A Parallel Classification Model for Marine Mammal Sounds Based on Multi-Dimensional Feature Extraction and Data Augmentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Description
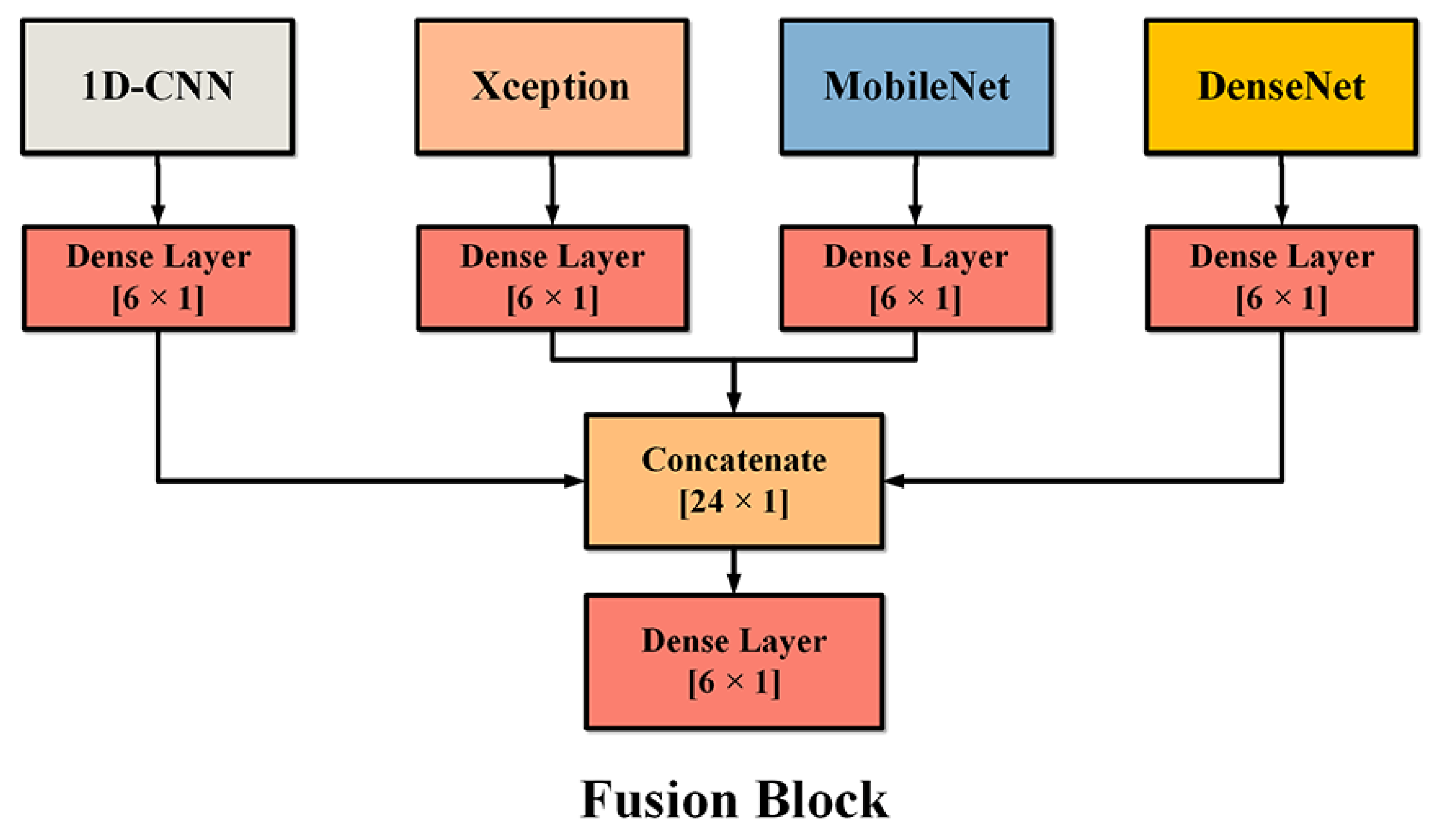
2.2. MDF-PNet Structure
2.3. Data Augmentation Method
2.4. Feature Extraction Algorithm
2.5. Transfer-Learning Method
2.6. Data Segmentation
3. Results
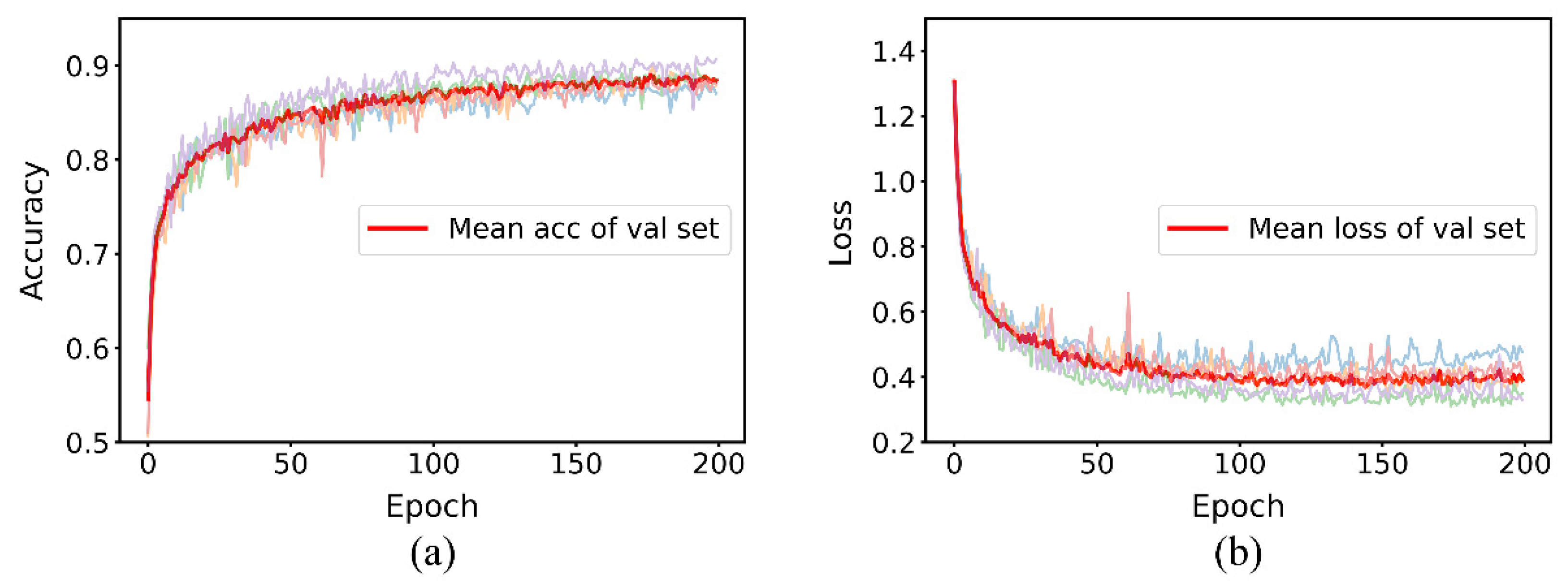
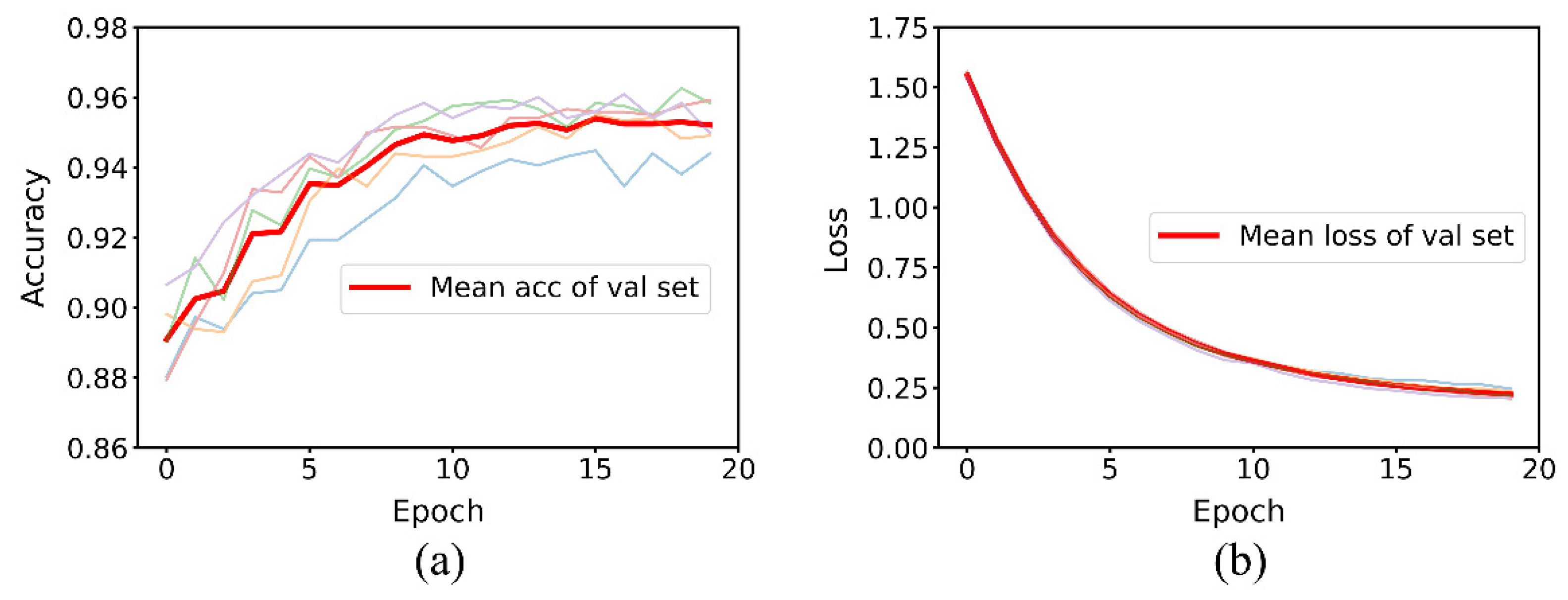
3.1. Performance Comparison between Baseline and MDF-PNet
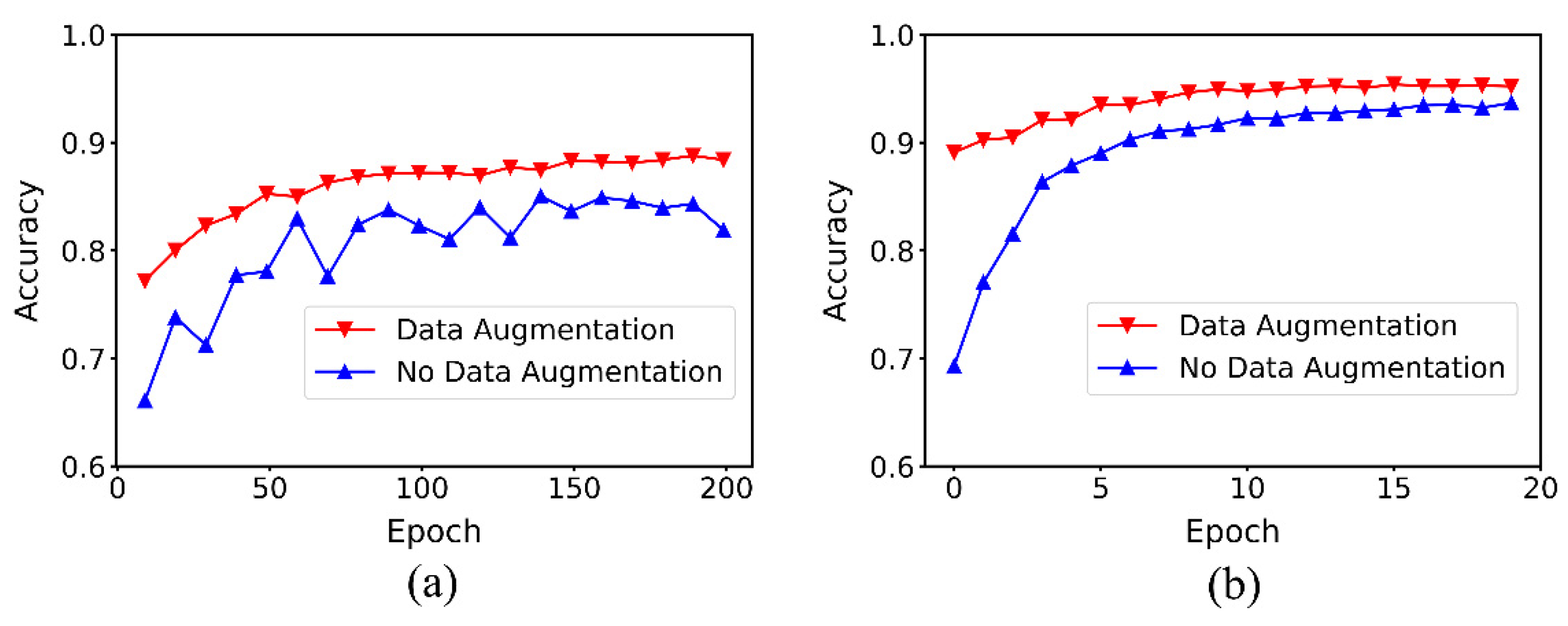
3.2. Effect of Data Augmentation on Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Williams, R.; Wright, A.J.; Ashe, E.; Blight, L.K.; Bruintjes, R.; Canessa, R.; Clark, C.W.; Cullis-Suzuki, S.; Dakin, D.T.; Erbe, C.; et al. Impacts of anthropogenic noise on marine life: Publication patterns, new discoveries, and future directions in research and management. Ocean. Coast. Manag. 2015, 115, 17–24. [Google Scholar] [CrossRef]
- Merchant, N.D.; Brookes, K.L.; Faulkner, R.C.; Bicknell, A.W.J.; Godley, B.J.; Witt, M.J. Underwater noise levels in UK waters. Sci. Rep. 2016, 6, 36942. [Google Scholar] [CrossRef]
- Blair, H.B.; Merchant, N.D.; Friedlaender, A.S.; Wiley, D.N.; Parks, S.E. Evidence for ship noise impacts on humpback whale foraging behaviour. Biol. Lett. 2016, 12, 20160005. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.A.; Hildebrand, J.A.; Wiggins, S.M. Increases in deep ocean ambient noise in the Northeast Pacific west of San Nicolas Island, California. J. Acoust. Soc. Am. 2006, 120, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Weilgart, L.S. A brief review of known effects of noise on marine mammals. Int. J. Comp. Psychol. 2007, 20, 159–168. [Google Scholar]
- Reeves, R.R. Marine Mammals: History of Exploitation. In Encyclopedia of Ocean Sciences; Elsevier: Amsterdam, The Netherlands, 2019; pp. 601–609. [Google Scholar] [CrossRef]
- Gervaise, C.; Simard, Y.; Aulanier, F.; Roy, N. Performance Study of Passive Acoustic Systems for Detecting North Atlantic Right Whales in Seaways: The Honguedo Strait in the Gulf of St. Lawrence; Department of Fisheries and Oceans, Government of Canada: Ottawa, ON, Canada, 2019. [Google Scholar]
- Mellinger, D.; Stafford, K.; Moore, S.; Dziak, R.; Matsumoto, H. An overview of fixed passive acoustic observation methods for cetaceans. Oceanography 2007, 20, 36–45. [Google Scholar] [CrossRef]
- Wang, D.; Garcia, H.; Huang, W.; Tran, D.D.; Jain, A.D.; Yi, D.H.; Gong, Z.; Jech, J.M.; Godø, O.R.; Makris, N.C.; et al. Vast assembly of vocal marine mammals from diverse species on fish spawning ground. Nature 2016, 531, 366–370. [Google Scholar] [CrossRef]
- Woods Hole Oceanographic Institution. Watkins Marine Mammal Sound Database. Available online: https://cis.whoi.edu/science/B/whalesounds/index.cfm (accessed on 1 March 2022).
- Rasmussen, M.H.; Koblitz, J.C.; Laidre, K.L. Buzzes and high-frequency clicks recorded from narwhals (Monodon monoceros) at their wintering ground. AquatMamm 2015, 41, 256–264. [Google Scholar] [CrossRef]
- Zubair, S.; Yan, F.; Wang, W. Dictionary learning based sparse coefficients for audio classification with max and average pooling. Digit. Signal Process. 2013, 23, 960–970. [Google Scholar] [CrossRef]
- Henaff, M.; Jarrett, K.; Kavukcuoglu, K.; LeCun, Y. Unsupervised Learning of Sparse Features for Scalable Audio Classification. In ISMIR; Citeseer: Princeton, NJ, USA, 2011; pp. 681–686. [Google Scholar]
- Trigeorgis, G.; Ringeval, F.; Brueckner, R.; Marchi, E.; Nicolaou, M.A.; Schuller, B.; Zafeiriou, S. Adieu Features? End-to-End Speech Emotion Recognition Using a Deep Convolutional Recurrent Network. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016; pp. 5200–5204. [Google Scholar] [CrossRef]
- Jiang, Y.; Bosch, N.; Baker, R.S.; Paquette, L.; Ocumpaugh, J.; Andres, J.M.A.L.; Moore, A.L.; Biswas, G. Expert Feature-Engineering vs. Deep Neural Networks: Which Is Better for Sensor-Free Affect Detection? In Artificial Intelligence in Education; Penstein Rosé, C., Martínez-Maldonado, R., Hoppe, H.U., Luckin, R., Mavrikis, M., Porayska-Pomsta, K., McLaren, B., du Boulay, B., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; pp. 198–211. [Google Scholar] [CrossRef]
- Ramaiah, V.S.; Rao, R.R. Multi-Speaker Activity Detection Using Zero Crossing Rates. In Proceedings of the 2016 International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India, 6–8 April 2016; pp. 23–26. [Google Scholar] [CrossRef]
- Shannon, B.J.; Paliwal, K.K. Feature extraction from higher-lag autocorrelation coefficients for robust speech recognition. Speech Commun. 2006, 48, 1458–1485. [Google Scholar] [CrossRef]
- Caetano, M.; Rodet, X. Improved Estimation of the Amplitude Envelope of Time-Domain Signals Using True Envelope Cepstral Smoothing. In Proceedings of the 2011 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Prague, Czech Republic, 22–27 May 2011; pp. 4244–4247. [Google Scholar] [CrossRef]
- Wisniewski, M.; Zielinski, T. Joint application of audio spectral envelope and tonality index in e-asthma monitoring system. IEEE J. Biomed. Health Inform. 2015, 19, 1009–1018. [Google Scholar] [CrossRef]
- Tzanetakis, G.; Cook, P. Musical genre classification of audio signals. IEEE Trans. Speech Audio Process. 2002, 10, 293–302. [Google Scholar] [CrossRef]
- Sharan, R.V.; Moir, T.J. Acoustic event recognition using cochleagram image and convolutional neural networks. Appl. Acoust. 2019, 148, 62–66. [Google Scholar] [CrossRef]
- Ogundile, O.O.; Usman, A.M.; Babalola, O.P.; Versfeld, D.J.J. Dynamic mode decomposition: A feature extraction technique based hidden Markov model for detection of Mysticetes’ vocalisations. Ecol. Inform. 2021, 63, 101306. [Google Scholar] [CrossRef]
- Dewi, S.P.; Prasasti, A.L.; Irawan, B. Analysis of LFCC Feature Extraction in Baby Crying Classification Using KNN. In Proceedings of the 2019 IEEE International Conference on Internet of Things and Intelligence System (IoTaIS), Bali, Indonesia, 5–7 November 2019; pp. 86–91. [Google Scholar] [CrossRef]
- Valero, X.; Alias, F. Gammatone cepstral coefficients: Biologically inspired features for non-speech audio classification. IEEE Trans. Multimed. 2012, 14, 1684–1689. [Google Scholar] [CrossRef]
- Ogundile, O.O.; Usman, A.M.; Babalola, O.P.; Versfeld, D.J.J. A hidden Markov model with selective time domain feature extraction to detect inshore Bryde’s whale short pulse calls. Ecol. Inform. 2020, 57, 101087. [Google Scholar] [CrossRef]
- Rakotomamonjy, A.; Gasso, G. Histogram of gradients of time-frequency representations for audio scene detection. IEEE/ACM Trans. Audio Speech Lang. Process. 2014, 23, 142–153. [Google Scholar] [CrossRef]
- Dennis, J.; Tran, H.D.; Chng, E.S. Image feature representation of the subband power distribution for robust sound event classification. IEEE Trans. Audio Speech Lang. Process. 2013, 21, 367–377. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ye, J. Acoustic Feature Extraction by Statistics Based Local Binary Pattern for Environmental Sound Classification. In Proceedings of the 2014 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Florence, Italy, 4–9 May 2014; pp. 3052–3056. [Google Scholar] [CrossRef]
- Padovese, B.; Frazao, F.; Kirsebom, O.S.; Matwin, S. Data augmentation for the classification of North Atlantic right whales upcalls. J. Acoust. Soc. Am. 2021, 149, 2520–2530. [Google Scholar] [CrossRef]
- Fletcher, H. Auditory patterns. Rev. Mod. Phys. 1940, 12, 47–65. [Google Scholar] [CrossRef]
- Mellinger, D.K.; Clark, C.W. Recognizing transient low-frequency whale sounds by spectrogram correlation. J. Acoust. Soc. Am. 2000, 107, 3518–3529. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, D. Detection and classification of right whale calls using an ‘edge’ detector operating on a smoothed spectrogram. Can. Acoust. 2004, 32, 39–47. [Google Scholar]
- Brown, J.C.; Miller, P.J.O. Automatic classification of killer whale vocalizations using dynamic time warping. J. Acoust. Soc. Am. 2007, 122, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Klinck, H.; Mellinger, D.K. The energy ratio mapping algorithm: A tool to improve the energy-based detection of odontocete echolocation clicks. J. Acoust. Soc. Am. 2011, 129, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Esfahanian, M.; Erdol, N.; Gerstein, E.; Zhuang, H. Two-stage detection of north Atlantic right whale upcalls using local binary patterns and machine learning algorithms. Appl. Acoust. 2017, 120, 158–166. [Google Scholar] [CrossRef]
- Kirsebom, O.S.; Frazao, F.; Simard, Y.; Roy, N.; Matwin, S.; Giard, S. Performance of a deep neural network at detecting North Atlantic right whale upcalls. J. Acoust. Soc. Am. 2020, 147, 2636–2646. [Google Scholar] [CrossRef]
- Shen, W.; Tu, D.; Yin, Y.; Bao, J. A new fusion feature based on convolutional neural network for pig cough recognition in field situations. Inf. Process. Agric. 2021, 8, 573–580. [Google Scholar] [CrossRef]
- Pentapati, H.; Vasamsetti, S.; Tenneti, M. MFCC for voiced part using VAD and GMM based gender recognition. AMA_B 2017, 60, 581–592. [Google Scholar] [CrossRef]
- Allen, J.A.; Murray, A.; Noad, M.J.; Dunlop, R.A.; Garland, E.C. Using self-organizing maps to classify humpback whale song units and quantify their similarity. J. Acoust. Soc. Am. 2017, 142, 1943–1952. [Google Scholar] [CrossRef]
- Ibrahim, A.K.; Zhuang, H.; Chérubin, L.M.; Schärer-Umpierre, M.T.; Erdol, N. Automatic classification of grouper species by their sounds using deep neural networks. J. Acoust. Soc. Am. 2018, 144, EL196–EL202. [Google Scholar] [CrossRef]
- Trawicki, M.B. Multispecies discrimination of whales (Cetaceans) using Hidden Markov Models (HMMS). Ecol. Inform. 2021, 61, 101223. [Google Scholar] [CrossRef]
- Mishachandar, B.; Vairamuthu, S. Diverse ocean noise classification using deep learning. Appl. Acoust. 2021, 181, 108141. [Google Scholar] [CrossRef]
- Lu, T.; Han, B.; Yu, F. Detection and classification of marine mammal sounds using AlexNet with transfer learning. Ecol. Inform. 2021, 62, 101277. [Google Scholar] [CrossRef]
- Dugan, P.J.; Clark, C.W.; LeCun, Y.A.; Van Parijs, S.M. DCL System Using Deep Learning Approaches for Land-Based or Ship-Based Real Time Recognition and Localization of Marine Mammals; Bioacoustics Research Program, Cornell University: Ithaca, NY, USA, 2015. [Google Scholar]
- Shiu, Y.; Palmer, K.J.; Roch, M.A.; Fleishman, E.; Liu, X.; Nosal, E.-M.; Helble, T.; Cholewiak, D.; Gillespie, D.; Klinck, H. Deep neural networks for automated detection of marine mammal species. Sci. Rep. 2020, 10, 607. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.N.; Harvey, M.; Harrell, L.; Jansen, A.; Merkens, K.P.; Wall, C.C.; Cattiau, J.; Oleson, E.M. A convolutional neural network for automated detection of humpback whale song in a diverse, long-term passive acoustic dataset. Front. Mar. Sci. 2021, 8, 607321. [Google Scholar] [CrossRef]
- Bianco, M.J.; Gerstoft, P.; Traer, J.; Ozanich, E.; Roch, M.A.; Gannot, S.; Deledalle, C.-A. Machine learning in acoustics: Theory and applications. J. Acoust. Soc. Am. 2019, 146, 3590–3628. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep Learning with Depth wise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- González-Hernández, F.R.; Sánchez-Fernández, L.P.; Suárez-Guerra, S.; Sánchez-Pérez, L.A. Marine mammal sound classification based on a parallel recognition model and octave analysis. Appl. Acoust. 2017, 119, 17–28. [Google Scholar] [CrossRef]
- Frasier, K.E.; Roch, M.A.; Soldevilla, M.S.; Wiggins, S.M.; Garrison, L.P.; Hildebrand, J.A. Automated classification of dolphin echolocation click types from the gulf of Mexico. PLoS Comput. Biol. 2017, 13, e1005823. [Google Scholar] [CrossRef]
- Usman, A.M.; Ogundile, O.O.; Versfeld, D.J.J. Review of automatic detection and classification techniques for cetacean vocalization. IEEE Access 2020, 8, 105181–105206. [Google Scholar] [CrossRef]
- Richardson, W.J.; Greene, J.; Malme, C.I.; Thomson, D.H. Marine Mammals and Noise; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar] [CrossRef]
- Jefferson, T.A.; Webber, M.A.; Pitman, R.L.; Gorter, U. Marine Mammals of the World: A Comprehensive Guide to Their Identification, 2nd ed.; Elsevier: London, UK; San Diego, CA, USA, 2015. [Google Scholar] [CrossRef]
- Sayigh, L.; Daher, M.A.; Allen, J.; Gordon, H.; Joyce, K.; Stuhlmann, C.; Tyack, P. The Watkins Marine Mammal Sound Database: An Online, Freely Accessible Resource. Proc. Mtgs. Acoust. 2016, 27, 040013. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications; NASA/ADS: Cambridge, MA, USA, 2017. [Google Scholar]
- Zhang, Z.; Sabuncu, M.R. Generalized cross entropy loss for training deep neural networks with noisy labels. Adv. Neural Inf. Process. Syst. 2018, 31, 8792–8802. [Google Scholar]
- Zhong, M.; Castellote, M.; Dodhia, R.; Lavista Ferres, J.; Keogh, M.; Brewer, A. Beluga whale acoustic signal classification using deep learning neural network models. J. Acoust. Soc. Am. 2020, 147, 1834–1841. [Google Scholar] [CrossRef] [PubMed]
- Prati, R.C.; Batista, G.E.A.P.A.; Silva, D.F. Class imbalance revisited: A new experimental setup to assess the performance of treatment methods. Knowl. Inf. Syst. 2015, 45, 247–270. [Google Scholar] [CrossRef]
- Taylor, L.; Nitschke, G. Improving Deep Learning with Generic Data Augmentation. In Proceedings of the 2018 IEEE Symposium Series on Computational Intelligence (SSCI), Bangalore, India, 18–21 November 2018; pp. 1542–1547. [Google Scholar] [CrossRef]
- Mushtaq, Z.; Su, S.-F.; Tran, Q.-V. Spectral images based environmental sound classification using CNN with meaningful data augmentation. Appl. Acoust. 2021, 172, 107581. [Google Scholar] [CrossRef]
- Zhou, X.; Garcia-Romero, D.; Duraiswami, R.; Espy-Wilson, C.; Shamma, S. Linear versus Mel Frequency Cepstral Coefficients for Speaker Recognition. In Proceedings of the 2011 IEEE Workshop on Automatic Speech Recognition & Understanding, Waikoloa, HI, USA, 11–15 December 2011; pp. 559–564. [Google Scholar] [CrossRef]
- Ye, Y.; Lao, L.; Yan, D.; Wang, R. Identification of weakly pitch-shifted voice based on convolutional neural network. Int. J. Digit. Multimed. Broadcast. 2020, 2020, 8927031. [Google Scholar] [CrossRef]
- Noda, J.J.; Travieso, C.M.; Sanchez-Rodriguez, D.; Dutta, M.K.; Singh, A. Using Bioacoustic Signals and Support Vector Machine for Automatic Classification of Insects. In Proceedings of the 2016 3rd International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 11–12 February 2016; pp. 656–659. [Google Scholar] [CrossRef]
- Qayyum, A.; Anwar, S.M.; Awais, M.; Majid, M. Medical image retrieval using deep convolutional neural network. Neurocomputing 2017, 266, 8–20. [Google Scholar] [CrossRef]
- Salman, A.; Maqbool, S.; Khan, A.H.; Jalal, A.; Shafait, F. Real-time fish detection in complex backgrounds using probabilistic background modelling. Ecol. Inform. 2019, 51, 44–51. [Google Scholar] [CrossRef]
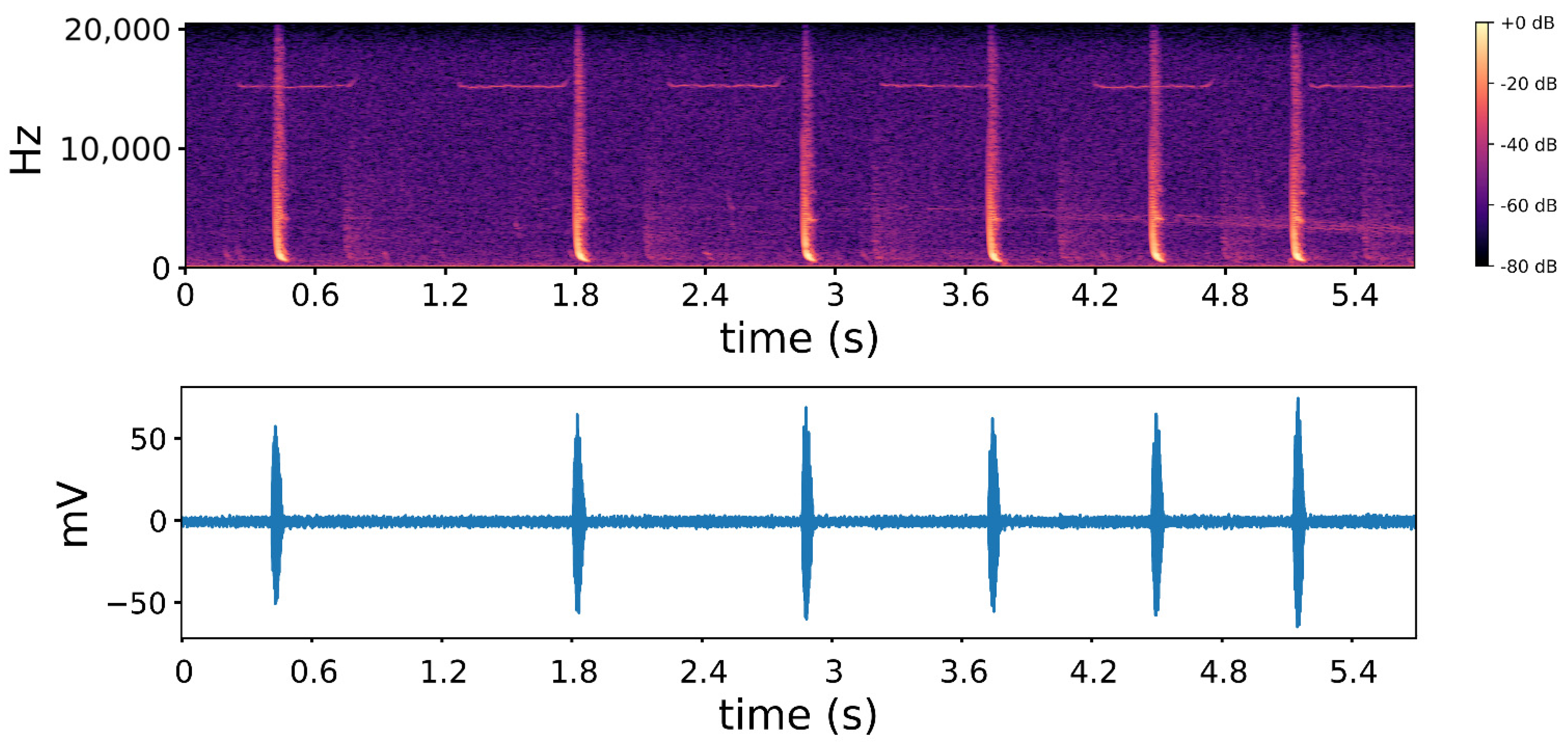







| Species Name | Abbreviation | Sample Size (No Augmentation) | Sample Size (Augmentation) | Sampling Rate (Hz) |
|---|---|---|---|---|
| Humpback whale | HW | 604 | 1208 | 5.12k–30k |
| Long-finned pilot whale | LFPW | 1213 | 1213 | 23.5k–166.6k |
| Pantropical spotted dolphin | PSD | 1034 | 1034 | 40.96k–192k |
| Sperm whale | SW | 1422 | 1422 | 10k–166.6k |
| Striped dolphin | SD | 681 | 1362 | 47.6k–192k |
| White-sided dolphin | WSD | 560 | 1120 | 60.6k–166.6k |
| Total | - | 5514 | 7359 | 5.12k–192k |
| Layers | Description |
|---|---|
| Input layer | Input tensor: 128 × 1 |
| {Conv + dropout + maxPooling1D} × 4 | {Convolution: 16 kernels (3 × 1, stride = 1) Pooling: (2 × 1, stride = 1) Activation function: ReLu} × 4 |
| Flatten + dropout | Convert the shape of tensor from 8 × 16 to 128 × 1 |
| fully-connected | Units = 128 |
| Fully-connected | Units = 6 |
| Softmax Layer | Output tensor: 6 × 1 |
| Model/Data | Mean Accuracy of Five Tests | Standard Deviation |
|---|---|---|
| Baseline model | 88.40% | 1.36% |
| MDF-PNet model | 95.21% | 0.65% |
| Species/Metrics | Precision (Baseline/MDF-PNet) (%) | Recall (Baseline/MDF-PNet) (%) | F1 Score (Baseline/MDF-PNet) (%) |
|---|---|---|---|
| HW | 94.19/99.61 | 95.67/99.61 | 94.92/99.61 |
| LFPW | 86.61/91.57 | 89.43/92.68 | 88.00/92.12 |
| PSD | 93.88/98.96 | 89.76/93.17 | 91.77/95.98 |
| SW | 78.24/88.84 | 78.90/90.72 | 78.57/89.77 |
| SD | 94.95/96.01 | 89.15/97.97 | 91.96/96.97 |
| WSD | 93.95/99.57 | 99.15/98.72 | 96.48/99.14 |
| Model | Mean Accuracy of Five Tests | Standard Deviation |
|---|---|---|
| Baseline + no data augmentation | 81.89% | 3.29% |
| Baseline + data augmentation | 88.40% | 1.36% |
| MDF-PNet + no data augmentation | 93.67% | 1.07% |
| MDF-PNet + data augmentation | 95.21% | 0.65% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, W.; Zhu, J.; Zhang, M.; Yang, Y. A Parallel Classification Model for Marine Mammal Sounds Based on Multi-Dimensional Feature Extraction and Data Augmentation. Sensors 2022, 22, 7443. https://doi.org/10.3390/s22197443
Cai W, Zhu J, Zhang M, Yang Y. A Parallel Classification Model for Marine Mammal Sounds Based on Multi-Dimensional Feature Extraction and Data Augmentation. Sensors. 2022; 22(19):7443. https://doi.org/10.3390/s22197443
Chicago/Turabian StyleCai, Wenyu, Jifeng Zhu, Meiyan Zhang, and Yong Yang. 2022. "A Parallel Classification Model for Marine Mammal Sounds Based on Multi-Dimensional Feature Extraction and Data Augmentation" Sensors 22, no. 19: 7443. https://doi.org/10.3390/s22197443
APA StyleCai, W., Zhu, J., Zhang, M., & Yang, Y. (2022). A Parallel Classification Model for Marine Mammal Sounds Based on Multi-Dimensional Feature Extraction and Data Augmentation. Sensors, 22(19), 7443. https://doi.org/10.3390/s22197443






