Classification of Motor Imagery EEG Signals Based on Data Augmentation and Convolutional Neural Networks
Abstract
:1. Introduction
2. Methods
2.1. Continuous Wavelet Transformation (CWT)
2.2. CNN
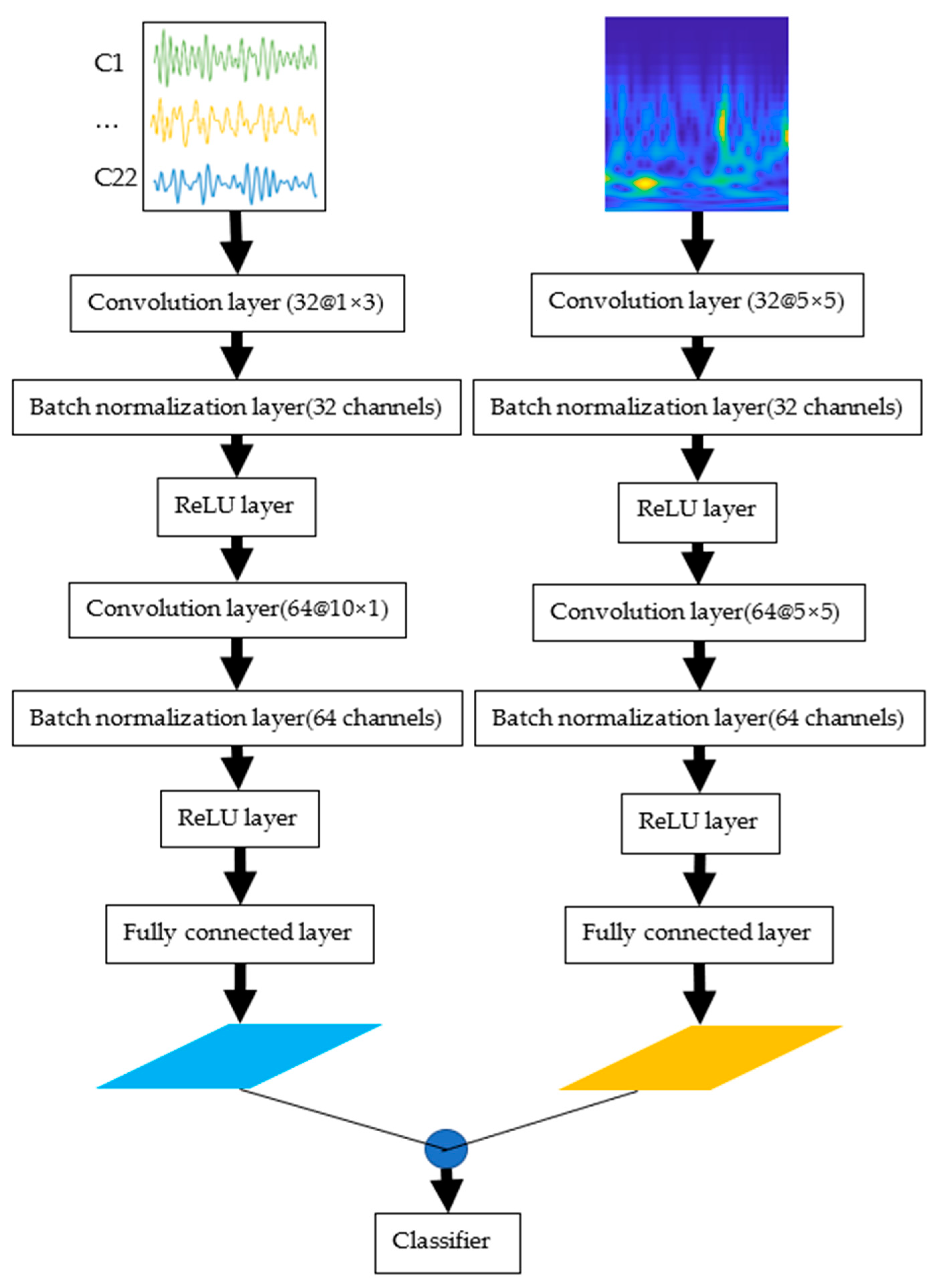
2.3. Proposed CNN Structure
2.4. Data Augmentation
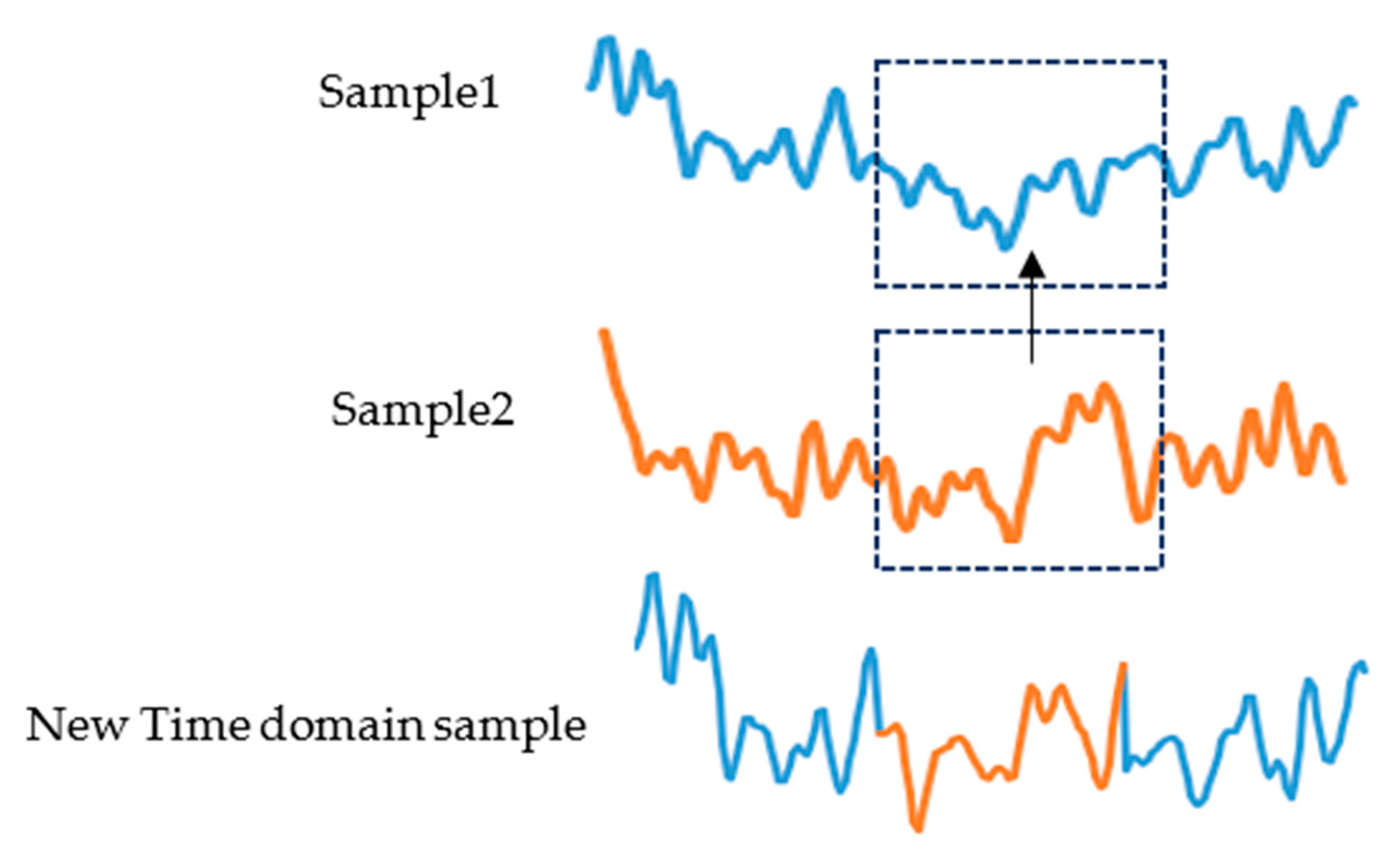
- Sample 1, sample 2, and sample 3 are three samples of the same class that were randomly selected. We randomly capture a period of 1 s of data from sample 2 to replace the data at the same time position in sample 1. Figure 1 shows a time domain sample generation. Do the same for the test set;
- The artificial time-domain EEG sample and sample 3 are divided into two frequency bands, 7–13 Hz and 14–30 Hz, after band-pass filtering, and then a frequency band of sample 3 is exchanged with the corresponding frequency band of the artificial time-domain EEG sample to reconstruct the time–frequency EEG signal.
3. Results
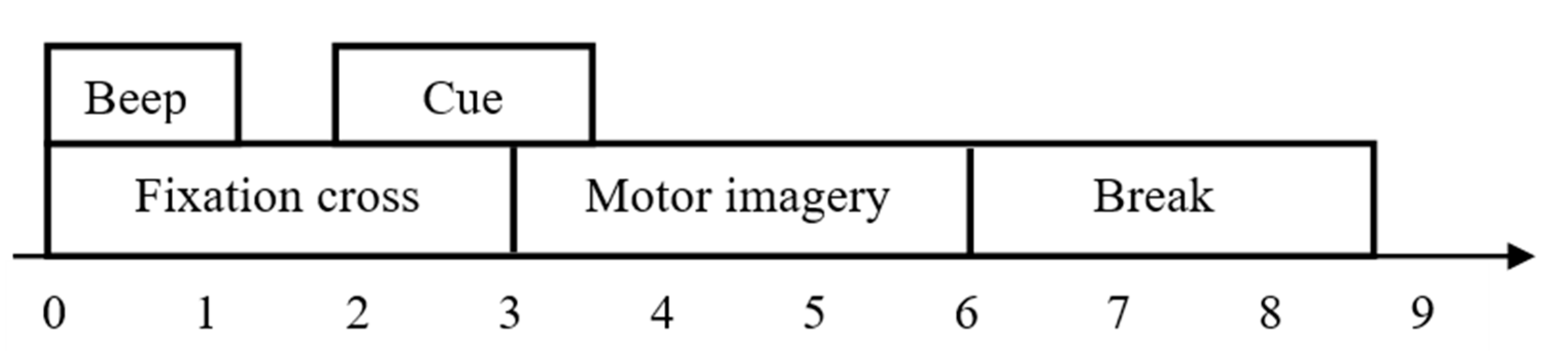
3.1. Database
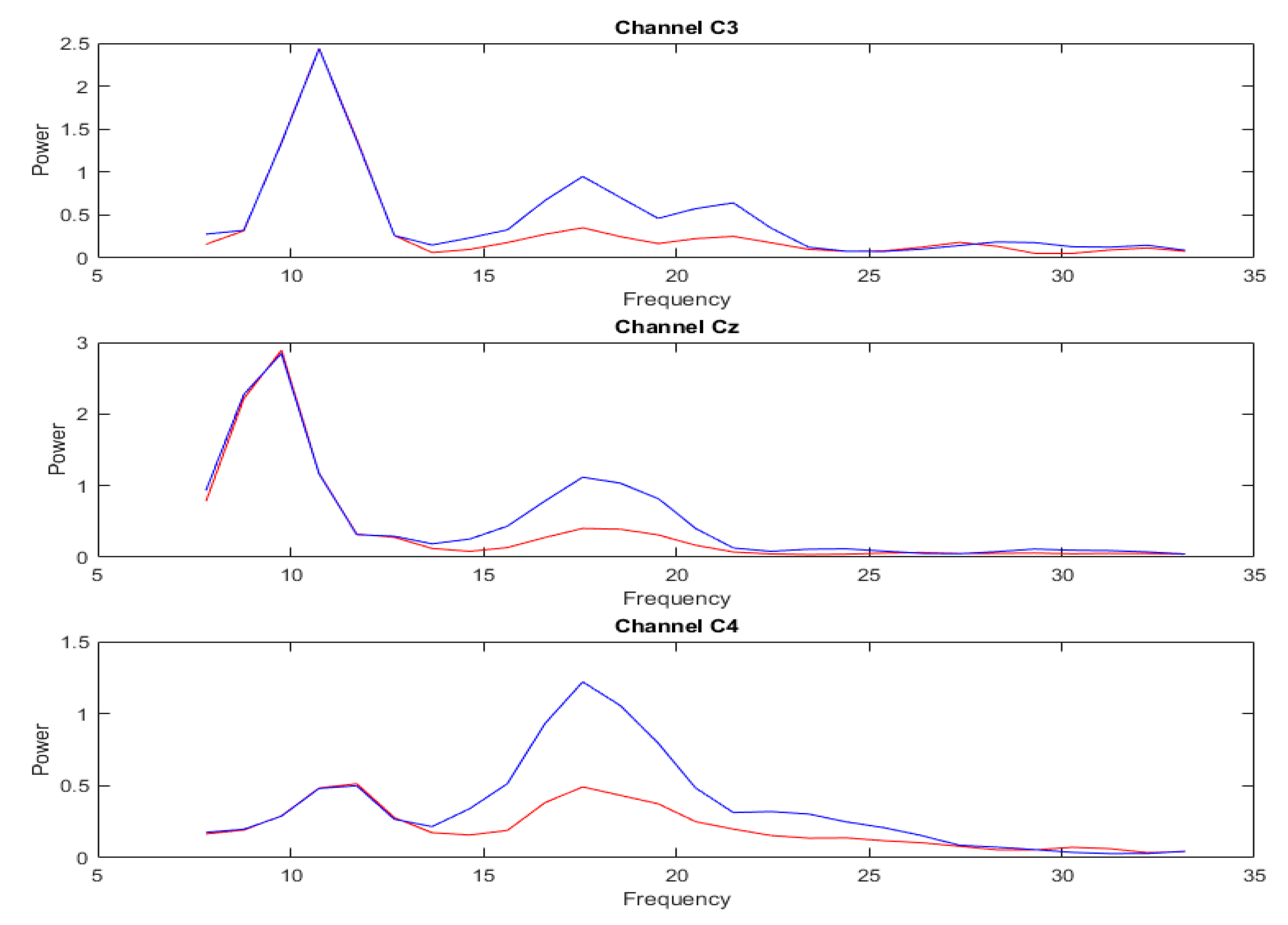
3.2. Analysis of DA Methods
3.3. Performance Evaluation Metrics
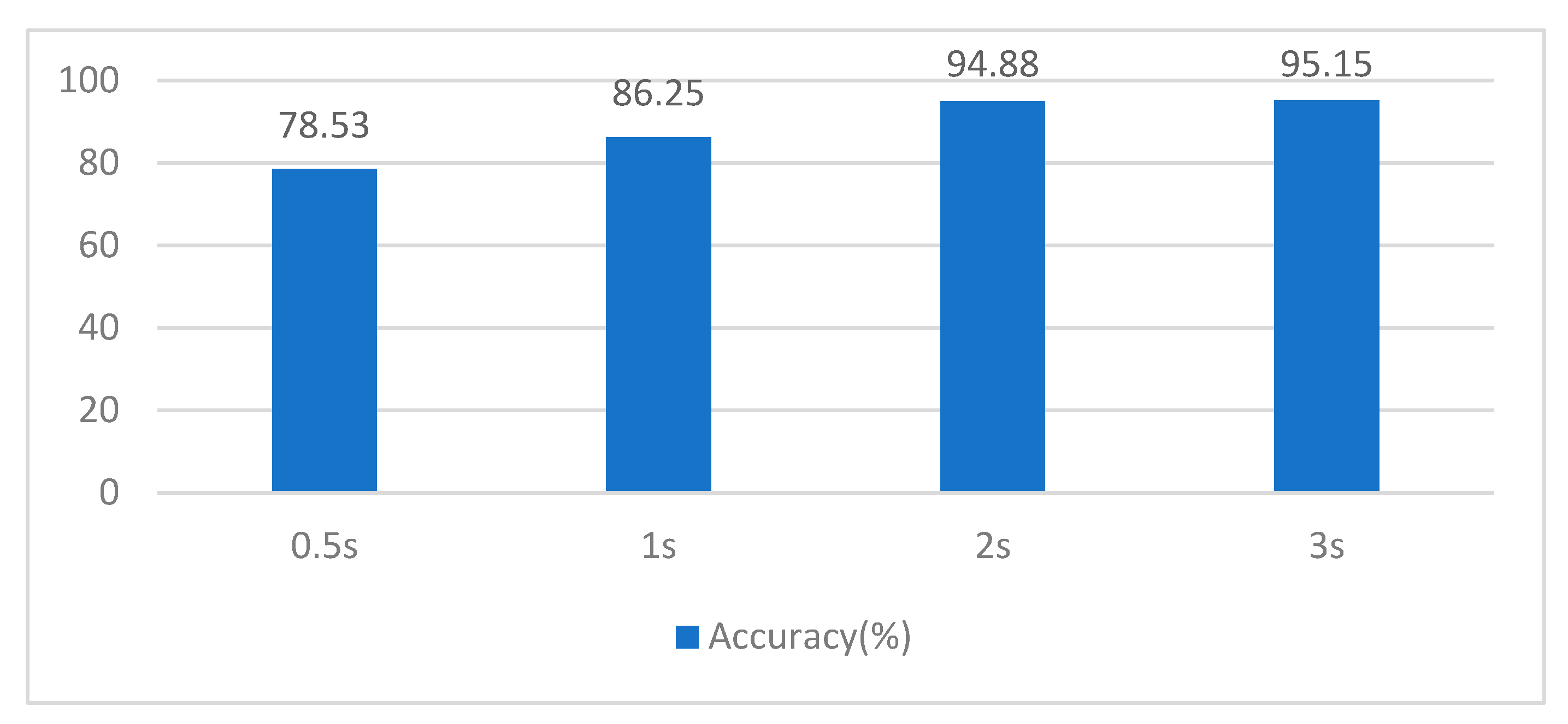
3.4. Determining the Length of Data Segments
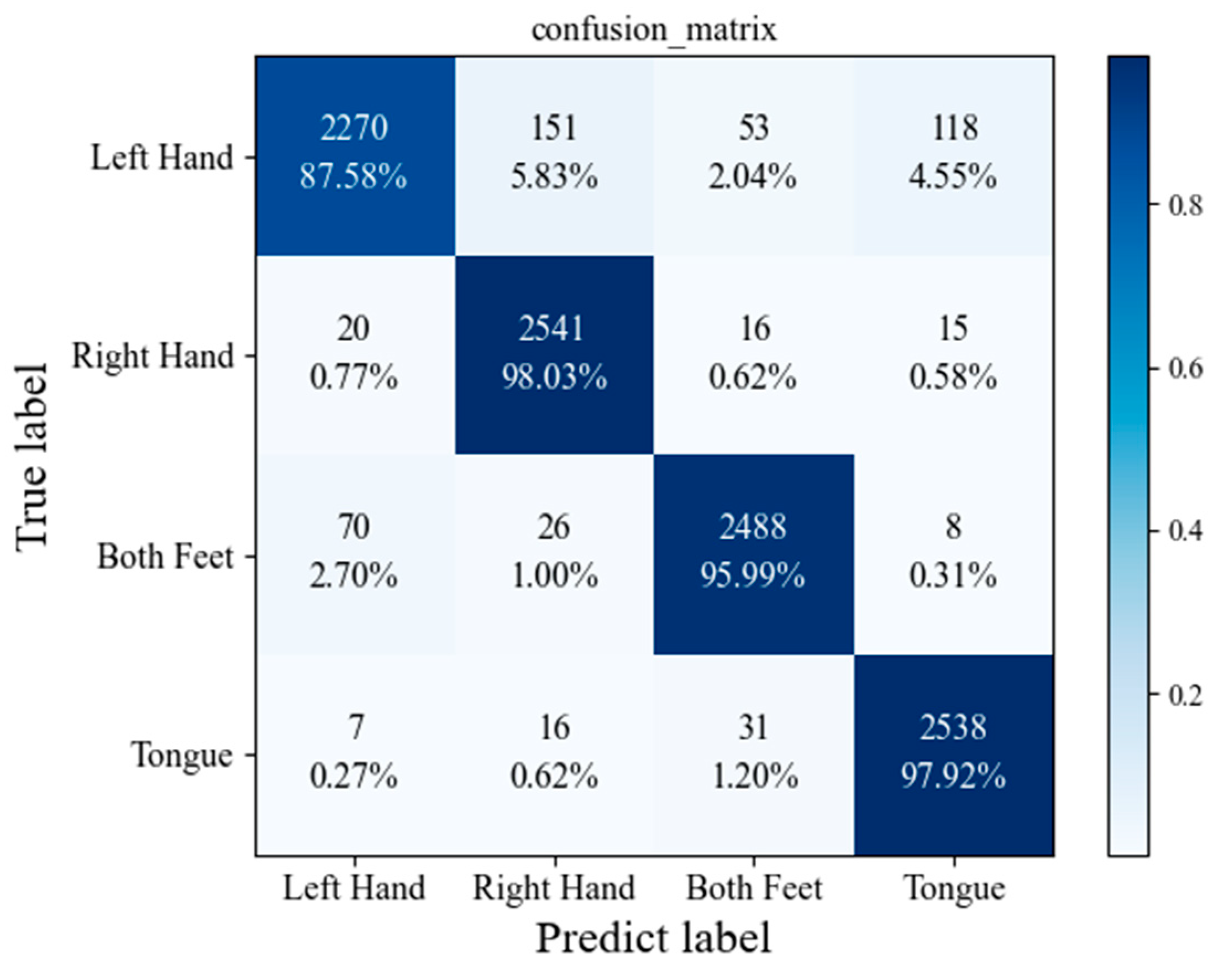
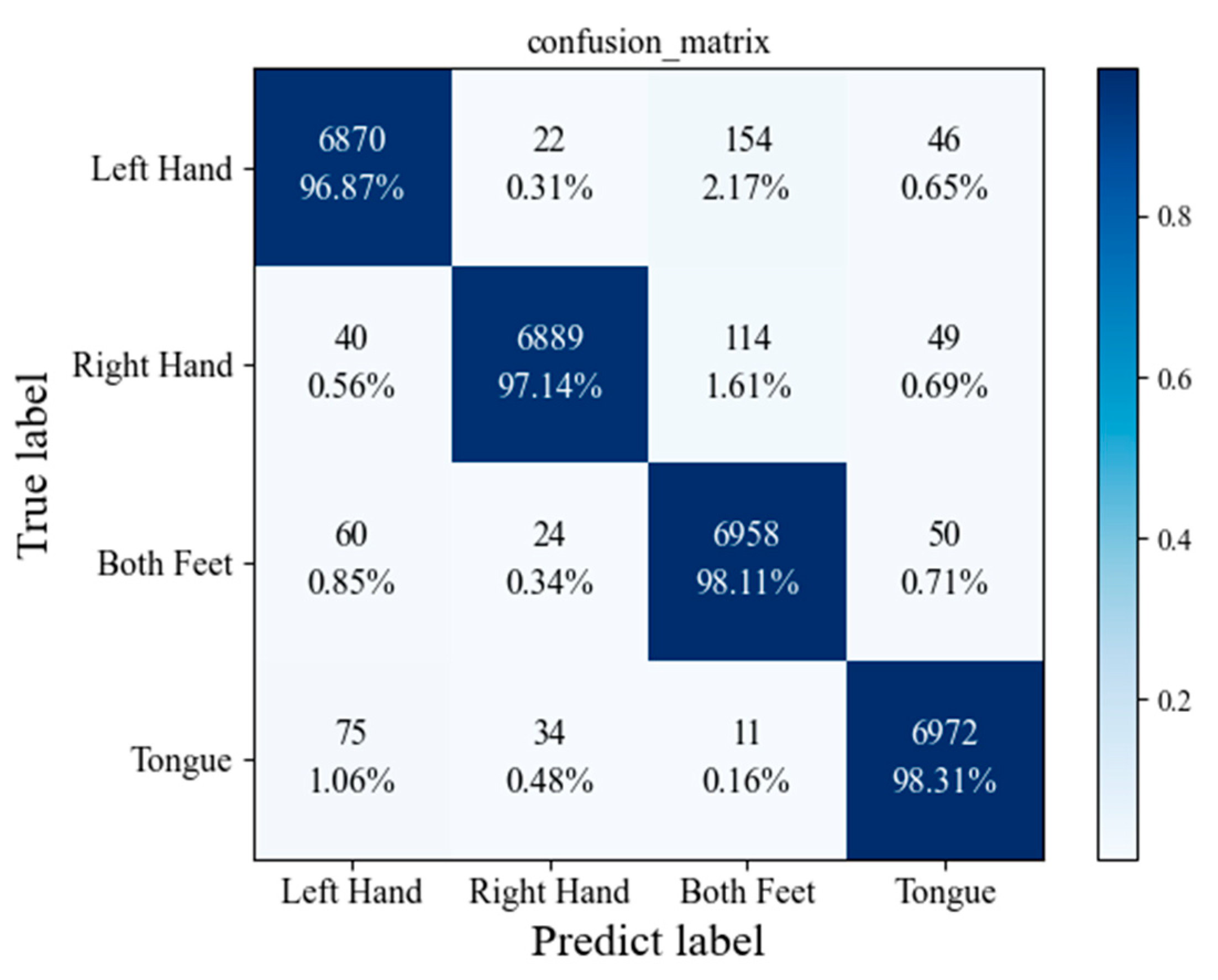
3.5. Performance of the Proposed Model
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curran, E.A.; Stokes, M.J. Learning to control brain activity: A review of the production and control of EEG components for driving brain–computer interface (BCI) systems. Brain Cogn. 2003, 51, 326–336. [Google Scholar] [PubMed]
- Zimmermann-Schlatter, A.; Schuster, C.; Puhan, M.A.; Siekierka, E.; Steurer, J. Efficacy of motor imagery in post-stroke rehabilitation: A systematic review. J. Neuroeng. Rehabil. 2008, 5, 8. [Google Scholar] [PubMed]
- Tong, Y.; Pendy Jr, J.T.; Li, W.A.; Du, H.; Zhang, T.; Geng, X.; Ding, Y. Motor imagery-based rehabilitation: Potential neural correlates and clinical application for functional recovery of motor deficits after stroke. Aging Dis. 2017, 8, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.K.; Chakrapani, M.; Kedambadi, R. Motor imagery training on muscle strength and gait performance in ambulant stroke subjects—A randomized clinical trial. J. Clin. Diagn. Res. JCDR 2016, 10, YC01. [Google Scholar] [PubMed]
- Shih, J.J.; Krusienski, D.J.; Wolpaw, J.R. Brain-computer interfaces in medicine. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2012; pp. 268–279. [Google Scholar]
- Ramoser, H.; Muller-Gerking, J.; Pfurtscheller, G. Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Trans. Rehabil. Eng. 2000, 8, 441–446. [Google Scholar] [CrossRef]
- Müller-Gerking, J.; Pfurtscheller, G.; Flyvbjerg, H. Designing optimal spatial filters for single-trial EEG classification in a movement task. Clin. Neurophysiol. 1999, 110, 787–798. [Google Scholar]
- Yang, H.; Sakhavi, S.; Ang, K.K.; Guan, C. On the use of convolutional neural networks and augmented CSP features for multi-class motor imagery of EEG signals classification. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2620–2623. [Google Scholar]
- Tabar, Y.R.; Halici, U. A novel deep learning approach for classification of EEG motor imagery signals. J. Neural Eng. 2016, 14, 016003. [Google Scholar] [CrossRef]
- Sakhavi, S.; Guan, C.; Yan, S. Learning temporal information for brain-computer interface using convolutional neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 5619–5629. [Google Scholar]
- Zhang, K.; Xu, G.; Han, Z.; Ma, K.; Zheng, X.; Chen, L.; Duan, N.; Zhang, S. Data Augmentation for Motor Imagery Signal Classification Based on a Hybrid Neural Network. Sensors 2020, 20, 4485. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Duan, K.; Fan, C.; Hu, C.; Wang, J. Automatic ocular artifacts removal in EEG using deep learning. Biomed. Signal Process. Control 2018, 43, 148–158. [Google Scholar] [CrossRef]
- Lashgari, E.; Liang, D.; Maoz, U. Data augmentation for deep-learning-based electroencephalography. J. Neurosci. Methods 2020, 346, 108885. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Shovon, T.H.; Al Nazi, Z.; Dash, S.; Hossain, M.F. Classification of motor imagery EEG signals with multi-input convolutional neural network by augmenting STFT. In Proceedings of the 2019 5th International Conference on Advances in Electrical Engineering (ICAEE), Dhaka, Bangladesh, 26–28 September 2019; pp. 398–403. [Google Scholar]
- Wang, F.; Zhong, S.-h.; Peng, J.; Jiang, J.; Liu, Y. Data augmentation for eeg-based emotion recognition with deep convolutional neural networks. In Proceedings of the International Conference on Multimedia Modeling, Bangkok, Thailand, 5–7 February 2018; pp. 82–93. [Google Scholar]
- Zhang, Z.; Duan, F.; Sole-Casals, J.; Dinares-Ferran, J.; Cichocki, A.; Yang, Z.; Sun, Z. A novel deep learning approach with data augmentation to classify motor imagery signals. IEEE Access 2019, 7, 15945–15954. [Google Scholar] [CrossRef]
- Pei, Y.; Luo, Z.; Yan, Y.; Yan, H.; Jiang, J.; Li, W.; Xie, L.; Yin, E. Data Augmentation: Using Channel-Level Recombination to Improve Classification Performance for Motor Imagery EEG. Front Hum Neurosci 2021, 15, 645952. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhu, L.-Z.; Wan, Z.-Y.; Lu, B.-L. Data augmentation for enhancing EEG-based emotion recognition with deep generative models. J. Neural Eng. 2020, 17, 056021. [Google Scholar]
- Kant, P.; Laskar, S.H.; Hazarika, J.; Mahamune, R. CWT Based transfer learning for motor imagery classification for brain computer interfaces. J. Neurosci. Methods 2020, 345, 108886. [Google Scholar] [CrossRef]
- Lee, H.K.; Choi, Y.-S. A convolution neural networks scheme for classification of motor imagery EEG based on wavelet time-frequecy image. In Proceedings of the 2018 International Conference on Information Networking (ICOIN), Chiang Mai, Thailand, 10–12 January 2018; pp. 906–909. [Google Scholar]
- Chaudhary, S.; Taran, S.; Bajaj, V.; Sengur, A. Convolutional neural network based approach towards motor imagery tasks EEG signals classification. IEEE Sens. J. 2019, 19, 4494–4500. [Google Scholar]
- Sethi, S.; Upadhyay, R.; Singh, H.S. Stockwell-common spatial pattern technique for motor imagery-based Brain Computer Interface design. Comput. Electr. Eng. 2018, 71, 492–504. [Google Scholar] [CrossRef]
- Roy, Y.; Banville, H.; Albuquerque, I.; Gramfort, A.; Falk, T.H.; Faubert, J. Deep learning-based electroencephalography analysis: A systematic review. J. Neural Eng. 2019, 16, 051001. [Google Scholar] [CrossRef]
- León, J.; Escobar, J.J.; Ortiz, A.; Ortega, J.; González, J.; Martín-Smith, P.; Gan, J.Q.; Damas, M. Deep learning for EEG-based Motor Imagery classification: Accuracy-cost trade-off. PLoS ONE 2020, 15, e0234178. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [PubMed]
- Xie, Y.; Oniga, S. A Review of Processing Methods and Classification Algorithm for EEG Signal. Carpathian J. Electron. Comput. Eng. 2020, 13, 23–29. [Google Scholar] [CrossRef]
- Liu, Y.; Gross, L.; Li, Z.; Li, X.; Fan, X.; Qi, W. Automatic building extraction on high-resolution remote sensing imagery using deep convolutional encoder-decoder with spatial pyramid pooling. IEEE Access 2019, 7, 128774–128786. [Google Scholar]
- Ouyang, Z.; Niu, J.; Liu, Y.; Guizani, M. Deep CNN-based real-time traffic light detector for self-driving vehicles. IEEE Trans. Mob. Comput. 2019, 19, 300–313. [Google Scholar]
- Yu, Y.; Samali, B.; Rashidi, M.; Mohammadi, M.; Nguyen, T.N.; Zhang, G. Vision-based concrete crack detection using a hybrid framework considering noise effect. J. Build. Eng. 2022, 61, 105246. [Google Scholar] [CrossRef]
- Xie, Y.; Oniga, S.; Majoros, T. Comparison of EEG Data Processing Using Feedforward and Convolutional Neural Network. In Proceedings of the Conference on Information Technology and Data Science 2020, Debrecen, Hungary, 6–8 November 2020; pp. 279–289. [Google Scholar]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain–computer interfaces: A 10 year update. J. Neural Eng. 2018, 15, 031005. [Google Scholar]
- Wang, Y.; Li, Y.; Song, Y.; Rong, X. The influence of the activation function in a convolution neural network model of facial expression recognition. Appl. Sci. 2020, 10, 1897. [Google Scholar]
- Xie, Y.; Majoros, T.; Oniga, S. FPGA-Based Hardware Accelerator on Portable Equipment for EEG Signal Patterns Recognition. Electronics 2022, 11, 2410. [Google Scholar] [CrossRef]
- McFarland, D.J.; Miner, L.A.; Vaughan, T.M.; Wolpaw, J.R. Mu and beta rhythm topographies during motor imagery and actual movements. Brain Topogr. 2000, 12, 177–186. [Google Scholar] [CrossRef]
- Shahid, S.; Sinha, R.K.; Prasad, G. Mu and beta rhythm modulations in motor imagery related post-stroke EEG: A study under BCI framework for post-stroke rehabilitation. Bmc Neurosci. 2010, 11, 1. [Google Scholar]
- Dai, G.; Zhou, J.; Huang, J.; Wang, N. HS-CNN: A CNN with hybrid convolution scale for EEG motor imagery classification. J. Neural Eng. 2020, 17, 016025. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Niu, Y.; Li, F.; Li, Y.; Fu, B.; Shi, G.; Dong, M. A parallel multiscale filter bank convolutional neural networks for motor imagery EEG classification. Front. Neurosci. 2019, 13, 1275. [Google Scholar] [PubMed]
- Li, F.; He, F.; Wang, F.; Zhang, D.; Xia, Y.; Li, X. A novel simplified convolutional neural network classification algorithm of motor imagery EEG signals based on deep learning. Appl. Sci. 2020, 10, 1605. [Google Scholar]
- Kim, S.K.; Kirchner, E.A.; Stefes, A.; Kirchner, F. Intrinsic interactive reinforcement learning–Using error-related potentials for real world human-robot interaction. Sci. Rep. 2017, 7, 17562. [Google Scholar]
- Le Guennec, A.; Malinowski, S.; Tavenard, R. Data augmentation for time series classification using convolutional neural networks. In Proceedings of the ECML/PKDD Workshop on Advanced Analytics and Learning on Temporal Data, Riva del Garda, Italy, 19 September 2016. [Google Scholar]
- Li, Y.; Huang, J.; Zhou, H.; Zhong, N. Human emotion recognition with electroencephalographic multidimensional features by hybrid deep neural networks. Appl. Sci. 2017, 7, 1060. [Google Scholar] [CrossRef]
- Tangermann, M.; Müller, K.-R.; Aertsen, A.; Birbaumer, N.; Braun, C.; Brunner, C.; Leeb, R.; Mehring, C.; Miller, K.J.; Mueller-Putz, G. Review of the BCI competition IV. Front. Neurosci. 2012, 6, 55. [Google Scholar]
- Freeman, W.J. Origin, structure, and role of background EEG activity. Part 2. Analytic phase. Clin. Neurophysiol. 2004, 115, 2089–2107. [Google Scholar] [CrossRef]
- Xu, G.; Shen, X.; Chen, S.; Zong, Y.; Zhang, C.; Yue, H.; Liu, M.; Chen, F.; Che, W. A deep transfer convolutional neural network framework for EEG signal classification. IEEE Access 2019, 7, 112767–112776. [Google Scholar] [CrossRef]
- Majidov, I.; Whangbo, T. Efficient classification of motor imagery electroencephalography signals using deep learning methods. Sensors 2019, 19, 1736. [Google Scholar] [CrossRef]
- Zhang, R.; Zong, Q.; Dou, L.; Zhao, X. A novel hybrid deep learning scheme for four-class motor imagery classification. J. Neural Eng. 2019, 16, 066004. [Google Scholar]
- Ma, X.; Wang, D.; Liu, D.; Yang, J. DWT and CNN based multi-class motor imagery electroencephalographic signal recognition. J. Neural Eng. 2020, 17, 016073. [Google Scholar] [CrossRef] [PubMed]


| Literature | Classifier | Contribution |
|---|---|---|
| VGGNet [45] | CNN |
|
| EEGNet [26] | CNN |
|
| HS-CNN [37] | CNN |
|
| PSO-CNN [46] | CNN |
|
| CNN-LSTM [47] | CNN+LSTM |
|
| DWT-CNN [48] | CNN |
|
| Subject No. | Acc (%) | |||||
|---|---|---|---|---|---|---|
| VGGNet | VGGNet_A | EEGNet | EEGNet_A | Our | Our_A | |
| 1 | 79.32 | 85.36 | 88.21 | 91.33 | 97.23 | 98.61 |
| 2 | 69.01 | 77.22 | 70.35 | 72.58 | 94.77 | 98.29 |
| 3 | 91.11 | 92.67 | 92.40 | 93.14 | 94.12 | 97.99 |
| 4 | 68.32 | 66.12 | 81.26 | 85.65 | 95.22 | 97.1 |
| 5 | 59.00 | 70.15 | 85.77 | 83.85 | 93.25 | 96.72 |
| 6 | 70.32 | 71.39 | 73.12 | 76.98 | 92.36 | 96.37 |
| 7 | 82.55 | 85.73 | 93.10 | 98.54 | 94.8 | 97.69 |
| 8 | 79.33 | 88.52 | 90.73 | 89.32 | 95.75 | 97.22 |
| 9 | 70.74 | 72.29 | 91.05 | 89.99 | 96.38 | 98.51 |
| Mean | 74.41 | 78.82 | 85.11 | 86.82 | 94.88 | 97.61 |
| Subject No. | Acc (%) | |||||
|---|---|---|---|---|---|---|
| HS-CNN | PSO-CNN | CNN-LSTM | DWT-CNN | Our | Our_A | |
| 1 | 90.07 | 93.30 | 98.82 | 98 | 97.23 | 98.61 |
| 2 | 80.28 | 84.59 | 98.64 | 98 | 94.77 | 98.29 |
| 3 | 97.08 | 91.68 | 96.92 | 95 | 94.12 | 97.99 |
| 4 | 89.66 | 84.55 | 96.50 | 96 | 95.22 | 97.1 |
| 5 | 97.04 | 86.54 | 92.75 | 86 | 93.25 | 96.72 |
| 6 | 87.04 | 76.92 | 91.84 | 92 | 92.36 | 96.37 |
| 7 | 92.14 | 94.03 | 95.07 | 94 | 94.8 | 97.69 |
| 8 | 98.51 | 93.20 | 95.25 | 93 | 95.75 | 97.22 |
| 9 | 92.31 | 92.24 | 99.23 | 98 | 96.38 | 98.51 |
| Mean | 91.57 | 85.56 | 96.13 | 94 | 94.88 | 97.61 |
| Metrics | Our | Our_A |
|---|---|---|
| Precision (%) | 94.93 | 97.62 |
| Recall (%) | 94.88 | 97.61 |
| F1-Score | 0.95 | 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Oniga, S. Classification of Motor Imagery EEG Signals Based on Data Augmentation and Convolutional Neural Networks. Sensors 2023, 23, 1932. https://doi.org/10.3390/s23041932
Xie Y, Oniga S. Classification of Motor Imagery EEG Signals Based on Data Augmentation and Convolutional Neural Networks. Sensors. 2023; 23(4):1932. https://doi.org/10.3390/s23041932
Chicago/Turabian StyleXie, Yu, and Stefan Oniga. 2023. "Classification of Motor Imagery EEG Signals Based on Data Augmentation and Convolutional Neural Networks" Sensors 23, no. 4: 1932. https://doi.org/10.3390/s23041932
APA StyleXie, Y., & Oniga, S. (2023). Classification of Motor Imagery EEG Signals Based on Data Augmentation and Convolutional Neural Networks. Sensors, 23(4), 1932. https://doi.org/10.3390/s23041932






