Advancing Brain Tumor Classification through Fine-Tuned Vision Transformers: A Comparative Study of Pre-Trained Models
Abstract
:1. Introduction
- ViT models can be effectively used for brain tumor image classification, even without any task-specific training.
- Fine-tuning of ViT models can significantly improve their performance on brain tumor images.
- A comprehensive evaluation of ViT models for brain tumor image classification, comparing them to existing approaches.
- The proposed models have the potential to improve the accuracy and efficiency of brain tumor image classification, which could lead to earlier diagnosis and treatment for patients.
2. Materials and Methods Section
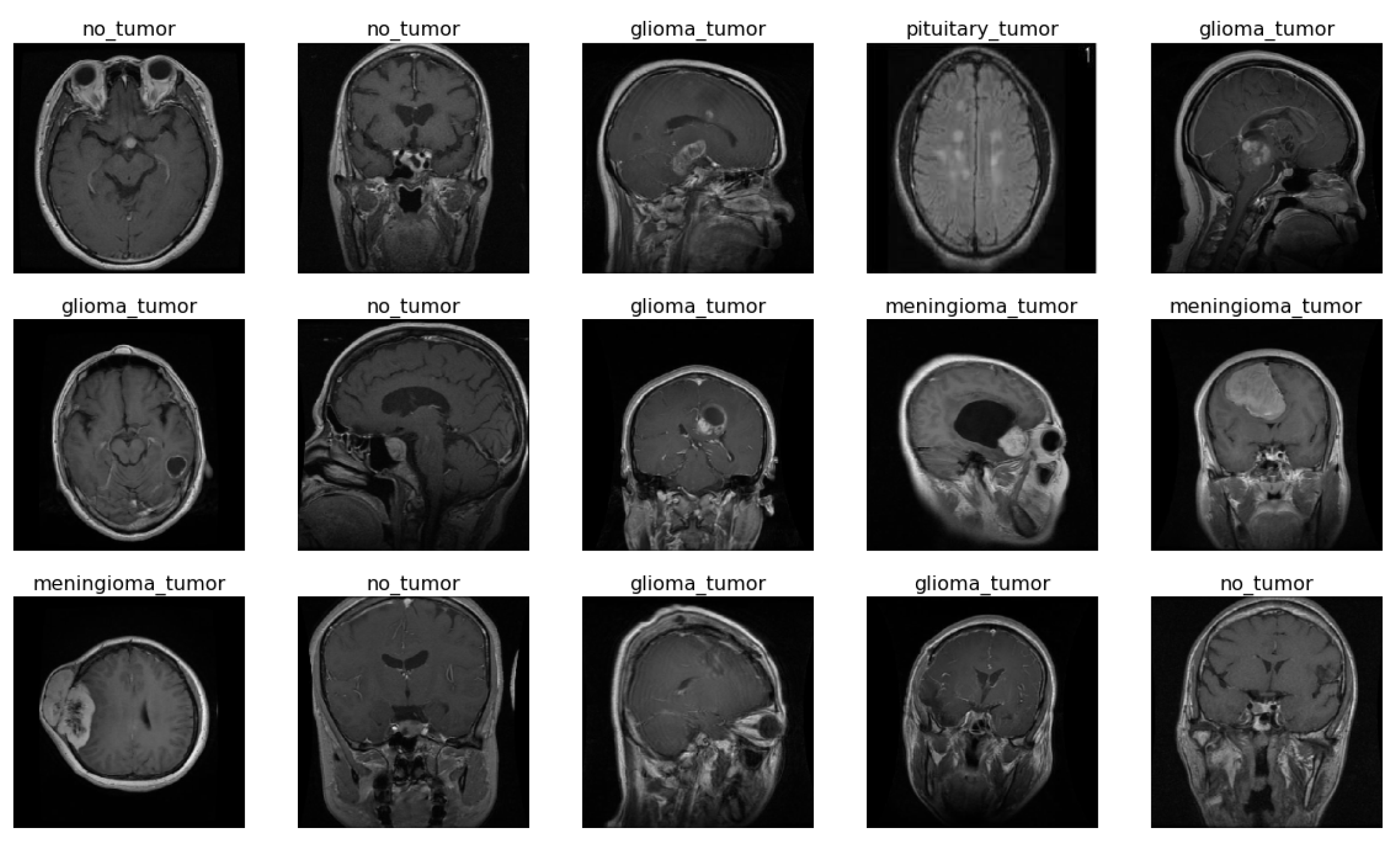
2.1. Dataset
2.2. Data Pre-Processing and Augmentation
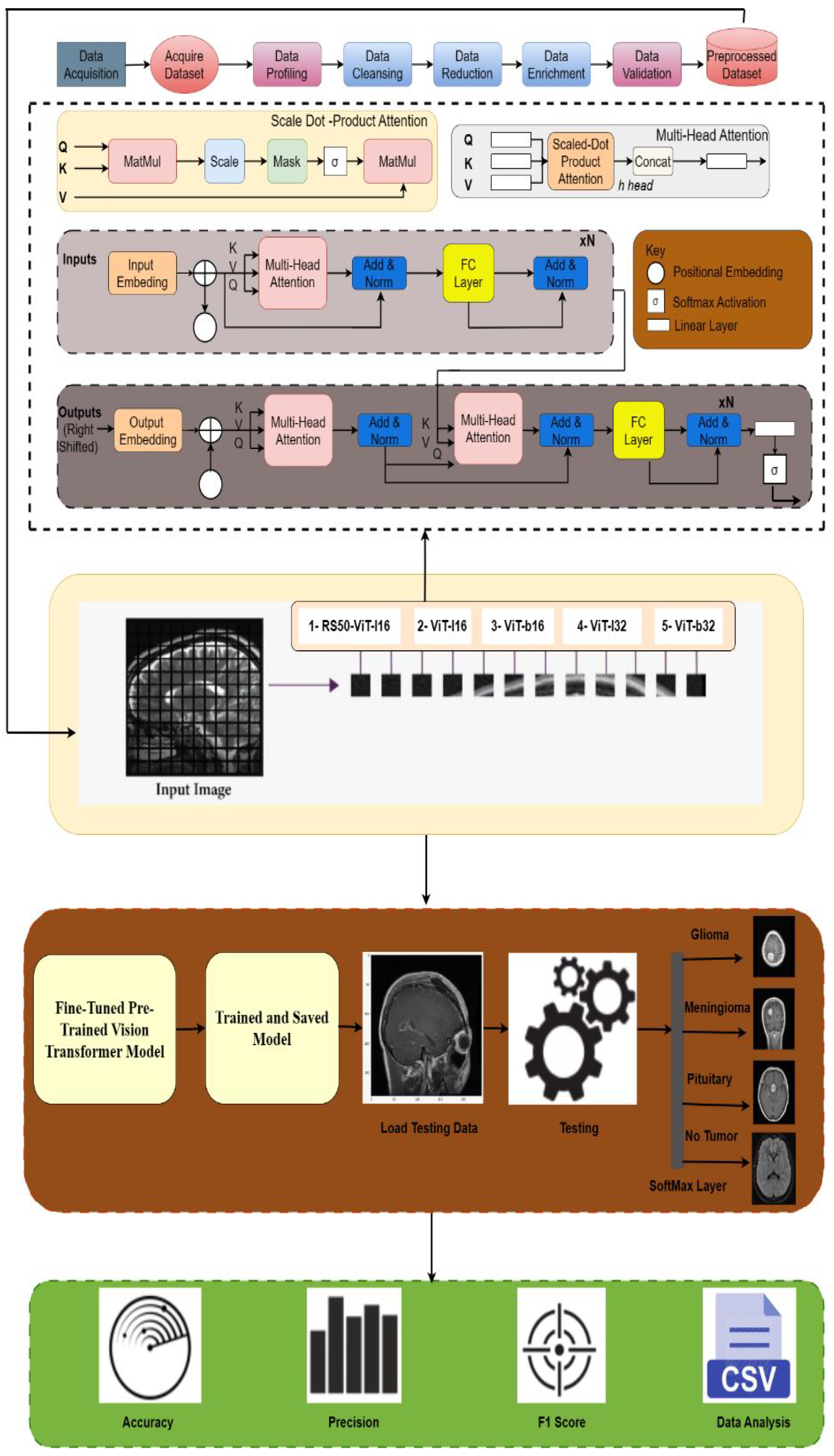
2.3. Vision Transformer Architecture
2.3.1. Key Components of ViT Architecture
2.3.2. Pre-Trained ViT Models
- R50-ViT-L16
- ViT-b16
- ViT-l16
- ViT-l32
- ViT-b32
2.3.3. Fine-Tuning of Pre-Trained Models
2.3.4. R50-ViT-l16 Model
2.3.5. ViT-b16 Model
2.3.6. ViT-l16 Model
2.3.7. ViT-l32 Model
2.3.8. ViT-b32 Model
3. Result
- NumPy is a library developed for effectively managing large-scale, multi-dimensional arrays and matrices containing numerical data. Its significance extends to image processing and tasks within the domain of machine learning.
- OpenCV serves as a specialized library dedicated to image processing and computer vision. It encompasses a diverse array of functions enabling tasks like image reading, writing, display, and manipulation.
- Scikit-learn emerges as a machine-learning library equipped with a variety of algorithms tailor-made for tasks encompassing image classification.
- Keras operates as a high-level neural network API meticulously crafted for Python and constructed atop the TensorFlow framework. It simplifies the creation and training of neural networks, particularly those designed for image classification purposes.
- ViT-keras is a library for using ViT models for image classification in Keras. ViT models are a type of neural network designed to be more efficient than CNNs for image classification.
- TensorFlow Hub is a library for using pre-trained TensorFlow models in Keras. It makes using pre-trained models for image classification easy without retraining them.
- ImageDataGenerator from Keras is a library that can perform data augmentation on images. This helps to improve the performance of models by preventing overfitting.
- panda is a library for data analysis in Python. It makes it easy to load, manipulate, and analyze data in a tabular format.
- sklearn.model_selection is a library for performing model selection in Python. It can be used to split data into a training set and a test set and evaluate models’ performances.
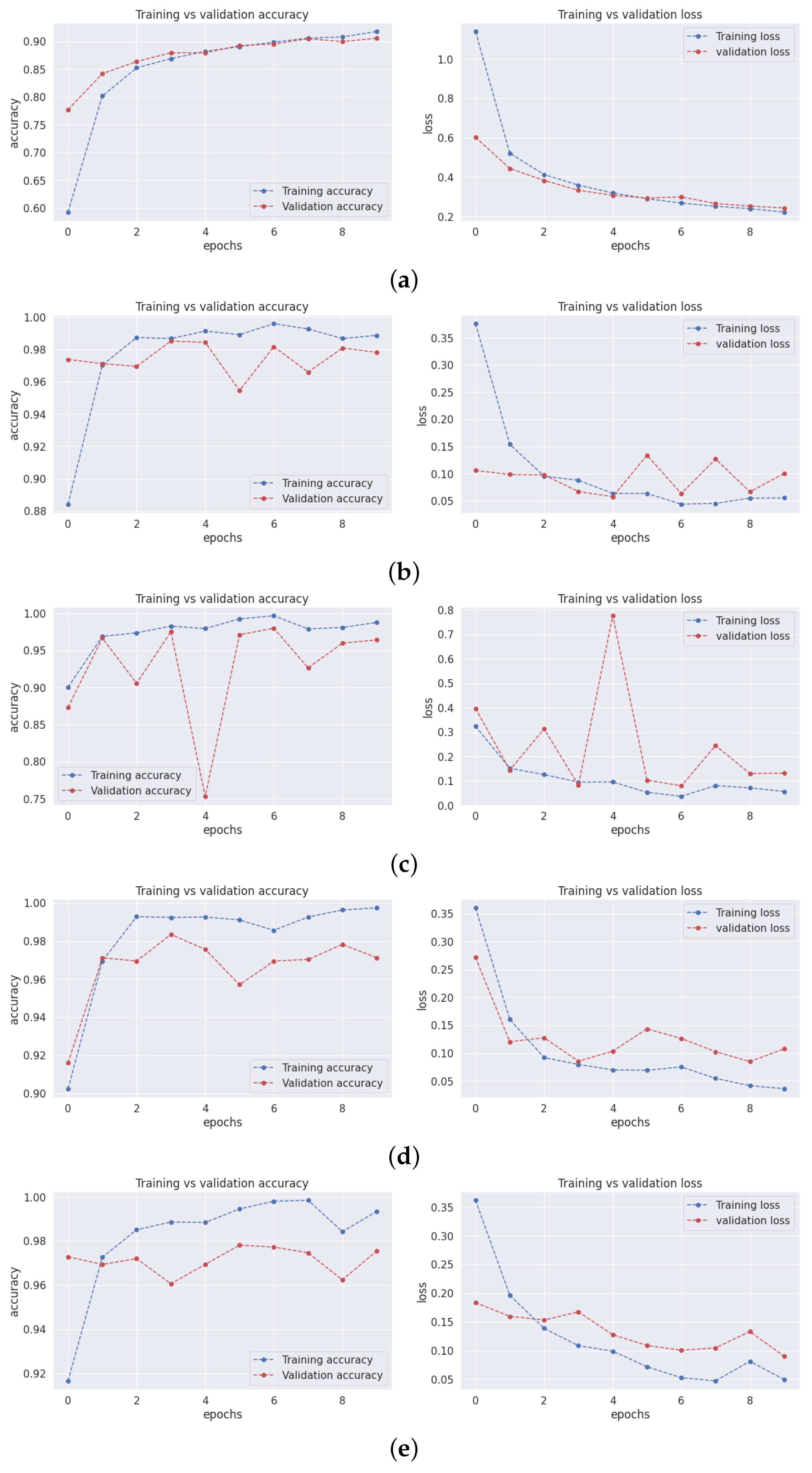
3.1. Training and Validation Accuracy and Loss
3.2. Confusion Matrix
- True Positive (TePe): This quadrant accounts for instances where the model accurately predicts the positive class.
- True Negative (TeNe): In this quadrant, the model precisely forecasts the negative class.
- False Positive (FePe): This quadrant pertains to situations where the model predicts the positive class, although the actual class is negative.
- False Negative (FeNe): This quadrant arises when the model forecasts the negative class, but the genuine class is positive.
- R50-ViT-l16
- −
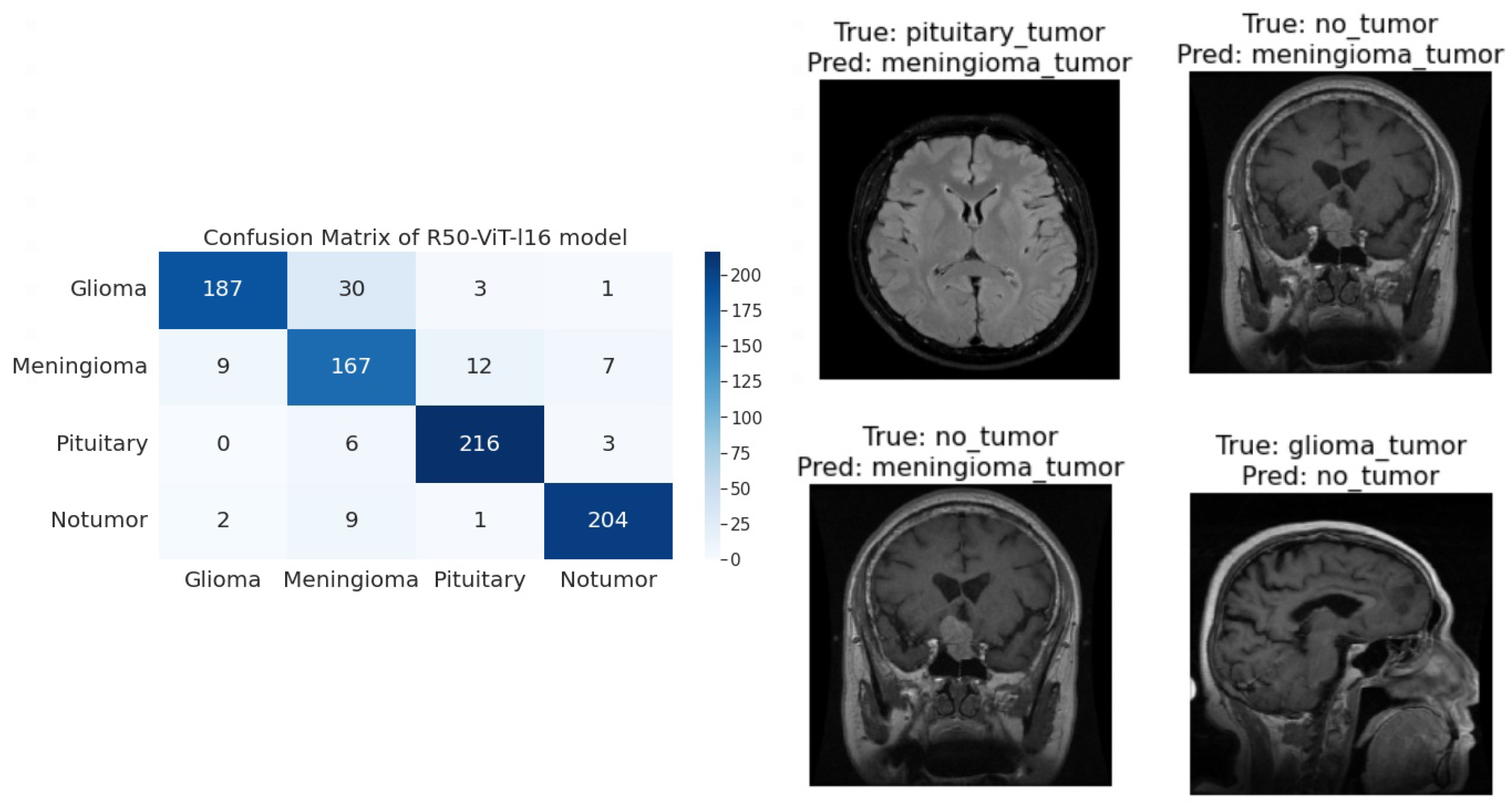
- For Glioma tumor, the model correctly classified 187 images with an accuracy of 85.25%. The model also misclassified 30 Glioma images as Meningioma, 3 Glioma images as Pituitary, and 1 Glioma image as No Tumor.
- −
- For Meningioma tumor, the model correctly classified 167 Meningioma images, which is an accuracy of 85.58%. The model also misclassified 9 Meningioma images as Glioma, 12 Meningioma images as Pituitary, and 7 Meningioma images as No Tumor.
- −
- For Pituitary tumors, the model correctly classified 216 Pituitary images, with an accuracy of 96%. The model also misclassified six Pituitary images as Glioma, three Pituitary images as Meningioma, and three Pituitary images as No Tumor.
- −
- For the No Tumor class, the model correctly classified 204 No Tumor images, with an accuracy of 94.74%. The model also misclassified two Tumor images as Glioma, nine No Tumor images as Meningioma, and one No Tumor image as Pituitary.
- −
- Overall, the model has an accuracy of 90.31%. Its confusion matrix and incorrect classification images are shown in Figure 4. The model performs the best for Pituitary, with an accuracy of 96%. The model performs the worst for Glioma, with an accuracy of 85.25%.
- ViT-b16
- −
- For the Glioma tumor, the model correctly classified 217 images, with an accuracy of 98.19%. The model also misclassified three Glioma images as Meningioma, one Glioma image as Pituitary, and one Glioma images as No Tumor.
- −
- For Meningioma tumor, the model correctly classified 182 Meningioma images, with an accuracy of 93.33%. The model also misclassified seven Meningioma images as Glioma, three Meningioma images as Pituitary, and three Meningioma images as No Tumor.
- −
- For Pituitary tumors, the model correctly classified 224 Pituitary images, with an accuracy of 99.56%. The model also misclassified one Pituitary image as Glioma, zero Pituitary images as Meningioma, and zero Pituitary images as No Tumor.
- −
- For the No Tumor class, the model correctly classified 216 No Tumor images, which is an accuracy of 100%. The model also misclassified zero No Tumor images as Glioma, zero No Tumor images as Meningioma, and zero No Tumor images as Pituitary.
- −
- Overall, the model has an accuracy of 97.89%. Its confusion matrix and incorrect classification images are shown in Figure 5. The model performs best for No Tumor, with an accuracy of 100%. The model performs worst for Meningioma, with an accuracy of 93.33%.
- ViT-l16
- −
- For Glioma tumor, the model correctly classified 215 images as Glioma, with an accuracy of 97.73%. The model misclassified six Glioma images as Meningioma, zero Glioma images as Pituitary, and zero Glioma images as No Tumor.
- −
- For Meningioma tumor, the model correctly classified 189 images as Meningioma, achieving an accuracy of 94.50%. The model misclassified two Meningioma images as Glioma, zero Meningioma images as Pituitary, and four Meningioma images as No Tumor.
- −
- For Pituitary tumor, the model correctly classified 225 images as Pituitary, resulting in a perfect accuracy of 100%. The model did not misclassify any Pituitary images as other classes.
- −
- For the No Tumor class, the model correctly classified 203 images as No Tumor, with an accuracy of 94.88%. The model misclassified 1 Tumor image as Glioma and 12 Tumor images as Meningioma.
- −
- Overall, the model achieved an accuracy of approximately 97.08%. Its confusion matrix and incorrect classification images are shown in Figure 6. It performed best for the Pituitary tumor with a perfect accuracy of 100%, while the lowest accuracy was observed for the Meningioma tumor with an accuracy of 94.50%.
- ViT-l32
- −
- For Glioma tumor, the model correctly classified 200 images as Glioma, with an accuracy of 89.60%. The model misclassified 2 Glioma images as Meningioma, 16 Glioma images as Pituitary, and 3 Glioma images as No Tumor.
- −
- For Meningioma tumor, the model correctly classified 169 images as Meningioma, achieving an accuracy of 89.47%. The model misclassified 7 Meningioma images as Glioma, 18 Meningioma images as Pituitary, and 1 Meningioma image as No Tumor.
- −
- For Pituitary tumor, the model correctly classified 225 images as Pituitary, resulting in a perfect accuracy of 100%. The model did not misclassify any Pituitary images as other classes.
- −
- For No Tumor class, The model correctly classified 216 images as No Tumor, with an accuracy of 100%. The model did not misclassify any No Tumor images as other classes.
- −
- Overall, the model achieved an accuracy of approximately 94.51%. Its confusion matrix and incorrect classification images are shown in Figure 7. It performed the best for Pituitary and No Tumor, with perfect accuracies of 100%, while the lowest accuracy was observed for Glioma with an accuracy of 89.47%.
- ViT-B32
- −
- For Glioma tumor, the model correctly classified 214 images as Glioma, an accuracy of 96.39%. The model misclassified seven Glioma images as Meningioma, zero Glioma images as Pituitary, and zero Glioma images as No Tumor.
- −
- For Meningioma tumor, the model correctly classified 191 images as Meningioma, achieving an accuracy of 96.43%. The model misclassified one Meningioma image as Glioma, one Meningioma image as Pituitary, and two Meningioma images as No Tumor.
- −
- For Pituitary tumor, the model correctly classified 224 images as Pituitary, resulting in a perfect accuracy of 100%. The model misclassified one Pituitary image as Glioma, zero Pituitary image as Meningioma, and zero Pituitary images as No Tumor.
- −
- For the No Tumor class, the model correctly classified 213 images as No Tumor, with an accuracy of 98.61%. The model misclassified three Tumor images as Glioma, zero No Tumor images as Meningioma, and zero No Tumor images as Pituitary.
- −
- Overall, the model achieved an accuracy of approximately 98.24%. Its confusion matrix and incorrect classification images are shown in Figure 8. it performed the best for Pituitary with perfect accuracies of 100%, while the lowest accuracy was observed for Glioma with an accuracy of 96.39%.
3.3. Statistical Values of All Classes
- Precision (Pn) Pn signifies the fraction of predicted positive instances that are truly positive.
- Recall (Rl) Rl denotes the fraction of actual positive instances that are correctly predicted as positive.
- F1 Score (Fe) The Fe is a harmonic mean of Pn and Rl, providing a measure of the model’s accuracy. A perfect Fe is 1.0, while a score of 0.0 is undesirable.
- Support (St) St corresponds to the number of instances in a class.
3.4. Comparison with Existing Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vankdothu, R.; Hameed, M.A.; Fatima, H. A brain tumor identification and classification using deep learning based on CNN-LSTM method. Comput. Electr. Eng. 2022, 101, 107960. [Google Scholar] [CrossRef]
- Asiri, A.A.; Ali, T.; Shaf, A.; Aamir, M.; Shoaib, M.; Irfan, M. A Novel Inherited Modeling Structure of Automatic Brain Tumor Segmentation from MRI. Comput. Mater. Contin. 2022, 73, 3983–4002. [Google Scholar] [CrossRef]
- Rosa, S.L.; Uccella, S. Pituitary tumors: Pathology and genetics. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Asiri, A.A.; Shaf, A.; Ali, T.; Aamir, M.; Irfan, M.; Alqahtani, S.; Mehdar, K.M.; Halawani, H.T.; Alghamdi, A.H.; Alshamrani, A.F.A.; et al. Brain tumor detection and classification using fine-tuned CNN with ResNet50 and U-Net model: A study on TCGA-LGG and TCIA dataset for MRI applications. Life 2023, 13, 1449. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.; Chakrabarty, A.; Gadekallu, T.R.; Alazab, M.; Piran, M.J. ViTs, ensemble model, and transfer learning leveraging Explainable AI for brain tumor detection and classification. IEEE J. Biomed. Health Inform. 2023. [Google Scholar] [CrossRef] [PubMed]
- Asif, S.; Wenhui, Y.; Jinhai, S.; Ain, Q.U.; Yueyang, Y.; Jin, H. Modeling a fine-tuned deep convolutional neural network for diagnosis of kidney diseases from CT images. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022. [Google Scholar]
- Tummala, S.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Classification of brain tumor from magnetic resonance imaging using ViTs ensembling. Curr. Oncol. 2022, 29, 7498–7511. [Google Scholar] [CrossRef]
- Ammari, S.; Pitre-Champagnat, S.; Dercle, L.; Chouzenoux, E.; Moalla, S.; Reuze, S.; Talbot, H.; Mokoyoko, T.; Hadchiti, J.; Diffetocq, S.; et al. Influence of magnetic field strength on magnetic resonance imaging radiomics features in brain imaging, an in vitro and in vivo study. Front. Oncol. 2021, 10, 541663. [Google Scholar] [CrossRef]
- Brindle, K.M.; Izquierdo-García, J.L.; Lewis, D.Y.; Mair, R.J.; Wright, A.J. Brain tumor imaging. J. Clin. Oncol. 2017, 35, 2432–2438. [Google Scholar] [CrossRef]
- Salmon, E.; Ir, C.B.; Hustinx, R. Pitfalls and limitations of PET/CT in brain imaging. In Seminars in Nuclear Medicine; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Vijithananda, S.M.; Jayatilake, M.L.; Hewavithana, B.; Gonçalves, T.; Rato, L.M.; Weerakoon, B.S.; Kalupahana, T.D.; Silva, A.D.; Dissanayake, K.D. Feature extraction from MRI ADC images for brain tumor classification using machine learning techniques. Biomed. Eng. Online 2022, 21, 52. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef]
- Thaha, M.M.; Kumar, K.P.M.; Murugan, B.S.; Dhanasekeran, S.; Vijayakarthick, P.; Selvi, A.S. Brain tumor segmentation using convolutional neural networks in MRI images. J. Med. Syst. 2019, 43, 294. [Google Scholar] [CrossRef]
- Arif, M.; Ajesh, F.; Shamsudheen, S.; Geman, O.; Izdrui, D.; Vicoveanu, D. Brain tumor detection and classification by MRI using biologically inspired orthogonal wavelet transform and deep learning techniques. J. Healthc. Eng. 2022, 2022, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Shu, H. Bitr-unet: A cnn-transformer combined network for mri brain tumor segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 7th International Workshop, BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, 27 September 2021; Part II. pp. 3–14. [Google Scholar]
- Kim, S.; Süsstrunk, S.; Salzmann, M. Volumetric Transformer Networks. arXiv 2020. [Google Scholar]
- Rajasree, R.; Columbus, C.C.; Shilaja, C. Multiscale-based multimodal image classification of brain tumor using deep learning method. Neural Comput. Appl. 2021, 33, 5543–5553. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Haldorai, A.; Yasmin, M.; Nayak, R.S. Brain tumor detection and classification using machine learning: A comprehensive survey. Complex Intell. Syst. 2022, 8, 3161–3183. [Google Scholar] [CrossRef]
- Vaswani, A. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Liu, Z.; Tong, L.; Chen, L.; Jiang, Z.; Zhou, F.; Zhang, Q.; Zhang, X.; Jin, Y.; Zhou, H. Deep learning based brain tumor segmentation: A survey. Complex Intell. Syst. 2023, 9, 1001–1026. [Google Scholar] [CrossRef]
- Huang, Z.; Du, X.; Chen, L.; Li, Y.; Liu, M.; Chou, Y.; Jin, L. Convolutional neural network based on complex networks for brain tumor image classification with a modified activation function. IEEE Access 2020, 8, 89281–89290. [Google Scholar] [CrossRef]
- Asiri, A.A.; Aamir, M.; Shaf, A.; Ali, T.; Zeeshan, M.; Irfan, M.; Alshamrani, K.A.; Alshamrani, H.A.; Alqahtani, F.F.; Alshehri, A.H. Block-Wise Neural Network for Brain Tumor Identification in Magnetic Resonance Images. Comput. Mater. Contin. 2022, 73, 5735–5753. [Google Scholar] [CrossRef]
- Muezzinoglu, T.; Baygin, N.; Tuncer, I.; Barua, P.D.; Baygin, M.; Dogan, S.; Tuncer, T.; Palmer, E.E.; Cheong, K.H.; Acharya, U.R. PatchResNet: Multiple patch division-based deep feature fusion framework for brain tumor classification using MRI images. J. Digit. Imaging 2023, 36, 973–987. [Google Scholar] [CrossRef]
- Aggarwal, M.; Tiwari, A.K.; Sarathi, M.P.; Bijalwan, A. An early detection and segmentation of Brain Tumor using Deep Neural Network. BMC Med. Inform. Decis. Mak. 2023, 23, 78. [Google Scholar] [CrossRef]
- Asiri, A.A.; Shaf, A.; Ali, T.; Aamir, M.; Usman, A.; Irfan, M.; Alshamrani, H.A.; Mehdar, K.M.; Alshehri, O.M.; Alqhtani, S.M. Multi-Level Deep Generative Adversarial Networks for Brain Tumor Classification on Magnetic Resonance Images. Intell. Autom. Soft Comput. 2023, 36, 127–143. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Asiri, A.A.; Shaf, A.; Ali, T.; Shakeel, U.; Irfan, M.; Mehdar, K.M.; Halawani, H.T.; Alghamdi, A.H.; Alshamrani, A.F.A.; Alqhtani, S.M. Exploring the power of deep learning: Fine-tuned ViT for accurate and efficient brain tumor detection in MRI scans. Diagnostics 2023, 13, 2094. [Google Scholar] [CrossRef] [PubMed]
- Toğaçar, M.; Ergen, B.; Cömert, Z. Tumor type detection in brain MR images of the deep model developed using hypercolumn technique, attention modules, and residual blocks. Med. Biol. Eng. Comput. 2021, 59, 57–70. [Google Scholar] [CrossRef]




| Class Name | Number of Images | Training | Testing |
|---|---|---|---|
| Glioma | 1321 | 1100 | 221 |
| Meningioma | 1339 | 1144 | 195 |
| No tumor | 1595 | 1241 | 216 |
| Pituitary | 1457 | 1370 | 225 |
| Total | 5712 | 4855 | 857 |
| (a) Model Vision_Transformer R50-vit-l16 | ||
|---|---|---|
| Layer (Type) | Output Shape | Parameters |
| keras_layer (KerasLayer) | (None, 2048) | 23,500,352 |
| Flatten | (None, 2048) | 0 |
| batch_normalization | (None, 2048) | 8192 |
| Dense | (None, 11) | 22,539 |
| batch_normalizatio | (None, 11) | 44 |
| Dense | (None, 4) | 48 |
| (b) Model Vision_Transformer vit_b16 | ||
| Layer (Type) | Output Shape | Parameters |
| vit-b16 (Functional) | (None, 768) | 85,798,656 |
| Flatten | (None, 768) | 0 |
| batch_normalization | (None, 768) | 3072 |
| Dense | (None, 11) | 8459 |
| batch_normalization | (None, 11) | 44 |
| Dense | (None, 4) | 48 |
| (c) Model Vision_Transformer vit_l16 | ||
| Layer (Type) | Output Shape | Parameters |
| vit-l16 (Functional) | (None, 1024) | 303,301,632 |
| Flatten | (None, 1024) | 0 |
| batch_normalization | (None, 1024) | 4096 |
| Dense | (None, 11) | 11,275 |
| batch_normalization | (None, 11) | 44 |
| Dense | (None, 4) | 48 |
| (d) Model Vision_Transformer vit_l32 | ||
| Layer (Type) | Output Shape | Parameters |
| vit-l32 (Functional) | (None, 1024) | 305,510,400 |
| Flatten | (None, 1024) | 0 |
| batch_normalization | (None, 1024) | 4096 |
| Dense | (None, 11) | 11,275 |
| batch_normalization | (None, 11) | 44 |
| Dense | (None, 4) | 48 |
| (e) Model Vision_Transformer vit_b32 | ||
| Layer (Type) | Output Shape | Parameters |
| vit-b32 (Functional) | (None, 768) | 87,455,232 |
| Flatten | (None, 768) | 0 |
| batch_normalization | (None, 768) | 3072 |
| Dense | (None, 11) | 8459 |
| batch_normalization | (None, 11) | 44 |
| Dense | (None, 4) | 48 |
| Model | Patch Size | Backbone | Hidden Units | ImageNet Accuracy |
|---|---|---|---|---|
| R50-ViT-l16 | 16 × 16 | ResNet-50 | 2048 | 92.50% |
| ViT-b16 | 16 × 16 | ViT-B16 | 1024 | 90.20% |
| ViT-l16 | 16 × 16 | ViT-L16 | 1280 | 92.30% |
| ViT-l32 | 16 × 16 | ViT-L32 | 2560 | 93.50% |
| ViT-b32 | 32 × 32 | ViT-B32 | 2048 | 91.10% |
| Model | Training Accuracy | Validation Accuracy | Training Loss | Validation Loss |
|---|---|---|---|---|
| R50-ViT-l16 | 0.98 | 0.90 | 0.03 | 0.05 |
| ViT-b16 | 0.99 | 0.97 | 0.01 | 0.10 |
| ViT-l16 | 0.98 | 0.97 | 0.08 | 0.15 |
| ViT-l32 | 1.00 | 0.94 | 0.04 | 0.12 |
| ViT-b32 | 0.98 | 0.98 | 0.04 | 0.12 |
| Tumor Category | R50-ViT-l16 | ViT_b16 | ViT_l16 | ViT_l32 | ViT_b32 |
|---|---|---|---|---|---|
| Pn: 0.94 | Pn: 0.97 | Pn: 0.99 | Pn: 0.97 | Pn: 0.99 | |
| Glioma Tumor | Rl: 0.85 | Rl: 0.98 | Rl: 0.97 | Rl: 0.90 | Rl: 0.97 |
| Fe: 0.89 | Fe: 0.98 | Fe: 0.98 | Fe: 0.93 | Fe: 0.98 | |
| Pn: 0.79 | Pn: 0.98 | Pn: 0.91 | Pn: 0.99 | Pn: 0.95 | |
| Meningioma Tumor | Rl: 0.86 | Rl: 0.93 | Rl: 0.97 | Rl: 0.87 | Rl: 0.98 |
| Fe: 0.82 | Fe: 0.96 | Fe: 0.94 | Fe: 0.92 | Fe: 0.96 | |
| Pn: 0.92 | Pn: 0.98 | Pn: 1.00 | Pn: 0.86 | Pn: 1.00 | |
| No Tumor | Rl: 0.94 | Rl: 1.00 | Rl: 0.94 | Rl: 1.00 | Rl: 0.99 |
| Fe: 0.93 | Fe: 0.99 | Fe: 0.97 | Fe: 0.93 | Fe: 0.99 | |
| Pn: 0.96 | Pn: 0.99 | Pn: 0.98 | Pn: 0.98 | Pn: 0.99 | |
| Pituitary Tumor | Rl: 0.96 | Rl: 1.00 | Rl: 1.00 | Rl: 1.00 | Rl: 1.00 |
| Fe: 0.96 | Fe: 0.99 | Fe: 0.99 | Fe: 0.99 | Fe: 0.99 | |
| Overall Accuracy | 90.31% | 97.89% | 97.08% | 94.51% | 98.24% |
| No | Classification Method | Accuracy |
|---|---|---|
| [21] | CCN based on complex networks | 95.49% |
| [22] | BW-VGG-19 | 98% |
| [23] | PatchResNet | 98.10% |
| [24] | Improved ResNet | 91.30% |
| [25] | GAN | 96% |
| [26] | GIST descriptor and ELM | 94.93% |
| [27] | FT-Vit | 98.13% |
| [28] | Hypercolumn technique, and Residual block | 97.69% |
| Our Proposed Approach | R50-ViT-l16 | 90.31% |
| ViT-b16 | 97.89% | |
| ViT-l16 | 97.08% | |
| ViT-l32 | 94.51% | |
| ViT-b32 | 98.24% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asiri, A.A.; Shaf, A.; Ali, T.; Pasha, M.A.; Aamir, M.; Irfan, M.; Alqahtani, S.; Alghamdi, A.J.; Alghamdi, A.H.; Alshamrani, A.F.A.; et al. Advancing Brain Tumor Classification through Fine-Tuned Vision Transformers: A Comparative Study of Pre-Trained Models. Sensors 2023, 23, 7913. https://doi.org/10.3390/s23187913
Asiri AA, Shaf A, Ali T, Pasha MA, Aamir M, Irfan M, Alqahtani S, Alghamdi AJ, Alghamdi AH, Alshamrani AFA, et al. Advancing Brain Tumor Classification through Fine-Tuned Vision Transformers: A Comparative Study of Pre-Trained Models. Sensors. 2023; 23(18):7913. https://doi.org/10.3390/s23187913
Chicago/Turabian StyleAsiri, Abdullah A., Ahmad Shaf, Tariq Ali, Muhammad Ahmad Pasha, Muhammad Aamir, Muhammad Irfan, Saeed Alqahtani, Ahmad Joman Alghamdi, Ali H. Alghamdi, Abdullah Fahad A. Alshamrani, and et al. 2023. "Advancing Brain Tumor Classification through Fine-Tuned Vision Transformers: A Comparative Study of Pre-Trained Models" Sensors 23, no. 18: 7913. https://doi.org/10.3390/s23187913
APA StyleAsiri, A. A., Shaf, A., Ali, T., Pasha, M. A., Aamir, M., Irfan, M., Alqahtani, S., Alghamdi, A. J., Alghamdi, A. H., Alshamrani, A. F. A., Alelyani, M., & Alamri, S. (2023). Advancing Brain Tumor Classification through Fine-Tuned Vision Transformers: A Comparative Study of Pre-Trained Models. Sensors, 23(18), 7913. https://doi.org/10.3390/s23187913











