body stiffness and hamstring injury in professional Australian rules footballers. Am. J. Sport. Med. 2010, 38, 2058–2064. [Google Scholar] [CrossRef] [PubMed]
- Amaral, J.L.; Sancho, A.G.; Faria, A.C.; Lopes, A.J.; Melo, P.L. Differential diagnosis of asthma and restrictive respiratory diseases by combining forced oscillation measurements, machine learning and neuro-fuzzy classifiers. Med. Biol. Eng. Comput. 2020, 58, 2455–2473. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Gu, F.; Wang, X.; Ma, S.; Wang, L. Interpretable Recognition for Dementia Using Brain Images. Front. Neurosci. 2021, 15, 748689. [Google Scholar] [CrossRef] [PubMed]
- Sabol, P.; Sinčák, P.; Hartono, P.; Kočan, P.; Benetinová, Z.; Blichárová, A.; Verbóová, L.; Štammová, E.; Sabolová-Fabianová, A.; Jašková, A. Explainable classifier for improving the accountability in decision-making for colorectal cancer diagnosis from histopathological images. J. Biomed. Inform. 2020, 109, 103523. [Google Scholar] [CrossRef]
- García-Pérez, P.; Lozano-Milo, E.; Landin, M.; Gallego, P.P. Machine Learning unmasked nutritional imbalances on the medicinal plant Bryophyllum sp. cultured in vitro. Front. Plant Sci. 2020, 11, 576177. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Groumpos, P.P.; Apostolopoulos, D.J. Advanced fuzzy cognitive maps: State-space and rule-based methodology for coronary artery disease detection. Biomed. Phys. Eng. Express 2021, 7, 045007. [Google Scholar] [CrossRef]
- Juang, C.F.; Wen, C.Y.; Chang, K.M.; Chen, Y.H.; Wu, M.F.; Huang, W.C. Explainable fuzzy neural network with easy-to-obtain physiological features for screening obstructive sleep apnea-hypopnea syndrome. Sleep Med. 2021, 85, 280–290. [Google Scholar] [CrossRef]
- Ding, L.; Zhang, X.y.; Wu, D.y.; Liu, M.l. Application of an extreme learning machine network with particle swarm optimization in syndrome classification of primary liver cancer. J. Integr. Med. 2021, 19, 395–407. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Alonso, J.M.; Islam, S.R.; Sultan, A.M.; Kwak, K.S. A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 2021, 11, 2660. [Google Scholar] [CrossRef]
- Kokkotis, C.; Ntakolia, C.; Moustakidis, S.; Giakas, G.; Tsaopoulos, D. Explainable machine learning for knee osteoarthritis diagnosis based on a novel fuzzy feature selection methodology. Phys. Eng. Sci. Med. 2022, 45, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Ntakolia, C.; Kokkotis, C.; Moustakidis, S.; Tsaopoulos, D. Identification of most important features based on a fuzzy ensemble technique: Evaluation on joint space narrowing progression in knee osteoarthritis patients. Int. J. Med. Inform. 2021, 156, 104614. [Google Scholar] [CrossRef] [PubMed]
- Burkart, N.; Huber, M.F. A survey on the explainability of supervised machine learning. J. Artif. Intell. Res. 2021, 70, 245–317. [Google Scholar] [CrossRef]
- Bucholc, M.; Ding, X.; Wang, H.; Glass, D.H.; Wang, H.; Prasad, G.; Maguire, L.P.; Bjourson, A.J.; McClean, P.L.; Todd, S.; et al. A practical computerized decision support system for predicting the severity of Alzheimer’s disease of an individual. Expert Syst. Appl. 2019, 130, 157–171. [Google Scholar] [CrossRef]
- Das, D.; Ito, J.; Kadowaki, T.; Tsuda, K. An interpretable machine learning model for diagnosis of Alzheimer’s disease. PeerJ 2019, 7, e6543. [Google Scholar] [CrossRef]
- Khan, I.U.; Aslam, N.; AlShedayed, R.; AlFrayan, D.; AlEssa, R.; AlShuail, N.A.; Al Safwan, A. A proactive attack detection for heating, ventilation, and air conditioning (HVAC) system using explainable extreme gradient boosting model (XGBoost). Sensors 2022, 22, 9235. [Google Scholar] [CrossRef]
- Calderón-Díaz, M.; Serey-Castillo, L.J.; Vallejos-Cuevas, E.A.; Espinoza, A.; Salas, R.; Macías-Jiménez, M.A. Detection of variables for the diagnosis of overweight and obesity in young Chileans using machine learning techniques. Procedia Comput. Sci. 2023, 220, 978–983. [Google Scholar] [CrossRef]

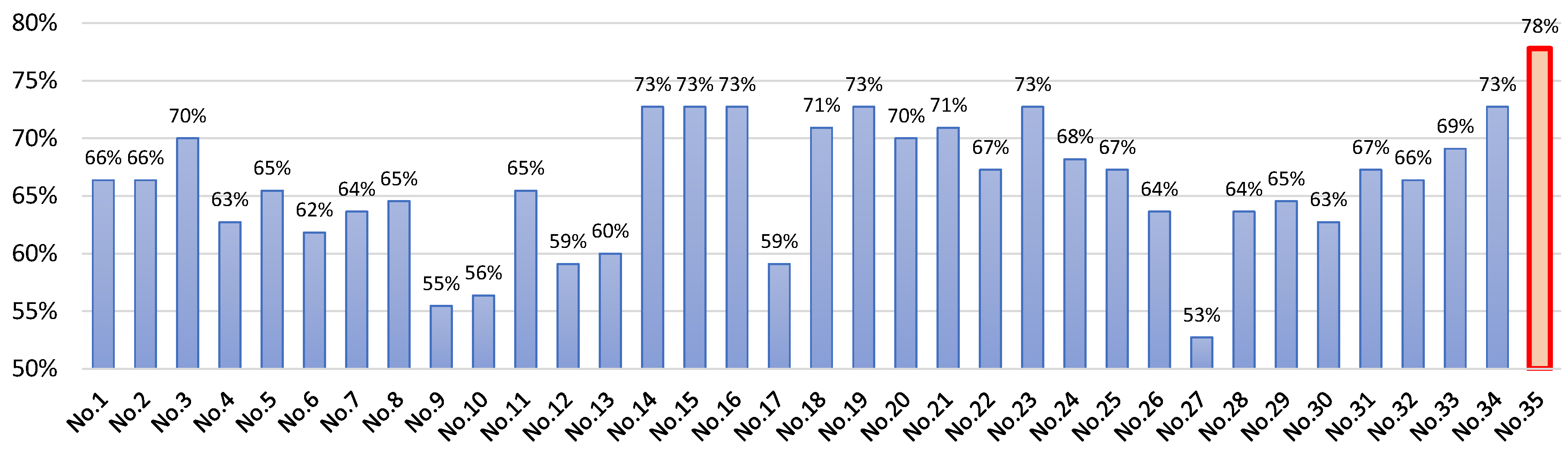
| No. | Model Name | Model Configuration | Model Description |
|---|---|---|---|
| No. 1 | Tree | 100 splits | |
| No. 2 | Tree | 20 splits | A flowchart-like structure where an internal node represents a feature, a branch represents a decision rule, and each leaf node represents an outcome. |
| No. 3 | Tree | 4 splits | |
| No. 4 | Linear discriminant | Full covariance structure | A statistical technique for binary and multiclass classification, which finds the linear combination of features that best separates classes. |
| No. 5 | Quadratic discriminant | Full covariance structure | A method similar to linear discriminant analysis, but it assumes that the features follow a Gaussian distribution and estimates the covariance between the classes. |
| No. 6 | Binary GLM Logistic Regression | Binomial distribution | Logistic regression with binary outcomes for estimating the probability of a binary outcome using a logistic function. |
| No. 7 | Efficient Logistic Regression | L2 regularization, alpha = 0.001, one-vs-one coding | A regression analysis similar to binary logistic regression but implemented efficiently to handle large datasets or high-dimensional data. |
| No. 8 | Efficient Linear SVM | L2 regularization, alpha = 0.001, one-vs-one coding | A supervised machine learning algorithm used for classification and regression analysis, which finds a hyperplane that best separates classes. |
| No. 9 | Gaussian Naive Bayes | Gaussian distribution | A probabilistic classifier, which assumes that the presence of a particular feature in a class is unrelated to the presence of other features. |
| No. 10 | Kernel Naive Bayes | Normal kernel, data standardization | A version of the naive Bayes classifier that can handle non-linear classification using kernel methods, transforming data into higher dimensions. |
| No. 11 | Linear SVM | Linear kernel, one-vs-one coding, data standardization | A supervised machine learning algorithm used for classification, which finds a hyperplane that best separates classes in a linearly separable dataset. |
| No. 12 | Quadratic SVM | Quadratic kernel, one-vs-one coding, data standardization | An extension of the SVM algorithm that uses a quadratic kernel to handle non-linearly separable data by mapping it into a higher-dimensional space. |
| No. 13 | Cubic SVM | Cubic kernel, one-vs-one coding, data standardization | An extension of the SVM algorithm that uses a cubic kernel to handle highly non-linearly separable data by mapping it into an even higher-dimensional space. |
| No. 14 | Fine Gaussian SVM | Kernel scale = 1.6, one-vs-one coding, data standardization | An SVM with a fine Gaussian kernel, suitable for datasets requiring high precision and accuracy. |
| No. 15 | Medium Gaussian SVM | kernel scale = 6.5, one-vs-one coding, data standardization | An SVM with a medium Gaussian kernel, suitable for datasets with moderate complexity and dimensionality. |
| No. 16 | Coarse Gaussian SVM | Kernel scale = 26, one-vs-one coding, data standardization | An SVM with a coarse Gaussian kernel, suitable for datasets with lower complexity and dimensionality. |
| No. | Model Name | Model Configuration | Model Description |
|---|---|---|---|
| No. 17 | Fine KNN | Number of neighbors = 1, Euclidean distance | A non-parametric classification algorithm, that classifies a data point based on the majority vote of its neighbors with a fine-tuned distance metric. |
| No. 18 | Medium KNN | Number of neighbors = 10, Euclidean distance | A non-parametric classification algorithm that classifies a data point based on the majority vote of its neighbors with a moderately adjusted distance metric. |
| No. 19 | Coarse KNN | Number of neighbors = 100, Euclidean distance | A non-parametric classification algorithm that classifies a data point based on the majority vote of its neighbors with a roughly adjusted distance metric. |
| No. 20 | Cosine KNN | Number of neighbors = 10, Euclidean distance | A variation of the K-nearest neighbor algorithm that computes the cosine similarity between data points to measure their similarity. |
| No. 21 | Cubic KNN | Number of neighbors = 10, Euclidean distance | A non-parametric classification algorithm that classifies a data point based on the majority vote of its neighbors with a cubic distance metric. |
| No. 22 | Weighted KNN | Number of neighbors = 10, Euclidean distance | A variant of the K-nearest neighbor algorithm that assigns weights to the contributions of the neighbors based on their distances. |
| No. 23 | Boosted Trees with AdaBoost ensemble | Decision tree learner, maximum splits = 20, learning rate=0.1 | An ensemble learning method that constructs a strong classifier by combining multiple weak classifiers such as decision trees using the AdaBoost algorithm. |
| No. 24 | Bagged trees with bag ensemble | Decision tree learner, maximum splits = 109, number of learners = 30 | An ensemble learning technique that combines multiple models such as decision trees to improve classification accuracy and stability. |
| No. 25 | Subspace discriminant ensemble | Discriminant learner, number of learners = 30, subspace dimension = 10 | An ensemble approach that combines multiple discriminant analysis models to improve the classification performance of the system. |
| No. 26 | Subspace KNN ensemble | Subspace ensemble method, decision tree learner, number of learners = 30, learning rate = 0.1 | An ensemble learning technique that combines multiple K-nearest neighbor models operating in different subspaces to improve classification accuracy. |
| No. 27 | RUSBoosted Trees | RUSBoost ensemble method, decision tree learner, number of learners = 30, learning rate = 0.1 | It is a variant of the AdaBoost algorithm that incorporates random under-sampling to address class imbalance, particularly in binary classification problems. |
| No. 28 | Neural Network | 1 layer, 10 neurons, 1k iterations | A network of interconnected nodes inspired by the structure of the human brain, capable of learning complex patterns and relationships within data. |
| No. 29 | Neural Network | 1 layer, 25 neurons, 1k iterations | |
| No. 30 | Neural Network | 1 layer, 100 neurons, 1k iterations | |
| No. 31 | Neural Network | 2 layers, 10 neuron, 1k iterations | |
| No. 32 | Neural Network | 3 layers, 10 neurons, 1k iterations | |
| No. 33 | SVM Kernel | SVM learner, lambda regularization = 0.01, one-vs-one coding, iteration limit = 1000 | A variant of the SVM algorithm that uses kernel methods to handle non-linear data by transforming it into a higher-dimensional space. |
| No. 34 | Logistic regression kernel | Logistic regression learner, lambda regularization = 0.01, one-vs-one coding, | A variant of logistic regression that uses kernel methods to handle non-linear data. |
| No. 35 | XGBoost | learning rate = 0.3, L2 regularization alpha = 0.001, sampling method = uniform | An optimized gradient-boosting library designed for speed and performance, effective for classification and regression. |
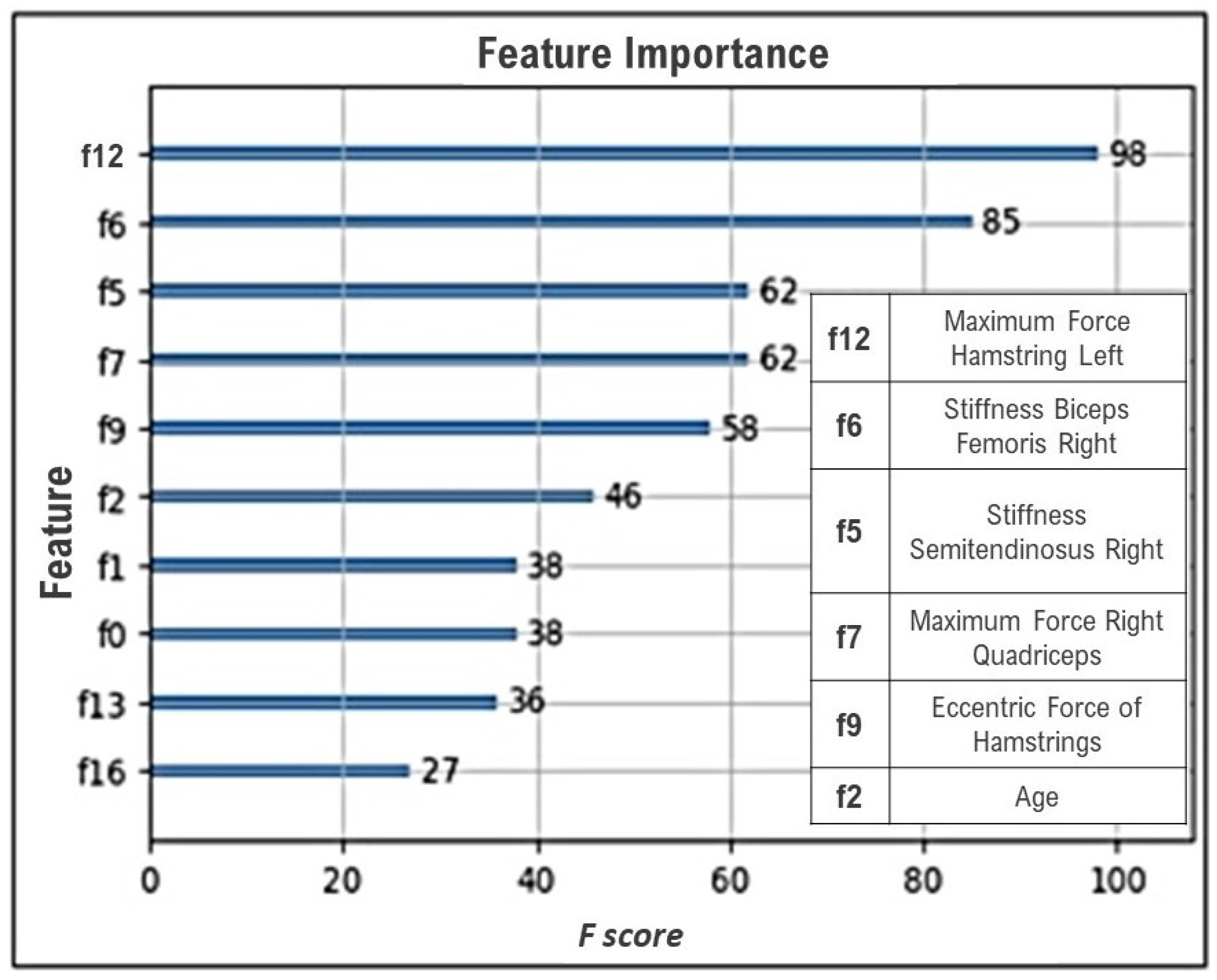
| Feature | Number of Repetitions |
|---|---|
| Maximum Force Hamstring Left | 28 |
| Stiffness Biceps Femoris Right | 28 |
| Stiffness Semitendinosus Right | 24 |
| Maximum Force Right Quadriceps | 21 |
| Eccentric Force of Hamstrings | 17 |
| Age | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calderón-Díaz, M.; Silvestre Aguirre, R.; Vásconez, J.P.; Yáñez, R.; Roby, M.; Querales, M.; Salas, R. Explainable Machine Learning Techniques to Predict Muscle Injuries in Professional Soccer Players through Biomechanical Analysis. Sensors 2024, 24, 119. https://doi.org/10.3390/s24010119
Calderón-Díaz M, Silvestre Aguirre R, Vásconez JP, Yáñez R, Roby M, Querales M, Salas R. Explainable Machine Learning Techniques to Predict Muscle Injuries in Professional Soccer Players through Biomechanical Analysis. Sensors. 2024; 24(1):119. https://doi.org/10.3390/s24010119
Chicago/Turabian StyleCalderón-Díaz, Mailyn, Rony Silvestre Aguirre, Juan P. Vásconez, Roberto Yáñez, Matías Roby, Marvin Querales, and Rodrigo Salas. 2024. "Explainable Machine Learning Techniques to Predict Muscle Injuries in Professional Soccer Players through Biomechanical Analysis" Sensors 24, no. 1: 119. https://doi.org/10.3390/s24010119
APA StyleCalderón-Díaz, M., Silvestre Aguirre, R., Vásconez, J. P., Yáñez, R., Roby, M., Querales, M., & Salas, R. (2024). Explainable Machine Learning Techniques to Predict Muscle Injuries in Professional Soccer Players through Biomechanical Analysis. Sensors, 24(1), 119. https://doi.org/10.3390/s24010119









