A Multifunctional Network with Uncertainty Estimation and Attention-Based Knowledge Distillation to Address Practical Challenges in Respiration Rate Estimation
Abstract
:1. Introduction
- 1.
- We propose an attention block embedded multitasking network (ATTMRNet), which estimates average and instantaneous RR. Crucially, we discuss and demonstrate the improvements in estimating RR brought about by the interpretation of the internal processes by the attention block.
- 2.
- We propose applying an MC-dropouts-based uncertainty estimation technique embedded in the same architecture, such that the estimates with high uncertainty can be discarded. Furthermore, we also elaborate on the trade-off between RR window rejection, error rate reduction, and the number of inference runs required.
- 3.
- We propose the application of an attention-based KD technique to optimize the inference time and parameter count, making it more viable for deployment on wearable edge devices.
- 4.
- We demonstrate the effectiveness of the entire framework, which includes the attention block, MC dropout, and KD, through extensive comparisons with previous state-of-the-art methods and baselines. We used the Incremental Running dataset (IR dataset), which was collected in-house, and the publicly available PPG-DaLiA dataset, both of which contain ambulatory activities that closely simulate real-world environments.
2. Related Work
2.1. Respiration Signal and RR Extraction
2.2. Machine Learning and Deep Learning Based Techniques
2.3. Attention-Based Techniques
2.4. Uncertainty Estimation Techniques
2.5. Knowledge Distillation Techniques
3. Dataset Description
3.1. PPG-DaLiA Dataset
3.2. Incremental Running Dataset
4. Methodology
4.1. Respiration Signal and Respiration Rate Extraction
4.2. Problem Formulation
4.2.1. Multitasking Functionality Problem Formulation
4.2.2. Problem Formulation for Attention Blocks
4.2.3. Uncertainty Estimation Using MC Dropouts
4.2.4. Problem Formulation for Knowledge Distillation
4.3. Model Architecture
4.3.1. The Encoder Block
4.3.2. The Decoder Block
4.3.3. IncResNet Block
4.3.4. Inception-Res Block
4.3.5. Design and Placement of Attention Block
4.3.6. Placement of MC Dropouts
4.3.7. Student Model Design for KD
4.4. Experimental and Evaluation Details
5. Results and Discussion
5.1. Comparison with Previously Developed Techniques
5.2. Utility of Attention Block
5.3. Evaluation of Predictions Based on Uncertainty
5.4. Application of Knowledge Distillation
5.5. Summary of Results and Discussion
- 1.
- The proposed ATTMRNet performed significantly better than traditional methods and other ML/DL-based techniques in terms of error scores, as shown in Table 2. In comparison to the state-of-the-art MRNet [14], the ATTMRNet had lower error. This was demonstrated by a 0.4% reduction in error for the average RR for PPG-DaLiA and 5.31% for the IR dataset. For instantaneous RR, the error reduction was 7.64% and 4% for the PPG-DaLiA and IR datasets, respectively.
- 2.
- 3.
- Using MC dropouts for uncertainty estimation reduced the error significantly by rejecting 3.8% of uncertain windows for PPG-DaLiA and 3.6% for the IR dataset.
- 4.
- The application of KD reduced the model’s parameter count by 49.5%. Consequently, a reduction in inference time by 36.89% was observed for the PPG-DaLiA and 39.39% for the IR dataset. Additionally, the application of KD also improved the final estimates’ accuracy when compared with the teacher model.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RR | Respiration rate |
| ECG | Electrocardiogram |
| PPG | Photoplethesmography |
| RRint | R–R-interval-based respiration signal |
| Rpeak | R-peak-based respiration signal |
| ADR | Accelerometer-derived respiration |
| DL | Deep learning |
| ML | Machine learning |
| LR | Learning rate |
| IT | Inference time |
| PC | Parameter count |
| MC | Monte Carlo |
| KD | Knowledge distillation |
References
- Nicolò, A.; Massaroni, C.; Passfield, L. Respiratory frequency during exercise: The neglected physiological measure. Front. Physiol. 2017, 8, 922. [Google Scholar] [CrossRef]
- Gilgen-Ammann, R.; Koller, M.; Huber, C.; Ahola, R.; Korhonen, T.; Wyss, T. Energy expenditure estimation from respiration variables. Sci. Rep. 2017, 7, 15995. [Google Scholar] [CrossRef]
- Charlton, P.H.; Birrenkott, D.A.; Bonnici, T.; Pimentel, M.A.; Johnson, A.E.; Alastruey, J.; Tarassenko, L.; Watkinson, P.J.; Beale, R.; Clifton, D.A. Breathing Rate Estimation from the Electrocardiogram and Photoplethysmogram: A Review. IEEE Rev. Biomed. Eng. 2018, 11, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.D. Estimating respiration rate using an accelerometer sensor. In Proceedings of the 8th International Conference on Computational Systems-Biology and Bioinformatics, Nha Trang City, Viet Nam, 7–8 December 2017; pp. 11–14. [Google Scholar]
- Pimentel, M.A.; Charlton, P.H.; Clifton, D.A. Probabilistic estimation of respiratory rate from wearable sensors. Smart Sens. Meas. Instrum. 2015, 15, 241–262. [Google Scholar] [CrossRef]
- Lee, H.; Lee, J.; Kwon, Y.; Kwon, J.; Park, S.; Sohn, R.; Park, C. Multitask Siamese Network for Remote Photoplethysmography and Respiration Estimation. Sensors 2022, 22, 5101. [Google Scholar] [CrossRef]
- Reiss, A.; Indlekofer, I.; Schmidt, P.; Van Laerhoven, K. Deep PPG: Large-scale heart rate estimation with convolutional neural networks. Sensors 2019, 19, 3079. [Google Scholar] [CrossRef]
- Birrenkott, D.A.; Pimentel, M.A.; Watkinson, P.J.; Clifton, D.A. A robust fusion model for estimating respiratory rate from photoplethysmography and electrocardiography. IEEE Trans. Biomed. Eng. 2018, 65, 2033–2041. [Google Scholar] [CrossRef]
- Kumar, A.K.; Ritam, M.; Han, L.; Guo, S.; Chandra, R. Deep learning for predicting respiratory rate from biosignals. Comput. Biol. Med. 2022, 144, 105338. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Xiang, W.; Atkinson, I. Determining respiratory rate from photoplethysmogram and electrocardiogram signals using respiratory quality indices and neural networks. PLoS ONE 2021, 16, e0249843. [Google Scholar] [CrossRef]
- Chan, A.M.; Ferdosi, N.; Narasimhan, R. Ambulatory respiratory rate detection using ECG and a triaxial accelerometer. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Osaka, Japan, 3–7 July 2013; pp. 4058–4061. [Google Scholar] [CrossRef]
- Bian, D.; Mehta, P.; Selvaraj, N. Respiratory Rate Estimation using PPG: A Deep Learning Approach. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Montreal, QC, Canada, 20–24 July 2020; pp. 5948–5952. [Google Scholar] [CrossRef]
- Ravichandran, V.; Murugesan, B.; Balakarthikeyan, V.; Ram, K.; Preejith, S.; Joseph, J.; Sivaprakasam, M. RespNet: A deep learning model for extraction of respiration from photoplethysmogram. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 5556–5559. [Google Scholar]
- Rathore, K.S.; Sricharan, V.; Preejith, S.; Sivaprakasam, M. MRNet—A Deep Learning Based Multitasking Model for Respiration Rate Estimation in Practical Settings. In Proceedings of the 2022 IEEE 10th International Conference on Serious Games and Applications for Health(SeGAH), Sydney, Australia, 10–12 August 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Orphanidou, C.; Bonnici, T.; Charlton, P.; Clifton, D.; Vallance, D.; Tarassenko, L. Signal-quality indices for the electrocardiogram and photoplethysmogram: Derivation and applications to wireless monitoring. IEEE J. Biomed. Health Inform. 2015, 19, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Li, J.; Zhang, Z.; Jiao, Z.; Gao, X. An Attention-Guided Deep Regression Model for Landmark Detection in Cephalograms. Lect. Notes Comput. Sci. (Incl. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2019, 11769 LNCS, 540–548. [Google Scholar] [CrossRef]
- Alzahrani, S.; Al-Bander, B.; Al-Nuaimy, W. Attention mechanism guided deep regression model for acne severity grading. Computers 2022, 11, 31. [Google Scholar] [CrossRef]
- Karlen, W.; Raman, S.; Ansermino, J.M.; Dumont, G.A. Multiparameter respiratory rate estimation from the photoplethysmogram. IEEE Trans. Biomed. Eng. 2013, 60, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Gal, Y.; Ghahramani, Z. Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the 33rd International Conference on Machine Learning, ICML 2016, New York City, NY, USA, 19–24 June 2016; Volume 3, pp. 1651–1660. [Google Scholar]
- Laves, M.H.; Ihler, S.; Fast, J.F.; Kahrs, L.A.; Ortmaier, T. Well-Calibrated Regression Uncertainty in Medical Imaging with Deep Learning. In Proceedings of the Proceedings of the Third Conference on Medical Imaging with Deep Learning, Montreal, QC, Canada, 6–8 July 2020; Volume 121, pp. 393–412. [Google Scholar]
- Laves, M.H.; Tölle, M.; Ortmaier, T. Uncertainty Estimation in Medical Image Denoising with Bayesian Deep Image Prior. Lect. Notes Comput. Sci. (Incl. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2020, 12443 LNCS, 81–96. [Google Scholar] [CrossRef]
- Alkhulaifi, A.; Alsahli, F.; Ahmad, I. Knowledge distillation in deep learning and its applications. PeerJ Comput. Sci. 2021, 7, e474. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, D.; Zhou, P.; Zhang, T. A survey of model compression and acceleration for deep neural networks. arXiv 2017, arXiv:1710.09282. [Google Scholar]
- Orphanidou, C. Derivation of respiration rate from ambulatory ECG and PPG using Ensemble Empirical Mode Decomposition: Comparison and fusion. Comput. Biol. Med. 2017, 81, 45–54. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, K.K.; Lata, O. Respiratory rate extraction from single-lead ECG using homomorphic fi ltering. Comput. Biol. Med. 2015, 59, 80–86. [Google Scholar] [CrossRef]
- Sarkar, S.; Bhattacherjee, S.; Pal, S. Extraction of respiration signal from ECG for respiratory rate estimation. IET Conf. Publ. 2015, 2015, 336–340. [Google Scholar] [CrossRef]
- Rajkumar, K.; Ramya, K. Respiration rate diagnosis using single lead ECG in real time. Glob. J. Med. Res. 2013, 13, 7–11. [Google Scholar]
- Liu, G.Z.; Guo, Y.W.; Zhu, Q.S.; Huang, B.Y.; Wang, L. Estimation of respiration rate from three-dimensional acceleration data based on body sensor network. Telemed. J. e-Health Off. J. Am. Telemed. Assoc. 2011, 17, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Jarchi, D.; Rodgers, S.J.; Tarassenko, L.; Clifton, D.A. Accelerometry-Based Estimation of Respiratory Rate for Post-Intensive Care Patient Monitoring. IEEE Sens. J. 2018, 18, 4981–4989. [Google Scholar] [CrossRef]
- Stankoski, S.; Kiprijanovska, I.; Mavridou, I.; Nduka, C.; Gjoreski, H.; Gjoreski, M. Breathing Rate Estimation from Head-Worn Photoplethysmography Sensor Data Using Machine Learning. Sensors 2022, 22, 2079. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.H.; Shuzan, M.N.I.; Chowdhury, M.E.; Reaz, M.B.I.; Mahmud, S.; Al Emadi, N.; Ayari, M.A.; Ali, S.H.M.; Bakar, A.A.A.; Rahman, S.M.; et al. Lightweight End-to-End Deep Learning Solution for Estimating the Respiration Rate from Photoplethysmogram Signal. Bioengineering 2022, 9, 558. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Khanh, T.L.B.; Dao, D.P.; Ho, N.H.; Yang, H.J.; Baek, E.T.; Lee, G.; Kim, S.H.; Yoo, S.B. Enhancing U-net with spatial-channel attention gate for abnormal tissue segmentation in medical imaging. Appl. Sci. 2020, 10, 5729. [Google Scholar] [CrossRef]
- Fort, S.; Hu, H.; Lakshminarayanan, B. Deep ensembles: A loss landscape perspective. arXiv 2019, arXiv:1912.02757. [Google Scholar]
- Qin, D.; Bu, J.J.; Liu, Z.; Shen, X.; Zhou, S.; Gu, J.J.; Wang, Z.H.; Wu, L.; Dai, H.F. Efficient Medical Image Segmentation Based on Knowledge Distillation. IEEE Trans. Med. Imaging 2021, 40, 3820–3831. [Google Scholar] [CrossRef]
- Zheng, Z.; Kang, G. Model Compression with NAS and Knowledge Distillation for Medical Image Segmentation. ACM Int. Conf. Proceeding Ser. 2021, 173–176. [Google Scholar] [CrossRef]
- Xu, P.; Kim, K.; Koh, J.; Wu, D.; Rim Lee, Y.; Young Park, S.; Young Tak, W.; Liu, H.; Li, Q. Efficient knowledge distillation for liver CT segmentation using growing assistant network. Phys. Med. Biol. 2021, 66, 235005. [Google Scholar] [CrossRef]
- Zagoruyko, S.; Komodakis, N. Paying more attention to attention: Improving the performance of convolutional neural networks via attention transfer. In Proceedings of the 5th International Conference on Learning Representations, ICLR 2017—Conference Track Proceedings, Toulon, France, 24–26 April 2017; pp. 1–13. [Google Scholar]
- Murugesan, B.; Vijayarangan, S.; Sarveswaran, K.; Ram, K.; Sivaprakasam, M. KD-MRI: A knowledge distillation framework for image reconstruction and image restoration in MRI workflow. In Proceedings of the Medical Imaging with Deep Learning (PMLR), Montréal, QC, Canada, 6–8 July 2020; pp. 515–526. [Google Scholar]
- Bruce, R.; Kusumi, F.; Hosmer, D. Fundamentals of clinical cardiology and recognition. Fundam. Clin. Cardiol. 1974, 88, 372–379. [Google Scholar]
- Akintola, A.A.; van de Pol, V.; Bimmel, D.; Maan, A.C.; van Heemst, D. Comparative analysis of the equivital EQ02 lifemonitor with holter ambulatory ECG device for continuous measurement of ECG, heart rate, and heart rate variability: A validation study for precision and accuracy. Front. Physiol. 2016, 7, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S.H.A. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Kratky, K.W. Estimation of breathing rate from respiratory sinus arrhythmia: Comparison of various methods. Ann. Biomed. Eng. 2008, 36, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Shu, Y.; Chen, G.; Ooi, B.C.; Wang, W.; Zhang, M. Effective and efficient dropout for deep convolutional neural networks. arXiv 2019, arXiv:1904.03392. [Google Scholar]
- Park, S.; Kwak, N. Analysis on the dropout effect in convolutional neural networks. In Proceedings of the Asian Conference on Computer Vision, Taipei, Taiwan, 20–24 November 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 189–204. [Google Scholar]
- Vijayarangan, S.; Vignesh, R.; Murugesan, B.; Preejith, S.; Joseph, J.; Sivaprakasam, M. RPnet: A Deep Learning approach for robust R Peak detection in noisy ECG. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 345–348. [Google Scholar]
- Cho, J.H.; Hariharan, B. On the efficacy of knowledge distillation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 4794–4802. [Google Scholar]



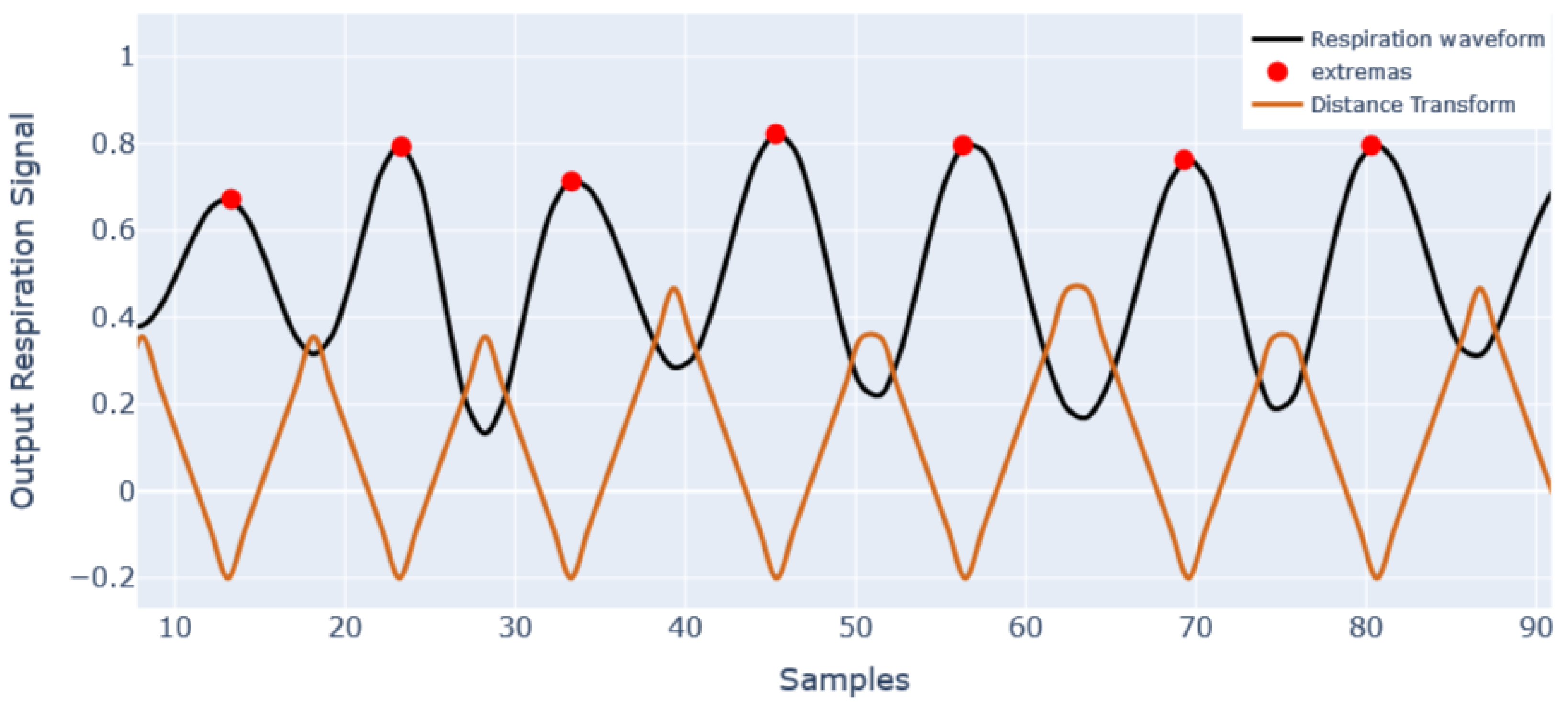
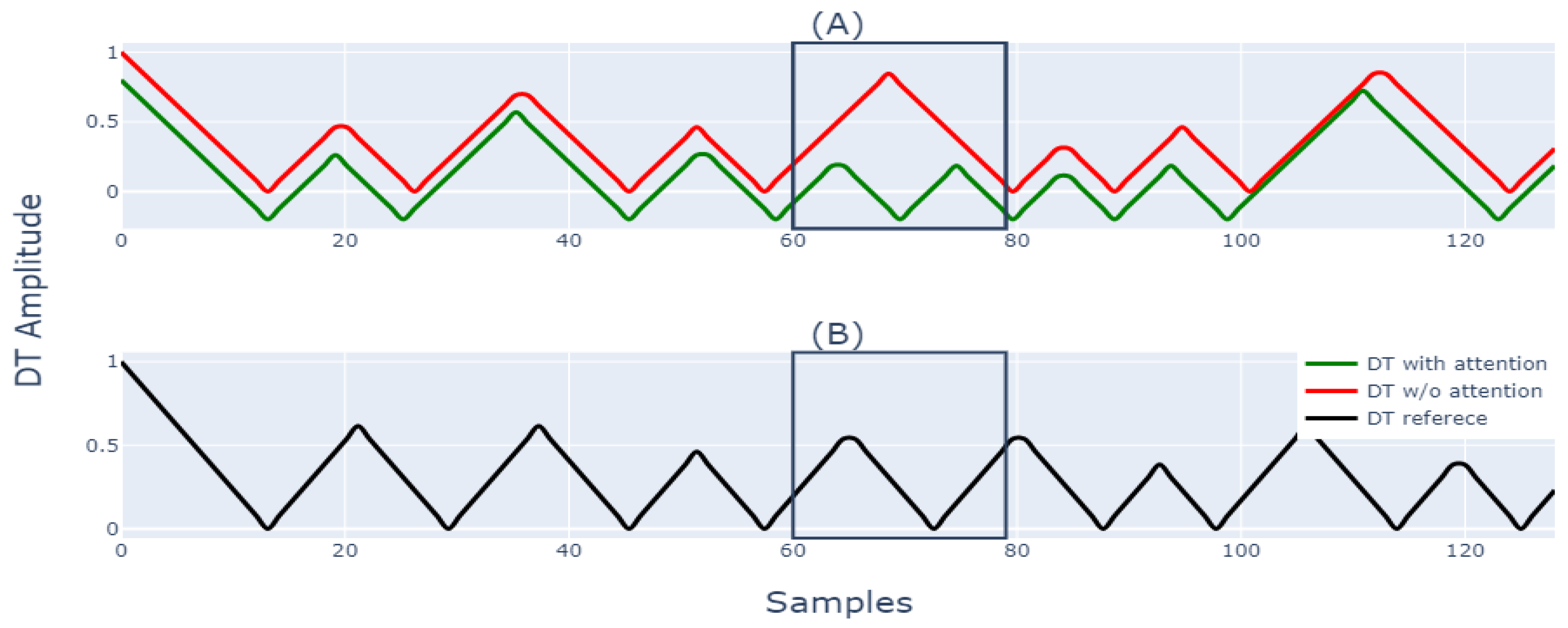



| STAGE | TIME (Min) | SPEED (km/h) | SLOPE (%) |
|---|---|---|---|
| 1 | 0 | 2.7 | 10 |
| 2 | 3 | 3 | 12 |
| 3 | 6 | 5.4 | 14 |
| 4 | 9 | 6.7 | 16 |
| 5 | 12 | 8 | 18 |
| 6 | 15 | 8.8 | 20 |
| 7 | 18 | 9.6 | 22 |
| 8 | 21 | 10.4 | 24 |
| 9 | 24 | 11.2 | 26 |
| 10 | 27 | 12 | 28 |
| DATASET | METHOD | AVG RR | INST RR | PC (M) | IT (s) | ||
|---|---|---|---|---|---|---|---|
| PPG-DaLiA | MAE | RMSE | MAE | RMSE | |||
| RRint | 3.26 | 4.13 | 3.54 | 4.5 | NA | ||
| Rpeak | 3.46 | 4.37 | 3.82 | 4.85 | NA | ||
| ADR | 3.15 | 3.96 | 3.5 | 4.35 | NA | ||
| SF | 2.94 | 3.88 | NA | NA | NA | 0.48 | |
| LREG | 3.46 | 4.7 | NA | NA | NA | ||
| CNN | 2.8 | 3.56 | NA | NA | 0.45 | 0.01 | |
| RN | 3.22 | 4.02 | 3.34 | 4.18 | 39.14 | 0.103 | |
| MRNet | 2.37 | 2.97 | 3.42 | 4.31 | 23.06 | 0.11 | |
| ATTMRNet | 2.36 | 2.97 | 3.16 | 3.97 | 24.82 | 0.11 | |
| IR DATASET | RRint | 4.35 | 5.4 | 4.18 | 5.24 | NA | |
| Rpeak | 3.92 | 4.99 | 3.86 | 4.94 | NA | ||
| ADR | 4.21 | 5.14 | 4.5 | 5.6 | NA | ||
| SF | 4.07 | 5.37 | NA | NA | NA | 0.15 | |
| LREG | 4.25 | 5.55 | NA | NA | NA | ||
| CNN | 3.17 | 3.99 | NA | NA | 0.45 | 0.01 | |
| RN | 3.57 | 4.46 | 3.85 | 4.5 | 39.14 | 0.12 | |
| MRNet | 2.82 | 3.57 | 3.54 | 4.39 | 23.06 | 0.12 | |
| ATTMRNet | 2.67 | 3.55 | 3.4 | 4.25 | 24.82 | 0.11 | |
| DATASET | INFERENCE SAMPLES | AVG RR | INST RR | % WINDOWS REJECTED | IT (s) | ||
|---|---|---|---|---|---|---|---|
| PPG-DaLiA | MAE | RMSE | MAE | RMSE | |||
| 5 | 2.27 | 2.88 | 3.73 | 4.8 | 3% | 8 | |
| 10 | 2.23 | 2.85 | 3 | 3.94 | 3.80% | 13.9 | |
| 15 | 2.23 | 2.84 | 3.04 | 3.88 | 4% | 20.32 | |
| 20 | 2.23 | 2.85 | 2.91 | 3.8 | 4% | 27.76 | |
| IR dataset | 5 | 2.45 | 3.27 | 3.63 | 4.68 | 5.12% | 7.16 |
| 10 | 2.39 | 3.24 | 2.97 | 4 | 3.60% | 12.08 | |
| 15 | 2.48 | 3.27 | 3.02 | 3.95 | 3.30% | 17.54 | |
| 20 | 2.48 | 3.27 | 3.03 | 3.94 | 2.90% | 23.06 | |
| DATASET | METHOD | AVG RR | INST RR | ||
|---|---|---|---|---|---|
| MAE | RMSE | MAE | RMSE | ||
| PPG-DaLiA | Without MC dropout | 2.36 | 2.97 | 3.16 | 3.97 |
| With MC dropout | 2.23 | 2.85 | 3.01 | 3.89 | |
| IR dataset | Without MC dropout | 2.67 | 3.55 | 3.4 | 4.25 |
| With MC dropout | 2.39 | 3.24 | 2.97 | 4 | |
| Number of Encoder and Decoder Levels () | Percentage Increment in Error for PPG-DaLiA Dataset | Percentage Increment in Error for IR Dataset | PC (M) |
|---|---|---|---|
| 1 | 16.45% | 8.07% | 10.91 |
| 2 | 4.76% | 7.3% | 11.09 |
| 3 | 2.16% | 2.69% | 12.53 |
| 4 | 2.45% | 2.69% | 16.97 |
| DATASET | METHOD | AVG RR | INTSANT RR | PC(M) | IT (s) | ||
|---|---|---|---|---|---|---|---|
| MAE | RMSE | MAE | RMSE | ||||
| PPG-DaLiA | Teacher model | 2.31 | 2.91 | 3.14 | 4.01 | 24.8 | 15.15 |
| Student model | 2.36 | 2.95 | 3.34 | 4.13 | 12.53 | 9.68 | |
| KD model | 2.22 | 2.87 | 2.81 | 3.66 | 12.53 | 9.56 | |
| IR dataset | Teacher model | 2.6 | 3.47 | 3.09 | 4.18 | 24.8 | 13.41 |
| Student model | 2.67 | 3.5 | 3.23 | 4.23 | 12.53 | 8.32 | |
| KD model | 2.38 | 3.17 | 2.81 | 3.73 | 12.53 | 8.14 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathore, K.S.; Vijayarangan, S.; SP, P.; Sivaprakasam, M. A Multifunctional Network with Uncertainty Estimation and Attention-Based Knowledge Distillation to Address Practical Challenges in Respiration Rate Estimation. Sensors 2023, 23, 1599. https://doi.org/10.3390/s23031599
Rathore KS, Vijayarangan S, SP P, Sivaprakasam M. A Multifunctional Network with Uncertainty Estimation and Attention-Based Knowledge Distillation to Address Practical Challenges in Respiration Rate Estimation. Sensors. 2023; 23(3):1599. https://doi.org/10.3390/s23031599
Chicago/Turabian StyleRathore, Kapil Singh, Sricharan Vijayarangan, Preejith SP, and Mohanasankar Sivaprakasam. 2023. "A Multifunctional Network with Uncertainty Estimation and Attention-Based Knowledge Distillation to Address Practical Challenges in Respiration Rate Estimation" Sensors 23, no. 3: 1599. https://doi.org/10.3390/s23031599
APA StyleRathore, K. S., Vijayarangan, S., SP, P., & Sivaprakasam, M. (2023). A Multifunctional Network with Uncertainty Estimation and Attention-Based Knowledge Distillation to Address Practical Challenges in Respiration Rate Estimation. Sensors, 23(3), 1599. https://doi.org/10.3390/s23031599







