Simulating the Effect of Gut Microbiome on Cancer Cell Growth Using a Microfluidic Device
Abstract
1. Introduction
2. Materials and Methods
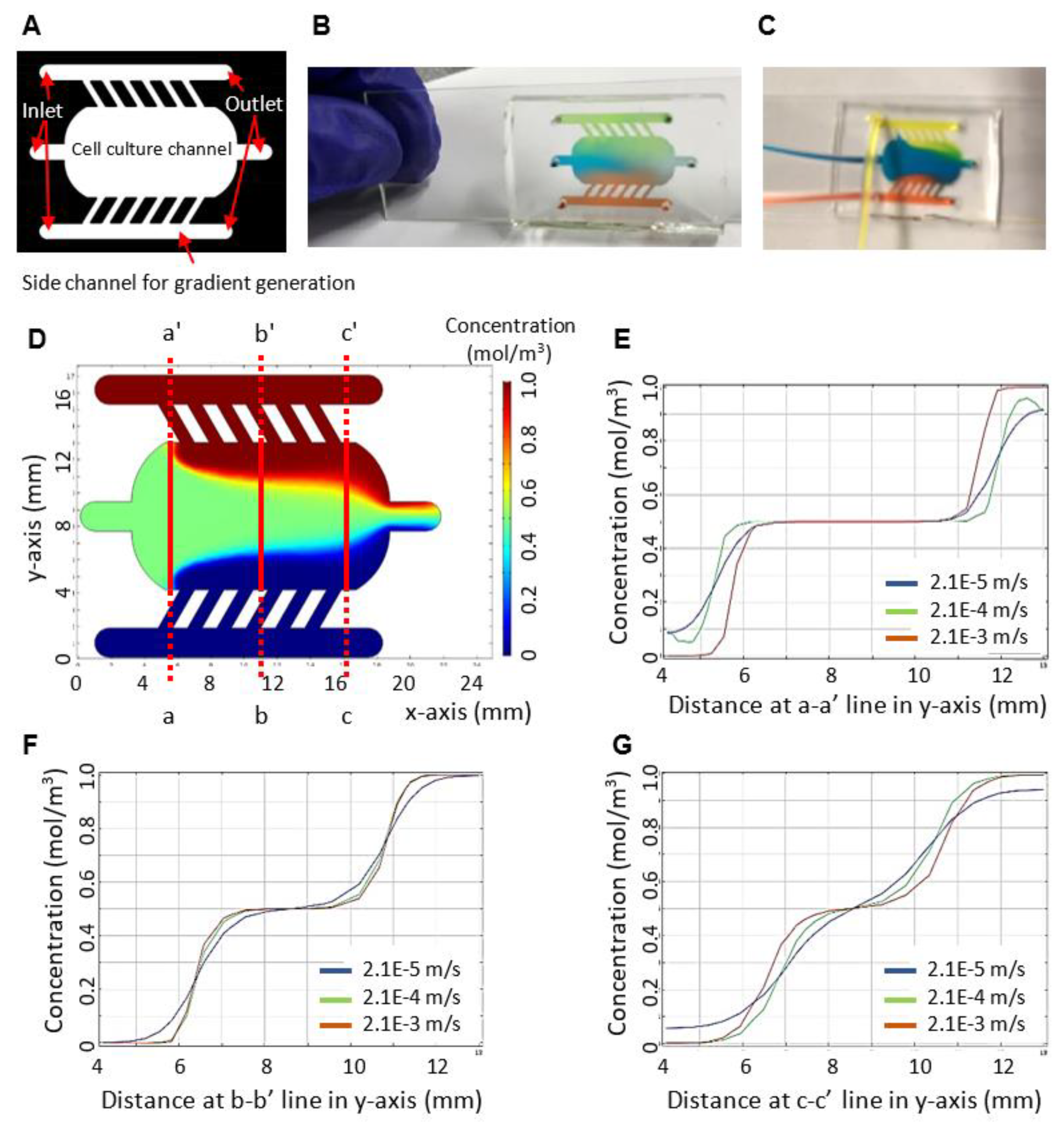
2.1. Fabrication of the Microfluidic Device
2.2. Co-Culture of Cancer Cells and Bacterial Cells
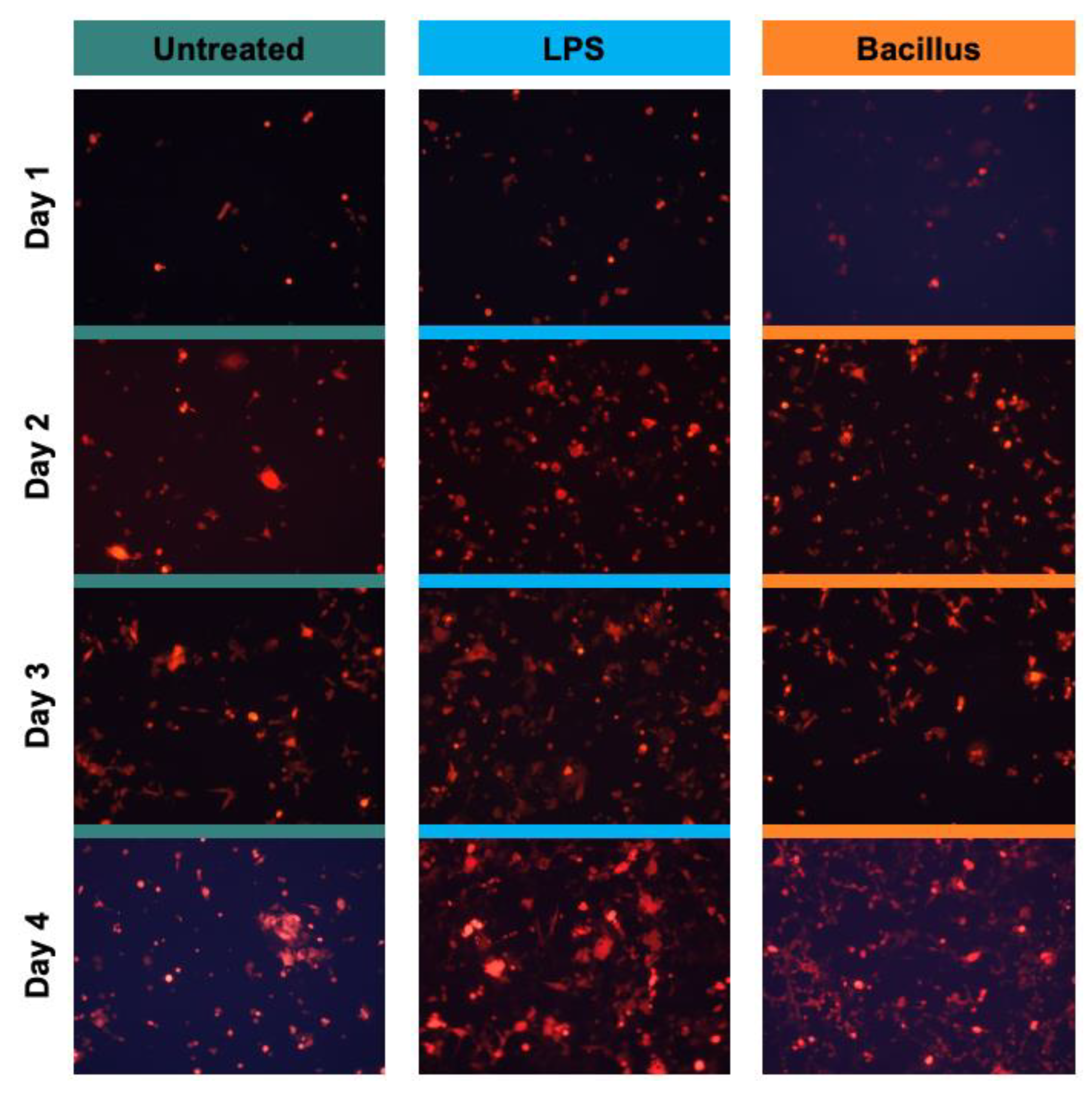
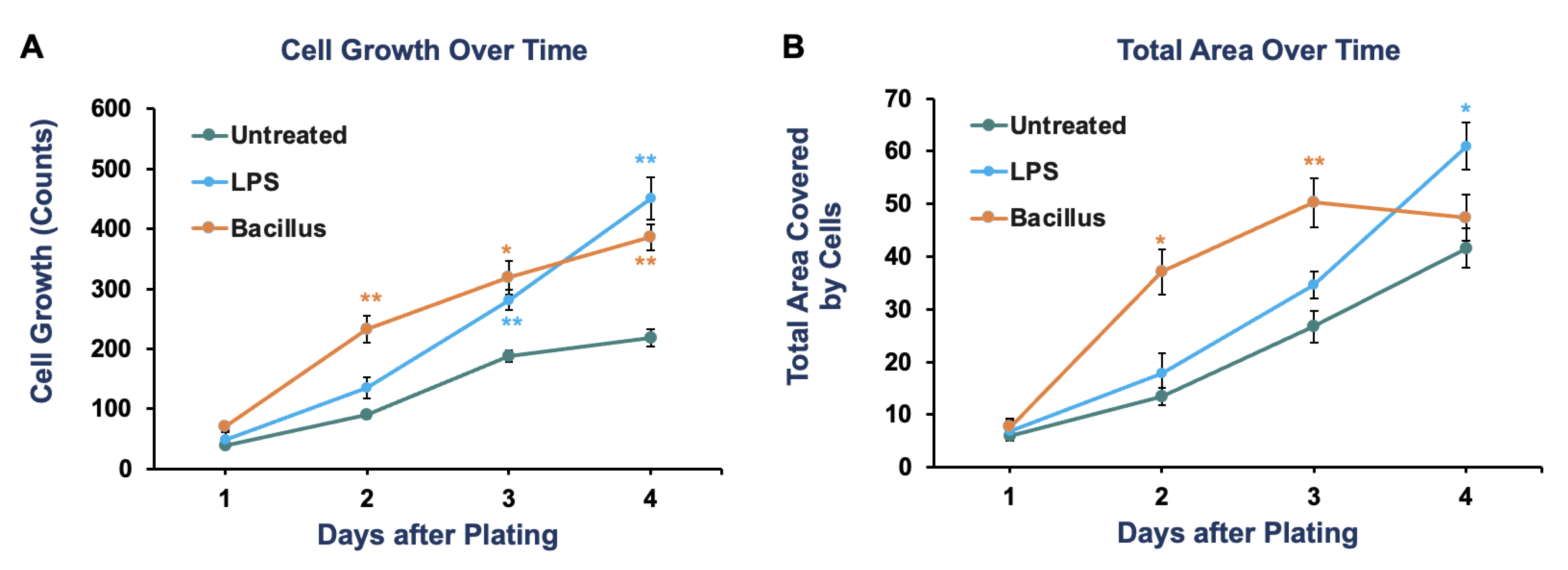
2.3. Quantification
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hartstra, A.V.; Bouter, K.E.; Bäckhed, F.; Nieuwdorp, M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care 2015, 38, 159–165. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.L.; O’Toole, P.W.; Shanahan, F. The Gut Microbiota in Causation, Detection, and Treatment of Cancer. Am. J. Gastroenterol. 2019, 114, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Galluzzi, L.; Viaud, S.; Vétizou, M.; Daillère, R.; Merad, M.; Kroemer, G. Cancer and the gut microbiota: An unexpected link. Sci. Transl. Med. 2015, 7, 271ps1. [Google Scholar] [CrossRef] [PubMed]
- Landman; Quevrain, E. Gut microbiota: Description, role and pathophysiologic implications. Rev. Med. Interne 2016, 37, 418–423. [Google Scholar] [CrossRef]
- Tibbs, T.N.; Lopez, L.R.; Arthur, J.C. The influence of the microbiota on immune development, chronic inflammation, and cancer in the context of aging. Microb. Cell 2019, 6, 324–334. [Google Scholar] [CrossRef]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- E Griffin, M.; Hespen, C.W.; Wang, Y.; Hang, H.C. Translation of peptidoglycan metabolites into immunotherapeutics. Clin. Transl. Immunol. 2019, 8, e1095. [Google Scholar] [CrossRef] [PubMed]
- Cancer microbiome. Cancer Cell 2022, 40, 1443–1447. [CrossRef]
- Ciernikova, S.; Sevcikova, A.; Stevurkova, V.; Mego, M. Tumor microbiome-an integral part of the tumor microenvironment. Front. Oncol. 2022, 12, 1063100. [Google Scholar] [CrossRef]
- Park, E.M.; Chelvanambi, M.; Bhutiani, N.; Kroemer, G.; Zitvogel, L.; Wargo, J.A. Targeting the gut and tumor microbiota in cancer. Nat. Med. 2022, 28, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Johnson, A.; Reimer, S.; Childres, R.; Cupp, G.; Kohs, T.C.L.; McCarty, O.J.T.; Kang, Y. The Applications and Challenges of the Development of In Vitro Tumor Microenvironment Chips. Cell. Mol. Bioeng. 2022, 16, 3–21. [Google Scholar] [CrossRef]
- Kang, Y.B.; Sodunke, T.R.; Lamontagne, J.; Cirillo, J.; Rajiv, C.; Bouchard, M.J.; Noh, M. Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol. Bioeng. 2015, 112, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.B.; Rawat, S.; Duchemin, N.; Bouchard, M.; Noh, M. Human Liver Sinusoid on a Chip for Hepatitis B Virus Replication Study. Micromachines 2017, 8, 27. [Google Scholar] [CrossRef]
- Imura, Y.; Asano, Y.; Sato, K.; Yoshimura, E. A microfluidic system to evaluate intestinal absorption. Anal Sci. 2009, 25, 1403–1407. [Google Scholar] [CrossRef]
- Gao, D.; Liu, H.; Lin, J.-M.; Wang, Y.; Jiang, Y. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab. Chip 2013, 13, 978–985. [Google Scholar] [CrossRef]
- Ramadan, Q.; Jing, L. Characterization of tight junction disruption and immune response modulation in a miniaturized Caco-2/U937 coculture-based in vitro model of the human intestinal barrier. Biomed. Microdevices 2016, 18, 11. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.-Y.; Lee, D.; Han, J.; Nguyen, N.-T.; Park, S.; Sung, J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 2017, 19, 37. [Google Scholar] [CrossRef]
- De Gregorio, V.; Corrado, B.; Sbrescia, S.; Sibilio, S.; Urciuolo, F.; Netti, P.; Imparato, G. Intestine-on-chip device increases ECM remodeling inducing faster epithelial cell differentiation. Biotechnol. Bioeng. 2020, 117, 556–566. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, J.; Choi, J.-H.; Bahinski, A.; Ingber, D.E. Co-culture of Living Microbiome with Microengineered Human Intestinal Villi in a Gut-on-a-Chip Microfluidic Device. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Strelez, C.; Ghaffarian, K.; Mumenthaler, S.M. Multiplexed imaging and effluent analysis to monitor cancer cell intravasation using a colorectal cancer-on-chip. STAR Protoc. 2021, 2, 100984. [Google Scholar] [CrossRef] [PubMed]
- Strelez, C.; Chilakala, S.; Ghaffarian, K.; Lau, R.; Spiller, E.; Ung, N.; Hixon, D.; Yoon, A.Y.; Sun, R.X.; Lenz, H.-J.; et al. Human colorectal cancer-on-chip model to study the microenvironmental influence on early metastatic spread. iScience 2021, 24, 102509. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Prantil-Baun, R.; Jiang, A.; Potla, R.; Mammoto, T.; Weaver, J.C.; Ferrante, T.C.; Kim, H.J.; Cabral, J.M.S.; Levy, O.; et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 2018, 9, 223. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Kim, H.J. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. USA 2018, 115, E10539–E10547. [Google Scholar] [CrossRef]
- Komen, J.; van Neerven, S.M.; Bossink, E.G.B.M.; de Groot, N.E.; Nijman, L.E.; Berg, A.V.D.; Vermeulen, L.; van der Meer, A.D. The Effect of Dynamic, In Vivo-like Oxaliplatin on HCT116 Spheroids in a Cancer-on-Chip Model Is Representative of the Response in Xenografts. Micromachines 2022, 13, 739. [Google Scholar] [CrossRef]
- Jeon, M.S.; Choi, Y.Y.; Mo, S.J.; Ha, J.H.; Lee, Y.S.; Lee, H.U.; Park, S.D.; Shim, J.-J.; Lee, J.-L.; Chung, B.G. Contributions of the microbiome to intestinal inflammation in a gut-on-a-chip. Nano Converg. 2022, 9, 8. [Google Scholar] [CrossRef]
- Mi, S.; Du, Z.; Xu, Y.; Wu, Z.; Qian, X.; Zhang, M.; Sun, W. Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening. Sci. Rep. 2016, 6, 35544. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.R.; Barata, D.; Teixeira, L.M.; Giselbrecht, S.; Reis, R.L.; Oliveira, J.M.; Truckenmüller, R.; Habibovic, P. Colorectal tumor-on-a-chip system: A 3D tool for precision onco-nanomedicine. Sci. Adv. 2019, 5, eaaw1317. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, V.; Sgambato, C.; Urciuolo, F.; Vecchione, R.; Netti, P.A.; Imparato, G. Immunoresponsive microbiota-gut-on-chip reproduces barrier dysfunction, stromal reshaping and probiotics translocation under inflammation. Biomaterials 2022, 286, 121573. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; Gresnigt, M.S.; Last, A.; Wollny, T.; Berlinghof, F.; Pospich, R.; Cseresnyes, Z.; Medyukhina, A.; Graf, K.; Gröger, M.; et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019, 220, 119396. [Google Scholar] [CrossRef]
- Virumbrales-Muñoz, M.; Ayuso, J.M.; Lacueva, A.; Ranđelović, T.; Livingston, M.K.; Beebe, D.J.; Oliván, S.; Pereboom, D.; Doblare, M.; Fernández, L.; et al. Enabling cell recovery from 3D cell culture microfluidic devices for tumour microenvironment biomarker profiling. Sci. Rep. 2019, 9, 6199. [Google Scholar] [CrossRef]
- Kang, Y.B.A.; Eo, J.; Mert, S.; Yarmush, M.L.; Usta, O.B. Metabolic Patterning on a Chip: Towards in vitro Liver Zonation of Primary Rat and Human Hepatocytes. Sci. Rep. 2018, 8, 8951. [Google Scholar] [CrossRef]
- Yang, Y.; Du, L.; Shi, D.; Kong, C.; Liu, J.; Liu, G.; Li, X.; Ma, Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat. Commun. 2021, 12, 6757. [Google Scholar] [CrossRef]
- Kang, Y.B.A.; Eo, J.; Bulutoglu, B.; Yarmush, M.L.; Usta, O.B. Progressive hypoxia-on-a-chip: An in vitro oxygen gradient model for capturing the effects of hypoxia on primary hepatocytes in health and disease. Biotechnol. Bioeng. 2020, 117, 763–775. [Google Scholar] [CrossRef]
- Panaro, M.A.; Carofiglio, V.; Acquafredda, A.; Cavallo, P.; Cianciulli, A. Anti-inflammatory effects of resveratrol occur via inhibition of lipopolysaccharide-induced NF-kappaB activation in Caco-2 and SW480 human colon cancer cells. Br. J. Nutr. 2012, 108, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.D.; Jiang, M.Z.; Zhao, Y.Y.; Li, X.; Lan, H.; Yang, W.Q.; Lai, Y. Effects of breviscapine on cerebral ischemia-reperfusion injury and intestinal flora imbalance by regulating the TLR4/MyD88/NF-kappaB signaling pathway in rats. J. Ethnopharmacol. 2023, 300, 115691. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mittal, E.; Cupp, G.; Kang, Y. Simulating the Effect of Gut Microbiome on Cancer Cell Growth Using a Microfluidic Device. Sensors 2023, 23, 1265. https://doi.org/10.3390/s23031265
Mittal E, Cupp G, Kang Y. Simulating the Effect of Gut Microbiome on Cancer Cell Growth Using a Microfluidic Device. Sensors. 2023; 23(3):1265. https://doi.org/10.3390/s23031265
Chicago/Turabian StyleMittal, Ekansh, Grace Cupp, and Youngbok (Abraham) Kang. 2023. "Simulating the Effect of Gut Microbiome on Cancer Cell Growth Using a Microfluidic Device" Sensors 23, no. 3: 1265. https://doi.org/10.3390/s23031265
APA StyleMittal, E., Cupp, G., & Kang, Y. (2023). Simulating the Effect of Gut Microbiome on Cancer Cell Growth Using a Microfluidic Device. Sensors, 23(3), 1265. https://doi.org/10.3390/s23031265




