Machine Learning Model Validated to Predict Outcomes of Liver Transplantation Recipients with Hepatitis C: The Romanian National Transplant Agency Cohort Experience
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Study Design: Definitions, Transplantation Techniques, and Follow-Up
2.3. Data Collection
2.4. Machine Learning Approach: Model Training
3. Results
3.1. Patients’ Characteristics
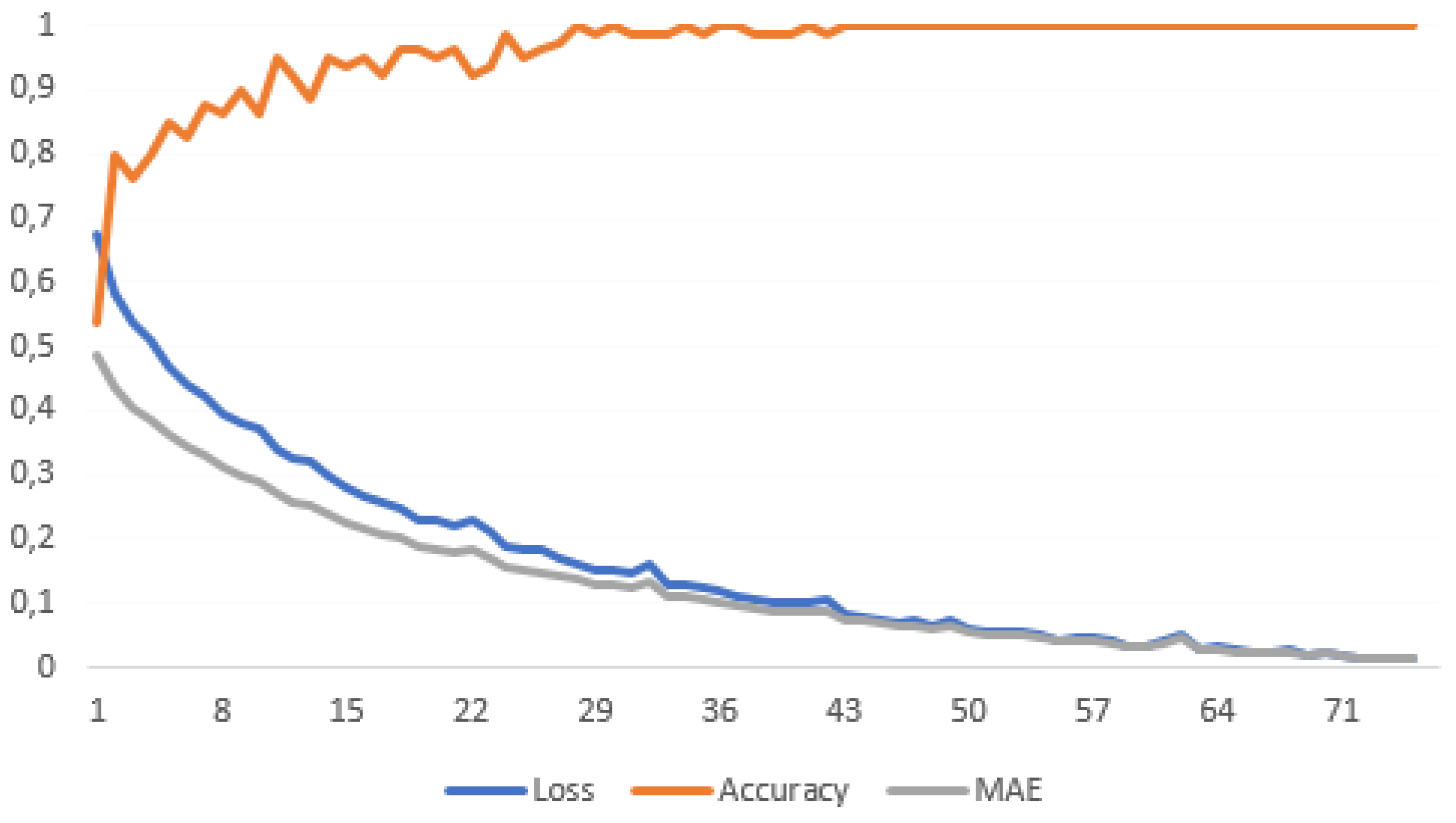
3.2. Training and Validation of the ML Model
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schweda, M.; Schicktanz, S. The “spare parts person”? Conceptions of the human body and their implications for public attitudes towards organ donation and organ sale. Philos. Ethic-Humanit. Med. 2009, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Quintin, J. Organ transplantation and meaning of life: The quest for self fulfilment. Med. Health Care Philos. 2013, 16, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Nordham, K.D.; Ninokawa, S. The history of organ transplantation. Bayl. Univ. Med Cent. Proc. 2022, 35, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Starzl, T.E.; Demetris, A.J. Liver transplantation: A 31-year perspective part I. Curr. Probl. Surg. 1990, 27, 55–116. [Google Scholar] [CrossRef] [PubMed]
- Tajana Filipec, K. Liver Transplantation in Patients with Hepatitis C. In Update on Hepatitis C.; Martina, S., Aleksandar, V., George, Y.W., Eds.; IntechOpen: Rijeka, Croatia, 2017; Chapter 3. [Google Scholar]
- Belga, S. Hepatitis C in non-hepatic solid organ transplant candidates and recipients: A new horizon. World J. Gastroenterol. 2016, 22, 1650–1663. [Google Scholar] [CrossRef]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Planas, R.; Ballesté, B.; Álvarez, M.A.; Rivera, M.; Montoliu, S.; Galeras, J.A.; Santos, J.; Coll, S.; Morillas, R.M.; Solà, R. Natural history of decompensated hepatitis C virus-related cirrhosis. A study of 200 patients. J. Hepatol. 2004, 40, 823–830. [Google Scholar] [CrossRef]
- Lange, C.M.; Zeuzem, S. Perspectives and challenges of interferon-free therapy for chronic hepatitis C. J. Hepatol. 2013, 58, 583–592. [Google Scholar] [CrossRef]
- Forns, X.; Garcı́a-Retortillo, M.; Serrano, T.; Feliu, A.; Suarez, F.; de la Mata, M.; Garcı́a-Valdecasas, J.C.; Navasa, M.; Rimola, A.; Rodés, J. Antiviral therapy of patients with decompensated cirrhosis to prevent recurrence of hepatitis C after liver transplantation. J. Hepatol. 2003, 39, 389–396. [Google Scholar] [CrossRef]
- Kim, W.R.; Terrault, N.A.; Pedersen, R.A.; Therneau, T.M.; Edwards, E.; Hindman, A.A.; Brosgart, C.L. Trends in Waiting List Registration for Liver Transplantation for Viral Hepatitis in the United States. Gastroenterology 2009, 137, 1680–1686. [Google Scholar] [CrossRef]
- Lin, M.V.; Chung, R. Recent FDA approval of sofosbuvir and simeprevir. Implications for current HCV treatment. Clin. Liver Dis. 2014, 3, 65–68. [Google Scholar] [CrossRef]
- Lawitz, E.; Sulkowski, M.S.; Ghalib, R.; Rodriguez-Torres, M.; Younossi, Z.M.; Corregidor, A.; DeJesus, E.; Pearlman, B.; Rabinovitz, M.; Gitlin, N.; et al. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: The COSMOS randomised study. Lancet 2014, 384, 1756–1765. [Google Scholar] [CrossRef]
- Lawitz, E.; Poordad, F.F.; Pang, P.S.; Hyland, R.H.; Ding, X.; Mo, H.; Symonds, W.T.; McHutchison, J.G.; Membreno, E.F. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): An open-label, randomised, phase 2 trial. Lancet 2014, 383, 515–523. [Google Scholar] [CrossRef]
- Everson, G.T.; Sims, K.D.; Rodriguez-Torres, M.; Hézode, C.; Lawitz, E.; Bourlière, M.; Loustaud-Ratti, V.; Rustgi, V.; Schwartz, H.; Tatum, H.; et al. Efficacy of an Interferon- and Ribavirin-Free Regimen of Daclatasvir, Asunaprevir, and BMS-791325 in Treatment-Naive Patients with HCV Genotype 1 Infection. Gastroenterology 2014, 146, 420–429. [Google Scholar] [CrossRef]
- Lens, S.; Gambato, M.; Londoño, M.-C.; Forns, X. Interferon-Free Regimens in the Liver-Transplant Setting. Semin. Liver Dis. 2014, 34, 58–71. [Google Scholar] [CrossRef]
- Lee, H.W.; Han, D.H.; Shin, H.J.; Lee, J.S.; Kim, S.U.; Park, J.Y.; Kim, D.Y.; Ahn, S.H.; Kim, B.K. Hepatocellular Carcinoma Risk According to Regimens for Eradication of Hepatitis C Virus; Interferon or Direct Acting Antivirals. Cancers 2020, 12, 3414. [Google Scholar] [CrossRef]
- Iliescu, E.L.; Mercan-Stanciu, A.; Toma, L.; Ioanitescu, E.S.; Dumitru, R.; Rusie, D. Hepatocellular carcinoma in the setting of Interferon-free treatment for chronic HCV hepatitis—Experience of a single center. Hepatoma Res. 2018, 4, 3. [Google Scholar] [CrossRef]
- Bolondi, G.; Mocchegiani, F.; Montalti, R.; Nicolini, D.; Vivarelli, M.; De Pietri, L. Predictive factors of short term outcome after liver transplantation: A review. World J. Gastroenterol. 2016, 22, 5936–5949. [Google Scholar] [CrossRef]
- Lee, Y.; Wang, J.J.; Zhu, Y.; Agopian, V.G.; Tseng, H.; Yang, J.D. Diagnostic Criteria and LI-RADS for Hepatocellular Carcinoma. Clin. Liver Dis. 2021, 17, 409–413. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Liver transplantation. J. Hepatol. 2016, 64, 433–485. [Google Scholar] [CrossRef]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- Piscaglia, F.; Cucchetti, A.; Benlloch, S.; Vivarelli, M.; Berenguer, J.; Bolondi, L.; Pinna, A.D.; Berenguer, M. Prediction of significant fibrosis in hepatitis C virus infected liver transplant recipients by artificial neural network analysis of clinical factors. Eur. J. Gastroenterol. Hepatol. 2006, 18, 1255–1261. [Google Scholar] [CrossRef]
- Sterling, R.K.; Lissen, E.; Clumeck, N.; Sola, R.; Correa, M.C.; Montaner, J.; Sulkowski, M.S.; Torriani, F.J.; Dieterich, D.T.; Thomas, D.L.; et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 2006, 43, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-D.; Lee, K.-S.; Kim, J.M.; Ryu, J.H.; Lee, J.-G.; Kim, B.-W.; Kim, D.-S. Artificial intelligence for predicting survival following deceased donor liver transplantation: Retrospective multi-center study. Int. J. Surg. 2022, 105, 106838. [Google Scholar] [CrossRef] [PubMed]
- Alter, M.J. Epidemiology of hepatitis C. Hepatology 1997, 26, 62S–65S. [Google Scholar] [CrossRef] [PubMed]
- Di Bisceglie, A.M. Hepatitis C and hepatocellular carcinoma. Hepatology 1997, 26, 34S–38S. [Google Scholar] [CrossRef]
- McDONALD, S.A.; Hutchinson, S.J.; Bird, S.M.; Mills, P.R.; Hayes, P.; Dillon, J.F.; Goldberg, D.J. Excess morbidity in the hepatitis C-diagnosed population in Scotland, 1991–2006. Epidemiol. Infect. 2011, 139, 344–353. [Google Scholar] [CrossRef]
- Neal, K.R.; Ramsay, S.; Thomson, B.J.; Irving, W.L. Excess mortality rates in a cohort of patients infected with the hepatitis C virus: A prospective study. Gut 2007, 56, 1098–1104. [Google Scholar] [CrossRef]
- Sweeting, M.J.; De Angelis, D.; Brant, L.J.; Harris, H.; Mann, A.G.; Ramsay, M.E. The burden of hepatitis C in England. J. Viral Hepat. 2007, 14, 570–576. [Google Scholar] [CrossRef]
- Delarocque-Astagneau, E.; Meffre, C.; Dubois, F.; Pioche, C.; Le Strat, Y.; Roudot-Thoraval, F.; Hillon, P.; Silvain, C.; Dhumeaux, D.; Desenclos, J.C.; et al. The impact of the prevention programme of hepatitis C over more than a decade: The French experience. J. Viral Hepat. 2010, 17, 435–443. [Google Scholar] [CrossRef]
- Pedersen, M.; Seetharam, A. Infections after Orthotopic Liver Transplantation. J. Clin. Exp. Hepatol. 2014, 4, 347–360. [Google Scholar] [CrossRef]
- Hernandez, M.D.P.; Martin, P.; Simkins, J. Infectious Complications After Liver Transplantation. Gastroenterol. Hepatol. 2015, 11, 741–753. [Google Scholar]
- Baganate, F.; Beal, E.W.; Tumin, D.; Azoulay, D.; Mumtaz, K.; Black, S.M.; Washburn, K.; Pawlik, T.M. Early mortality after liver transplantation: Defining the course and the cause. Surgery 2018, 164, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Burra, P.; De Martin, E.; Gitto, S. Influence of Age and Gender Before and After Liver Transplantation. Liver Transplant. 2013, 19, 122–134. [Google Scholar] [CrossRef]
- Egawa, H.; Ohdan, H.; Saito, K. Current Status of ABO-incompatible Liver Transplantation. Transplantation 2023, 107, 313–325. [Google Scholar] [CrossRef] [PubMed]
| Numerical Variables | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Standard Deviation | Median | Median Absolute Deviation | Min | Max | |||||
| Age | 47.93 | 8.74 | 48 | 5.19 | 12 | 68 | ||||
| Age at diagnosis | 37.89 | 8.26 | 40 | 1.48 | 2 | 51 | ||||
| MELD-Na | 16.35 | 6.63 | 16 | 5.93 | 5 | 37 | ||||
| AFP (ng/mL) | 127.03 | 358.30 | 10.43 | 10.27 | 0.01 | 2000 | ||||
| Categorical variables | ||||||||||
| Categories definition | Number of occurrences of each category | |||||||||
| 1 | 2 | 3 | 4 | 5 | ||||||
| Sex | 1 (female), 2 (male) | 35 | 45 | - | - | - | ||||
| AB0 blood type | 1 (0), 2(A), 3 (B), 4 (AB) | 16 | 43 | 12 | 9 | - | ||||
| Rh group | 1 (−), 2 (+) | 10 | 70 | - | - | - | ||||
| The diagnosis that prompted LT | 1 (hepatitis C cirrhosis), 2 (hepatitis C cirrhosis and HCC), 3 (coinfection of HCV, hepatitis B virus, and hepatitis D virus), 4 (HCC associated with the coinfection of HCV, hepatitis B virus, and hepatitis D virus) | 55 | 23 | 1 | 1 | - | ||||
| Total bilirubin (mg/dL) | 1 (0.2–1.20), 2 (1.2–4), 3 (4–8), 4 (>8) | 16 | 39 | 22 | 3 | - | ||||
| Platelet count (×103/μL) | 1 (0–20) 2 (20–40) 3 (40–80) 4 (80–150) 5 (150–400) | 9 | 28 | 27 | 11 | 4 | ||||
| Albumin (g/dL) | 1 (≤2.8), 2 (2.8–3.5), 3 (≥3.5) | 5 | 50 | 25 | - | - | ||||
| INR | 1 (˂1.7), 2 (1.7–2.2), 3 (˃ 2.2) | 5 | 56 | 19 | - | - | ||||
| Pre-transplant antiviral treatment | 1 (none), 2 (interferon), 3 (interferon free) | 1 | 68 | 11 | - | - | ||||
| Liver re-transplantation | 1 (No), 2 (Yes) | 74 | 6 | - | - | - | ||||
| Ascites | 1 (No), 2 (Yes) | 20 | 60 | - | - | - | ||||
| Postoperative complications | 1 (No), 2 (Yes) | 58 | 22 | - | - | - | ||||
| Numerical Variables | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Standard Deviation | Median | Median Absolute Deviation | Min | Max | |||||
| Age | 47.93 | 47.1 | 10.24 | 47 | 12.60 | 32 | ||||
| Age at diagnosis | 37.89 | 36.2 | 9.81 | 36.5 | 7.41 | 20 | ||||
| MELD-Na | 16.35 | 17.9 | 6.47 | 15 | 5.19 | 11 | ||||
| AFP (ng/mL) | 127.03 | 53 | 69.61 | 2 | 0.01 | 0.1 | ||||
| Categorical variables | ||||||||||
| Categories definition | Number of occurrences of each category | |||||||||
| 1 | 2 | 3 | 4 | 5 | ||||||
| Sex | 1 (female), 2 (male) | 4 | 6 | - | - | - | ||||
| AB0 blood type | 1 (0), 2(A), 3 (B), 4 (AB) | 3 | 2 | 3 | 2 | - | ||||
| Rh group | 1 (−), 2 (+) | 4 | 6 | - | - | - | ||||
| The diagnosis that prompted LT | 1 (hepatitis C cirrhosis), 2 (hepatitis C cirrhosis and HCC), 3 (coinfection of HCV, hepatitis B virus, and hepatitis D virus), 4 (HCC associated with the coinfection of HCV, hepatitis B virus, and hepatitis D virus) | 6 | 4 | 0 | 0 | - | ||||
| Total bilirubin (mg/dL) | 1 (0.2–1.20), 2 (1.2–4), 3 (4–8), 4 (>8) | 2 | 2 | 3 | 3 | - | ||||
| Platelet count (×103/μL) | 1 (0–20) 2 (20–40) 3 (40–80) 4 (80–150) 5 (150–400) | 2 | 3 | 3 | 2 | - | ||||
| Albumin (g/dL) | 1 (≤2.8), 2 (2.8–3.5), 3 (≥3.5) | 3 | 5 | 2 | - | - | ||||
| INR | 1 (˂1.7), 2 (1.7–2.2), 3 (˃ 2.2) | 3 | 5 | 2 | - | - | ||||
| Pre-transplant antiviral treatment | 1 (none), 2 (interferon), 3 (interferon free) | 1 | 4 | 5 | - | - | ||||
| Liver re-transplantation | 1 (No), 2 (Yes) | 8 | 2 | - | - | - | ||||
| Ascites | 1 (No), 2 (Yes) | 5 | 5 | - | - | - | ||||
| Postoperative complications | 1 (No), 2 (Yes) | 6 | 4 | - | - | - | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zabara, M.L.; Popescu, I.; Burlacu, A.; Geman, O.; Dabija, R.A.C.; Popa, I.V.; Lupascu, C. Machine Learning Model Validated to Predict Outcomes of Liver Transplantation Recipients with Hepatitis C: The Romanian National Transplant Agency Cohort Experience. Sensors 2023, 23, 2149. https://doi.org/10.3390/s23042149
Zabara ML, Popescu I, Burlacu A, Geman O, Dabija RAC, Popa IV, Lupascu C. Machine Learning Model Validated to Predict Outcomes of Liver Transplantation Recipients with Hepatitis C: The Romanian National Transplant Agency Cohort Experience. Sensors. 2023; 23(4):2149. https://doi.org/10.3390/s23042149
Chicago/Turabian StyleZabara, Mihai Lucian, Irinel Popescu, Alexandru Burlacu, Oana Geman, Radu Adrian Crisan Dabija, Iolanda Valentina Popa, and Cristian Lupascu. 2023. "Machine Learning Model Validated to Predict Outcomes of Liver Transplantation Recipients with Hepatitis C: The Romanian National Transplant Agency Cohort Experience" Sensors 23, no. 4: 2149. https://doi.org/10.3390/s23042149
APA StyleZabara, M. L., Popescu, I., Burlacu, A., Geman, O., Dabija, R. A. C., Popa, I. V., & Lupascu, C. (2023). Machine Learning Model Validated to Predict Outcomes of Liver Transplantation Recipients with Hepatitis C: The Romanian National Transplant Agency Cohort Experience. Sensors, 23(4), 2149. https://doi.org/10.3390/s23042149








