MTPA_Unet: Multi-Scale Transformer-Position Attention Retinal Vessel Segmentation Network Joint Transformer and CNN
Abstract
:1. Introduction
- A TPA module is proposed to replace the MSA structure in the traditional Transformer, which not only considers the relationship between long-distance pixels but also focuses on the acquisition of blood vessel pixel position information. The network model is adapted to the fine segmentation task of retinal blood vessels with a small number of samples.
- The MTPA_Unet network model is proposed, and the Transformer and convolutional neural network are combined to design and apply it to the retinal blood vessel segmentation task. MTPA_Unet can alleviate the limitations exhibited by CNN in modeling long-term dependencies and achieve higher retinal vessel segmentation accuracy.
- Perform ablation experiments and comparative experiments on three datasets, DRIVE, CHASE DB1 and STARE, and analyze the results. The results show that the network model proposed in this paper achieves better vessel segmentation performance.
2. Related Work
2.1. Related Work of CNN on Image Segmentation
2.2. Related Work of Transformer in Computer Vision
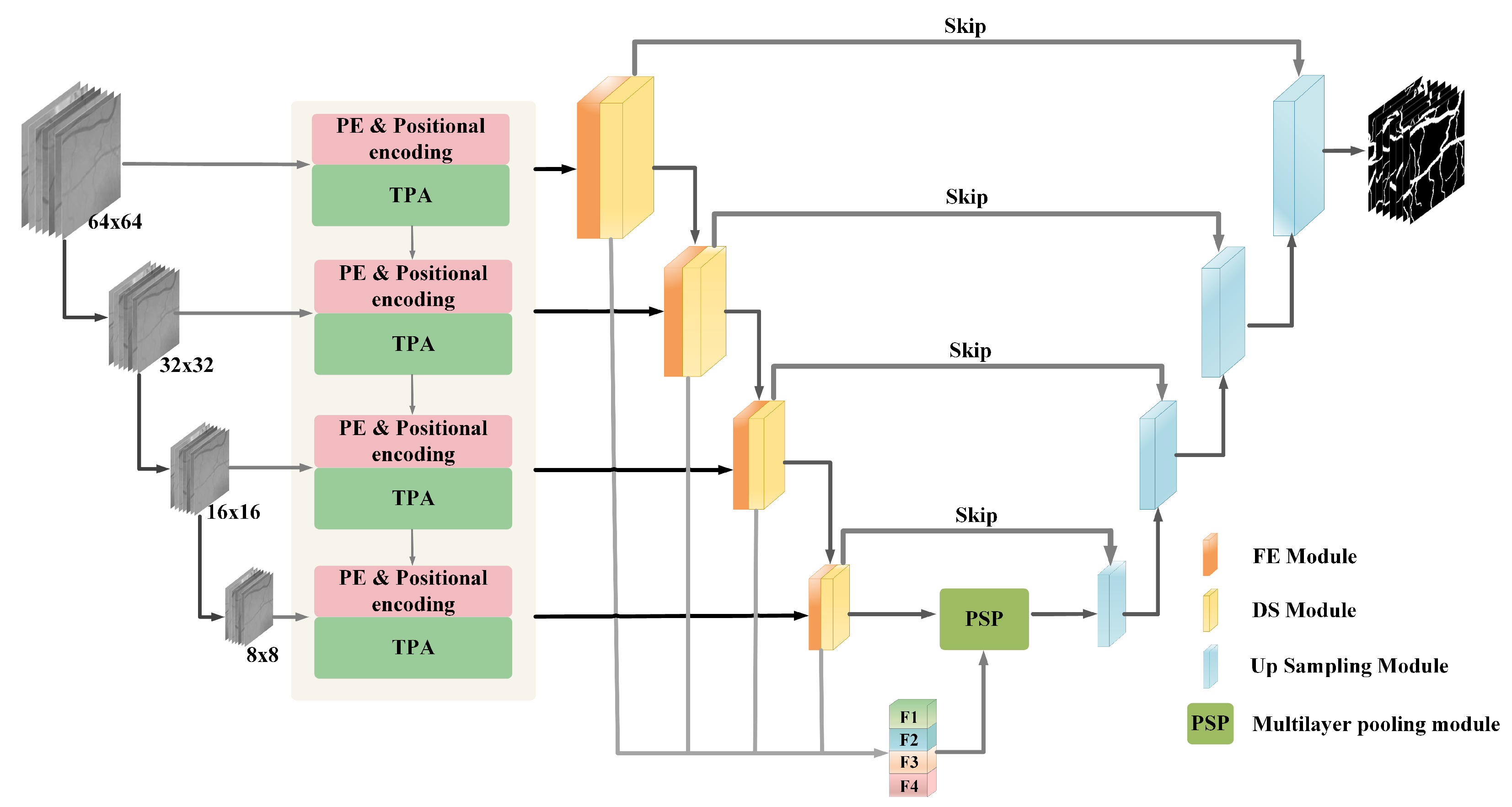
3. Muti-Transformer-Position Attention_Unet Method
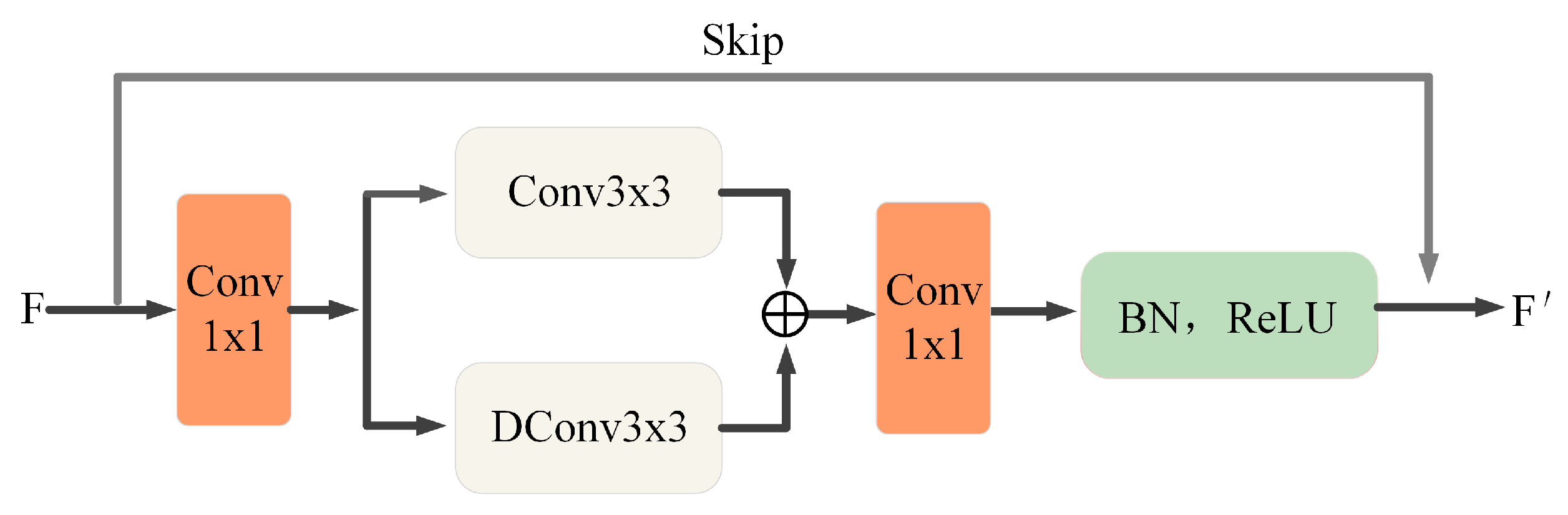
3.1. Encoder Block
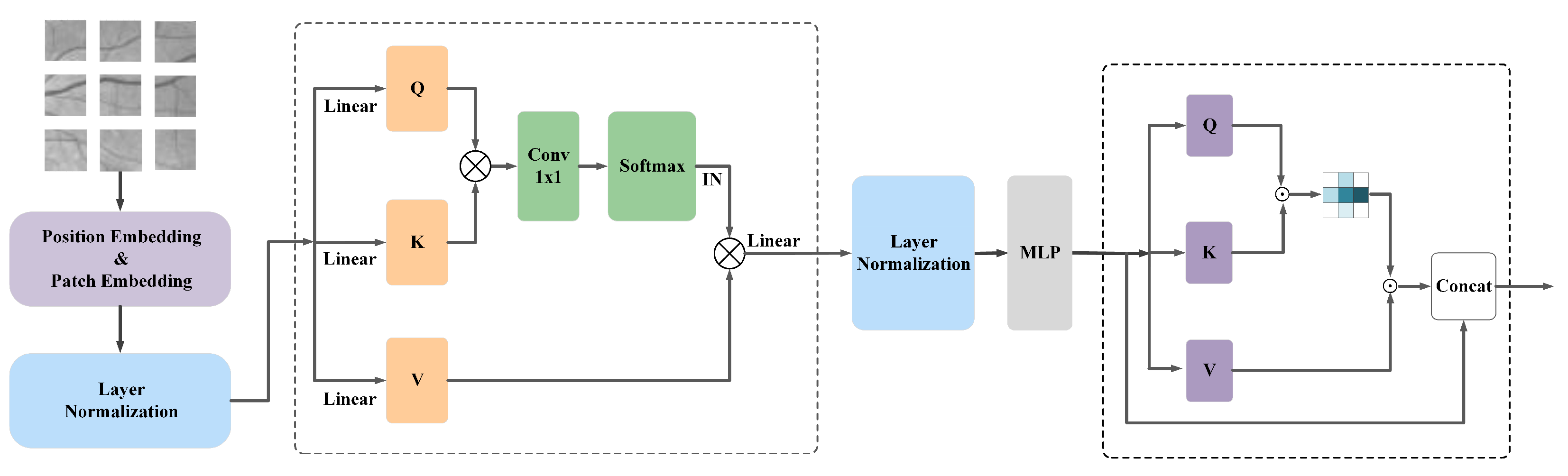
3.2. Transformer-Position Attention Module
3.2.1. Structure Description
3.2.2. Description of Location Attention Structure
3.3. Loss Function
4. Dataset and Evaluation Criteria
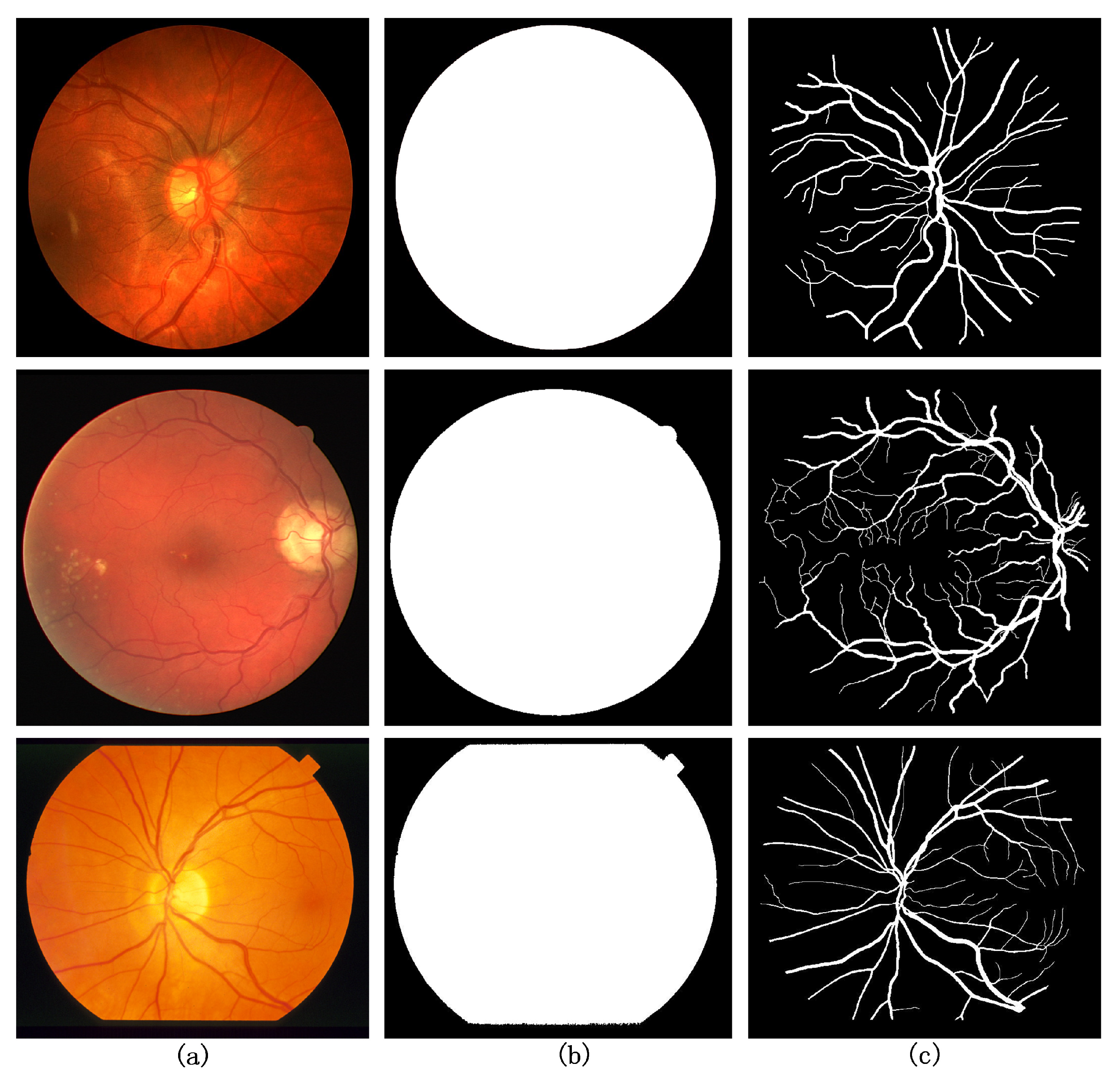
4.1. Dataset
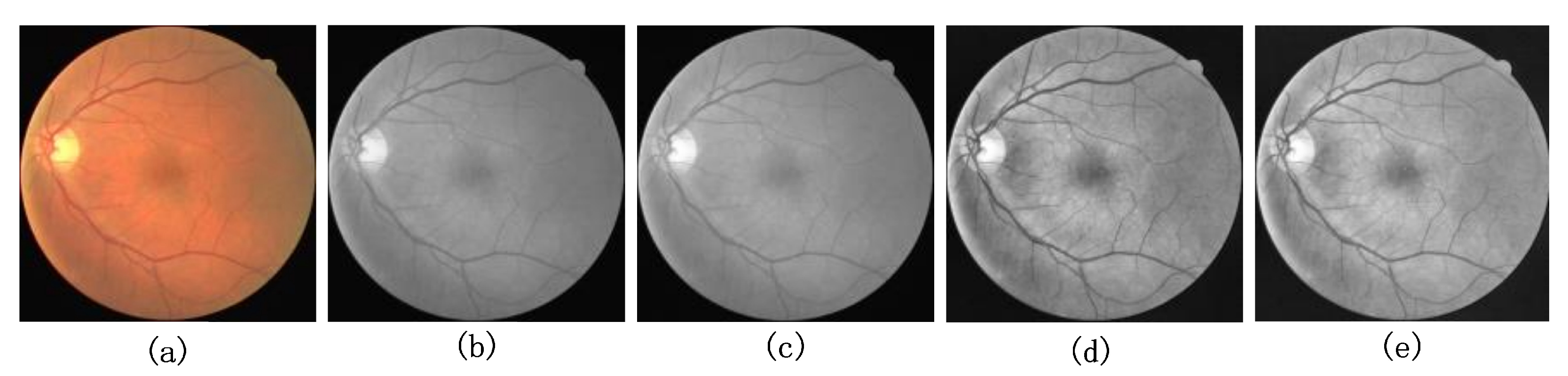
4.2. Image Preprocessing
4.3. Experimental Evaluation Metrics
5. Experimental Results and Analysis
5.1. Experimental Environment and Parameter Settings
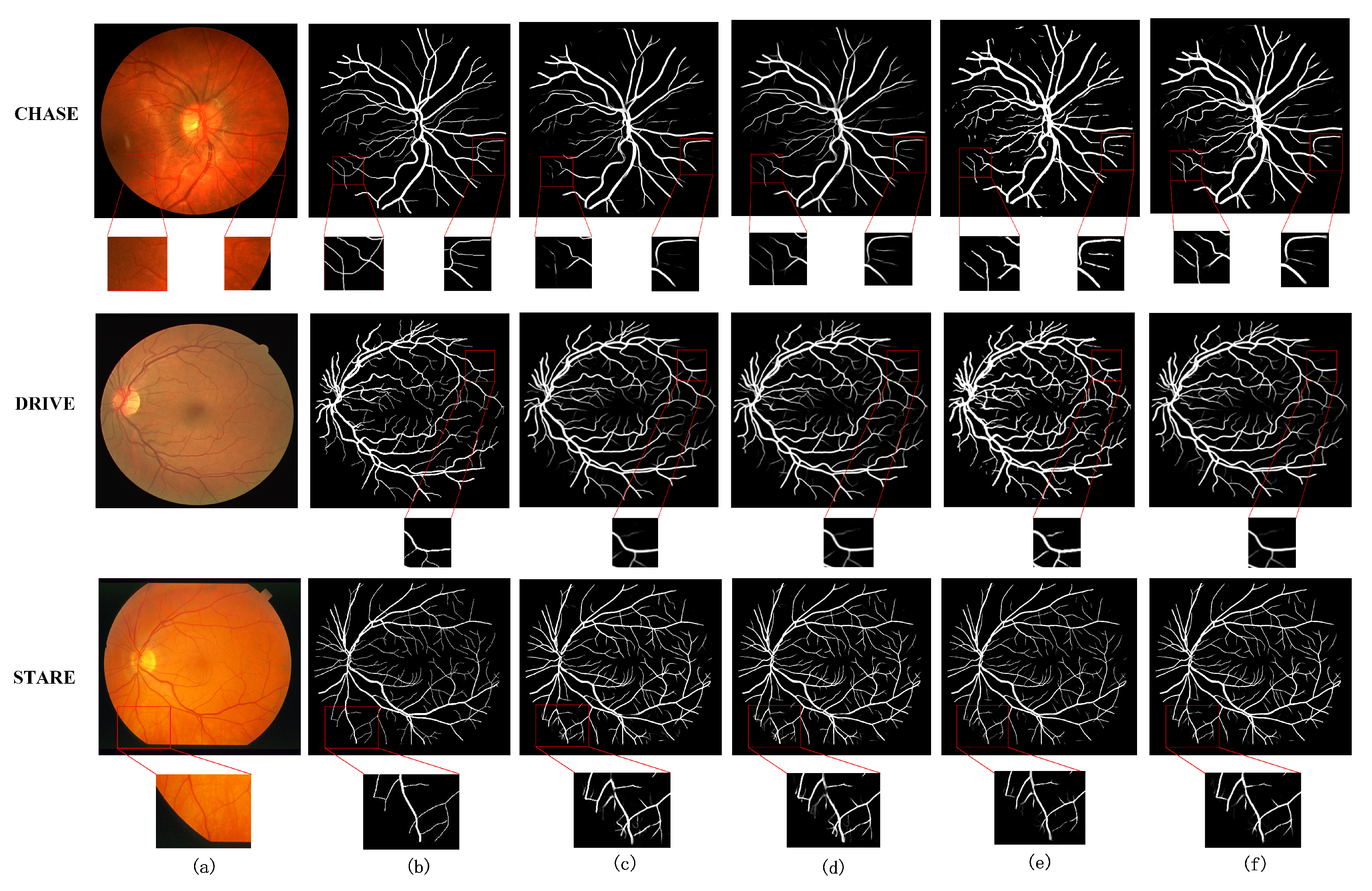
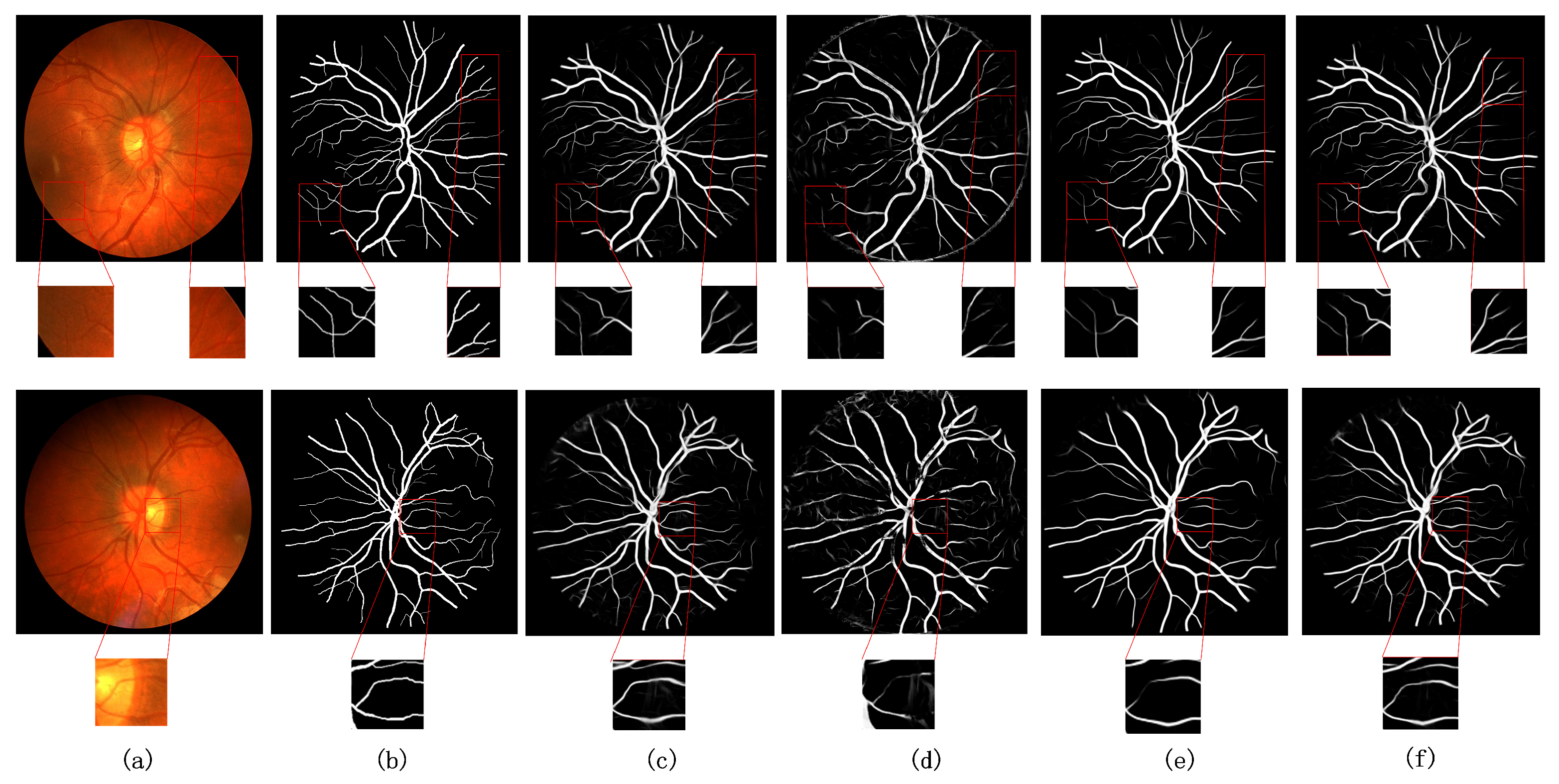
5.2. Experimental Comparison of Ablation Structures
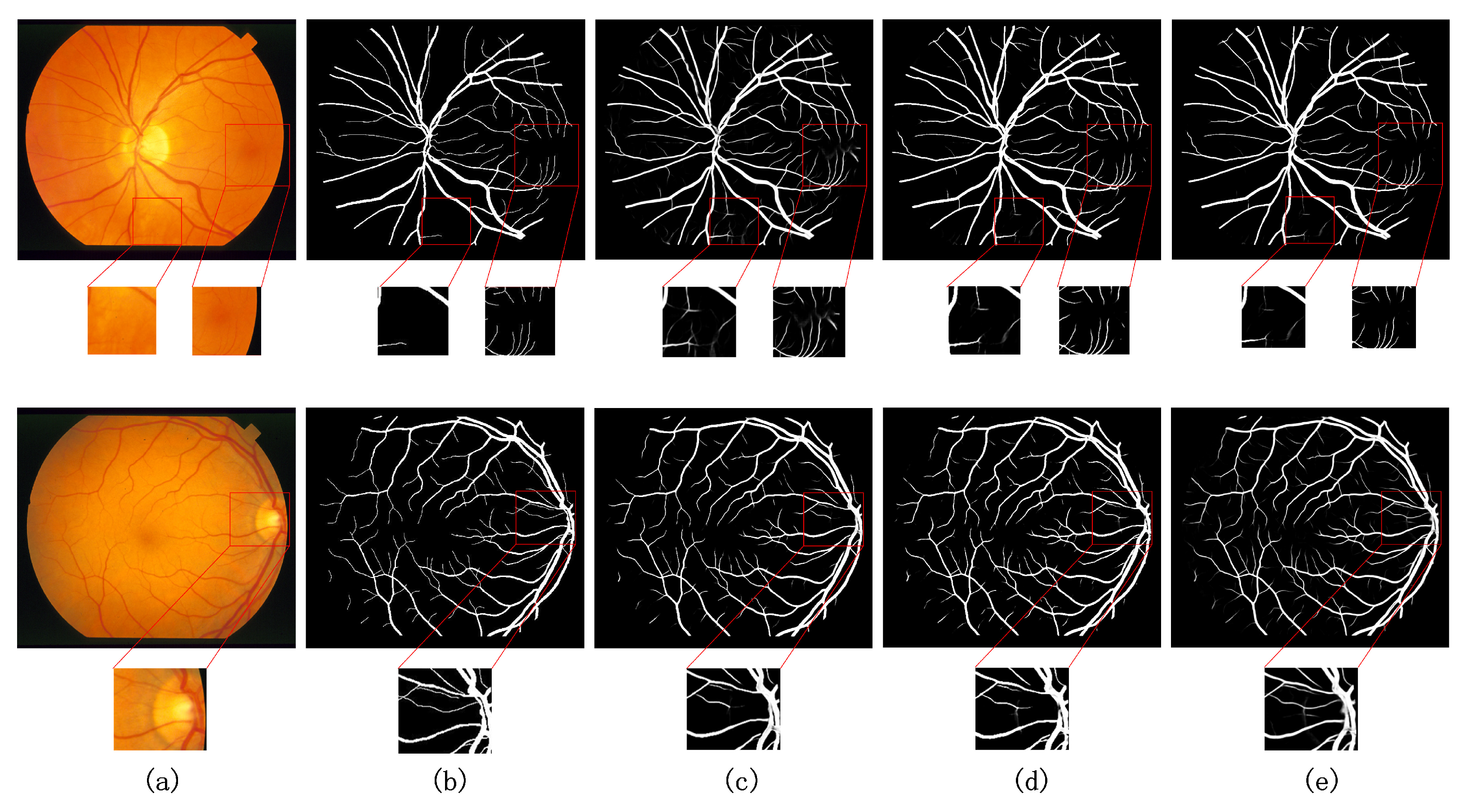
5.3. Comparison with Existing Models
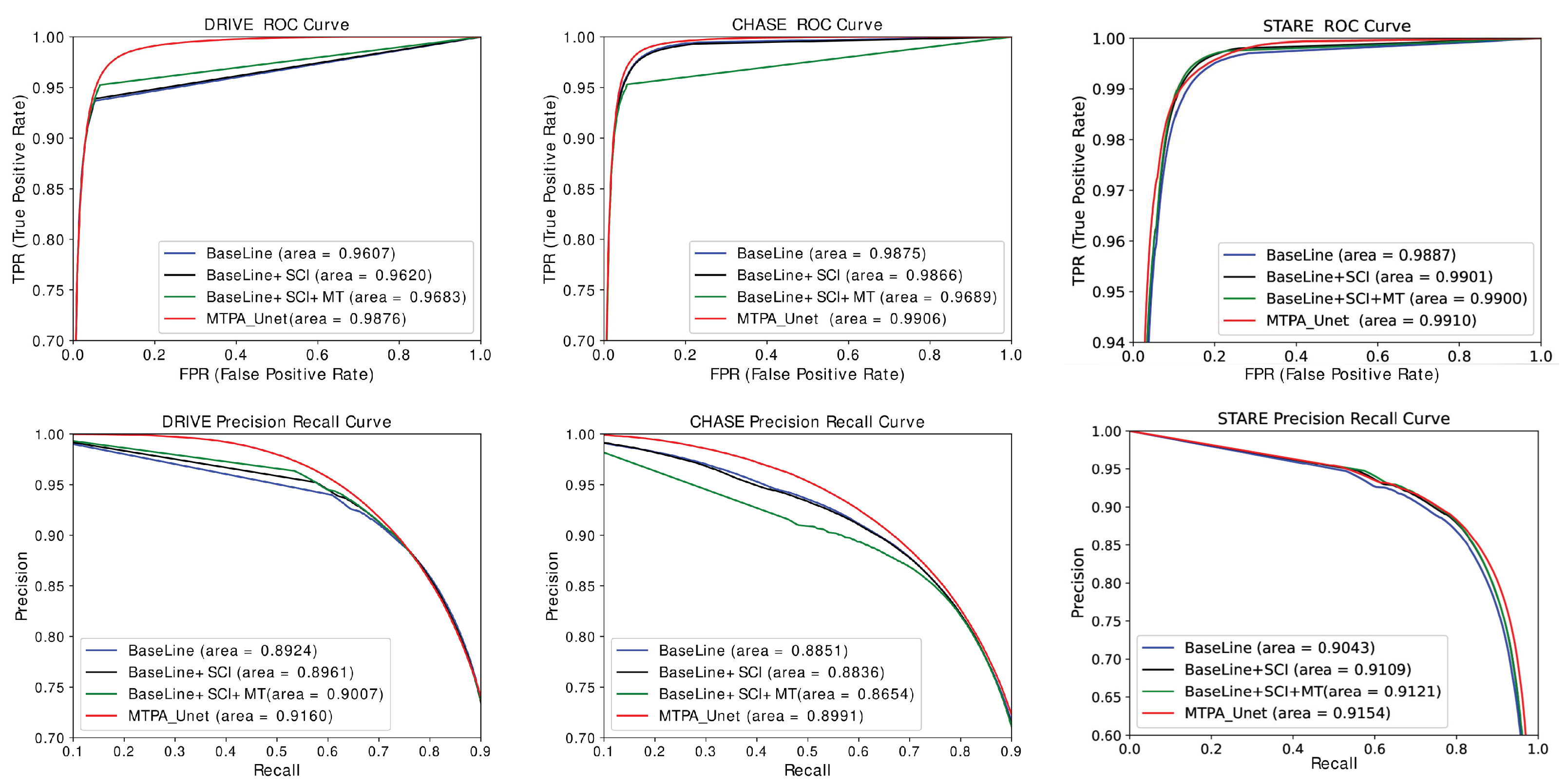
5.4. Analysis of the Number of Model Parameters and Evaluation of ROC Curves
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| MTPA_UNet | Multi-scale Transformer-Position Attention_Unet |
| CNN | Convolutional Neural Networks |
| TPA | Transformer-Position Attention |
| FE | Extraction Module |
| DS | Downsampling Module |
References
- Fan, Z.; Wei, J.; Zhu, G.; Mo, J.; Li, W. ENAS U-Net: Evolutionary Neural Architecture Search for Retinal Vessel Segmentation. arXiv 2020, arXiv:2001.06678. [Google Scholar]
- Oshitari, T. Diabetic retinopathy: Neurovascular disease requiring neuroprotective and regenerative therapies. Neural Regen. Res. 2022, 17, 795. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Klein, B.E.; Klein, R.; Jun, G.; Lee, K.E.; Iyengar, S.K. Genome-wide linkage study of retinal vessel diameters in the Beaver Dam Eye Study. Hypertension 2006, 47, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Cunha-Vaz, J. The blood-retinal barrier in the management of retinal disease: EURETINA award lecture. Ophthalmologica 2017, 237, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. Blood vessel segmentation of fundus images by major vessel extraction and subimage classification. IEEE J. Biomed. Health Inform. 2014, 19, 1118–1128. [Google Scholar]
- Shah, S.A.A.; Shahzad, A.; Khan, M.A.; Lu, C.K.; Tang, T.B. Unsupervised Method for Retinal Vessel Segmentation based on Gabor Wavelet and Multiscale Line Detector. IEEE Access 2019, 7, 167221–167228. [Google Scholar] [CrossRef]
- Jainish, G.R.; Jiji, G.W.; Infant, P.A. A novel automatic retinal vessel extraction using maximum entropy based EM algorithm. Multimed. Tools Appl. 2020, 79, 22337–22353. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Gu, R.; Wang, G.; Song, T.; Huang, R.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. CA-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging 2020, 40, 699–711. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Swizterland, 2018; pp. 3–11. [Google Scholar]
- Wang, B.; Qiu, S.; He, H. Dual encoding u-net for retinal vessel segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2019; pp. 84–92. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; pp. 327–331. [Google Scholar]
- Hu, R.; Singh, A. Transformer is all you need: Multimodal multitask learning with a unified transformer. arXiv 2021, arXiv:2102.10772. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Online, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 568–578. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Chen, B.; Liu, Y.; Zhang, Z.; Lu, G.; Zhang, D. Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. arXiv 2021, arXiv:2107.05274. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical transformer: Gated axial-attention for medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2021; pp. 36–46. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Huang, Z.; Wang, X.; Huang, L.; Huang, C.; Wei, Y.; Liu, W. Ccnet: Criss-cross attention for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 603–612. [Google Scholar]
- Zhu, Z.; Xu, M.; Bai, S.; Huang, T.; Bai, X. Asymmetric non-local neural networks for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 593–602. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3146–3154. [Google Scholar]
- Lian, S.; Li, L.; Lian, G.; Xiao, X.; Luo, Z.; Li, S. A global and local enhanced residual u-net for accurate retinal vessel segmentation. IEEE ACM Trans. Comput. Biol. Bioinform. 2019, 18, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Fan, Y. ACEnet: Anatomical context-encoding network for neuroanatomy segmentation. Med. Image Anal. 2021, 70, 101991. [Google Scholar] [CrossRef]
- Zhang, Y.; He, M.; Chen, Z.; Hu, K.; Li, X.; Gao, X. Bridge-Net: Context-involved U-net with patch-based loss weight mapping for retinal blood vessel segmentation. Expert Syst. Appl. 2022, 195, 116526. [Google Scholar] [CrossRef]
- Tan, Y.; Yang, K.F.; Zhao, S.X.; Li, Y.J. Retinal Vessel Segmentation with Skeletal Prior and Contrastive Loss. IEEE Trans. Med. Imaging 2022. [Google Scholar] [CrossRef]
- Arsalan, M.; Haider, A.; Choi, J.; Park, K.R. Diabetic and Hypertensive Retinopathy Screening in Fundus Images Using Artificially Intelligent Shallow Architectures. J. Pers. Med. 2021, 12, 7. [Google Scholar] [CrossRef]
- Arsalan, M.; Haider, A.; Lee, Y.W.; Park, K.R. Detecting retinal vasculature as a key biomarker for deep Learning-based intelligent screening and analysis of diabetic and hypertensive retinopathy. Expert Syst. Appl. 2022, 200, 117009. [Google Scholar] [CrossRef]
- Yin, P.; Cai, H.; Wu, Q. DF-Net: Deep fusion network for multi-source vessel segmentation. Inf. Fusion 2022, 78, 199–208. [Google Scholar] [CrossRef]
- d’Ascoli, S.; Touvron, H.; Leavitt, M.; Morcos, A.; Biroli, G.; Sagun, L. Convit: Improving vision Transformers with soft convolutional inductive biases. In Proceedings of the ICLR 2021, Online, 3–7 May 2021. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision 2021, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Huang, S.; Li, J.; Xiao, Y.; Shen, N.; Xu, T. RTNet: Relation Transformer Network for Diabetic Retinopathy Multi-lesion Segmentation. IEEE Trans. Med. Imaging 2022, 41, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Heo, B.; Yun, S.; Han, D.; Chun, S.; Choe, J.; Oh, S.J. Rethinking spatial dimensions of vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision 2021, Montreal, BC, Canada, 11–17 October 2021; pp. 11936–11945. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. arXiv 2021, arXiv:2105.05537. [Google Scholar]
- Gao, Y.; Zhou, M.; Liu, D.; Metaxas, D. A Multi-scale Transformer for Medical Image Segmentation: Architectures, Model Efficiency, and Benchmarks. arXiv 2022, arXiv:2203.00131. [Google Scholar]
- Zhang, Q.; Yang, Y.B. Rest: An efficient transformer for visual recognition. Adv. Neural Inf. Process. Syst. 2021, 34, 15475–15485. [Google Scholar]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Owen, C.G.; Rudnicka, A.R.; Mullen, R.; Barman, S.A.; Monekosso, D.; Whincup, P.H.; Ng, J.; Paterson, C. Measuring retinal vessel tortuosity in 10-year-old children: Validation of the computer-assisted image analysis of the retina(CAIAR) program. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2004–2010. [Google Scholar] [CrossRef] [Green Version]
- Hoover, A.D.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Zhang, H.; Tan, N.; Chen, L. Automatic retinal blood vessel segmentation based on fully convolutional neural networks. Symmetry 2019, 11, 1112. [Google Scholar] [CrossRef] [Green Version]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Wu, H.; Wang, W.; Zhong, J.; Lei, B.; Wen, Z.; Qin, J. SCS-Net: A Scale and Context Sensitive Network for Retinal Vessel Segmentation. Med. Image Anal. 2021, 70, 102025. [Google Scholar] [CrossRef]
- Azzopardi, G.; Strisciuglio, N.; Vento, M.; Petkov, N. Trainable COSFIRE filters for vessel delineation with application to retinal images. Med. Image Anal. 2015, 19, 46–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.; Cheng, Y. Automatic extraction of retinal blood vessel based on matched filtering and local entropy thresholding. In Proceedings of the 2015 8th International Conference on Biomedical Engineering and Informatics (BMEI), Shenyang, China, 14–16 October 2015; pp. 62–67. [Google Scholar]
- Chen, G.; Chen, M.; Li, J.; Zhang, E. Retina image vessel segmentation using a hybrid CGLI level set method. BioMed Res. Int. 2017, 2017, 1263056. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Szemenyei, M.; Yi, Y.; Zhou, W.; Bian, H. Residual Spatial Attention Network for Retinal Vessel Segmentation. In International Conference on Neural Information Processing; Springer: Cham, Switzerland, 2020; pp. 509–519. [Google Scholar]
- Lv, Y.; Ma, H.; Li, J.; Liu, S. Attention guided u-net with atrous convolution for accurate retinal vessels segmentation. IEEE Access 2020, 8, 32826–32839. [Google Scholar] [CrossRef]
- Tomar, N.K.; Jha, D.; Riegler, M.A.; Johansen, H.D.; Johansen, D.; Rittscher, J.; Halvorsen, P.; Ali, S. FANet: A Feedback Attention Network for Improved Biomedical Image Segmentation. arXiv 2021, arXiv:2103.17235. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Fang, Z.; Wei, Z.; Cai, Q.; Gao, Y. SAT-Net: A side attention network for retinal image segmentation. Appl. Intell. 2021, 51, 5146–5156. [Google Scholar] [CrossRef]
- Wang, W.; Zhong, J.; Wu, H.; Wen, Z.; Qin, J. Rvseg-net: An efficient feature pyramid cascade network for retinal vessel segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2020; pp. 796–805. [Google Scholar]
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl.-Based Syst. 2019, 178, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Fang, Y.; Huang, H.; Xu, X.; Wang, J.; Lai, X. Automatic Retinal Vessel Segmentation Based on an Improved U-Net Approach. Sci. Program. 2021, 2021, 5520407. [Google Scholar] [CrossRef]
- Li, L.; Verma, M.; Nakashima, Y.; Nagahara, H.; Kawasaki, R. Iternet: Retinal image segmentation utilizing structural redundancy in vessel networks. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision 2020, Snowmass Village, CO, USA, 1–5 March 2020; pp. 3656–3665. [Google Scholar]



| Model | Accuracy | Specificity | Sensitivity | Dice | AUC_ROC |
|---|---|---|---|---|---|
| U-Net [10] | 0.9531 | 0.9820 | 0.7537 | 0.8142 | - |
| BaseLine | 0.9705 | 0.9865 | 0.8060 | 0.8266 | 0.9728 |
| BaseLine+SCI | 0.9702 | 0.9841 | 0.8278 | 0.8289 | 0.9766 |
| BaseLine+SCI+MT | 0.9705 | 0.9848 | 0.8245 | 0.8300 | 0.9785 |
| MTPA_Unet (w/o pre) | 0.9702 | 0.9856 | 0.8116 | 0.8275 | 0.9875 |
| MTPA_Unet | 0.9718 | 0.9836 | 0.8410 | 0.8318 | 0.9877 |
| Model | Accuracy | Specificity | Sensitivity | Dice | AUC_ROC |
|---|---|---|---|---|---|
| U-Net [10] | 0.9578 | 0.9701 | 0.8288 | 0.7783 | - |
| BaseLine | 0.9759 | 0.9857 | 0.8324 | 0.8136 | 0.9884 |
| BaseLine+SCI | 0.9761 | 0.9861 | 0.8296 | 0.8143 | 0.9878 |
| BaseLine+SCI+MT | 0.9757 | 0.9848 | 0.8434 | 0.8139 | 0.9861 |
| MTPA_Unet (w/o pre) | 0.9758 | 0.9857 | 0.8299 | 0.8122 | 0.9900 |
| MTPA_Unet | 0.9762 | 0.9858 | 0.8437 | 0.8164 | 0.9905 |
| Image | Accuracy | Specificity | Sensitivity | Dice | AUC_ROC |
|---|---|---|---|---|---|
| 0 | 0.9742 | 0.9825 | 0.8856 | 0.8483 | 0.9908 |
| 1 | 0.9753 | 0.9812 | 0.8916 | 0.8276 | 0.9892 |
| 2 | 0.9805 | 0.9859 | 0.8964 | 0.8468 | 0.9931 |
| 3 | 0.9694 | 0.9787 | 0.8525 | 0.8052 | 0.9866 |
| 4 | 0.9715 | 0.9824 | 0.8621 | 0.8455 | 0.9886 |
| 5 | 0.9778 | 0.9831 | 0.9061 | 0.8503 | 0.9921 |
| 6 | 0.9732 | 0.9744 | 0.9602 | 0.8521 | 0.9934 |
| 7 | 0.9775 | 0.9799 | 0.9484 | 0.8634 | 0.9941 |
| 8 | 0.9827 | 0.9872 | 0.9302 | 0.8948 | 0.9958 |
| 9 | 0.9755 | 0.9799 | 0.9257 | 0.8591 | 0.9923 |
| 10 | 0.9811 | 0.9876 | 0.8962 | 0.8713 | 0.9947 |
| 11 | 0.9812 | 0.9828 | 0.9620 | 0.8881 | 0.9954 |
| 12 | 0.9805 | 0.9864 | 0.9205 | 0.8939 | 0.9947 |
| 13 | 0.9803 | 0.9883 | 0.8997 | 0.8923 | 0.9946 |
| 14 | 0.9787 | 0.9871 | 0.8906 | 0.8783 | 0.9929 |
| 15 | 0.9684 | 0.9814 | 0.8542 | 0.8468 | 0.9879 |
| 16 | 0.9761 | 0.9864 | 0.8707 | 0.8671 | 0.9915 |
| 17 | 0.9866 | 0.9935 | 0.8571 | 0.8664 | 0.9926 |
| 18 | 0.9839 | 0.9903 | 0.8417 | 0.8191 | 0.9922 |
| 19 | 0.9721 | 0.9825 | 0.8254 | 0.7974 | 0.9883 |
| Average | 0.9773 | 0.9841 | 0.8938 | 0.8557 | 0.9920 |
| Model | Accuracy | Specificity | Sensitivity | Dice | AUC_ROC |
|---|---|---|---|---|---|
| U-Net [10] | 0.9690 | 0.9842 | 0.8270 | 0.8373 | - |
| BaseLine | 0.9747 | 0.9833 | 0.8701 | 0.8379 | 0.9901 |
| BaseLine+SCI | 0.9754 | 0.9834 | 0.8772 | 0.8425 | 0.9905 |
| BaseLine+SCI+ MT | 0.9766 | 0.9835 | 0.8802 | 0.8434 | 0.9908 |
| MTPA_Unet | 0.9773 | 0.9841 | 0.8938 | 0.8557 | 0.9920 |
| Type | Methods | Year | Acc | Sp | Se | Dice | |
|---|---|---|---|---|---|---|---|
| Non-learning based methods | Azzopardi et al. [48] | 2015 | 0.9442 | 0.9704 | 0.7655 | - | |
| Miao et al. [49] | 2015 | 0.9597 | 0.9748 | 0.7481 | - | ||
| Chen et al. [50] | 2017 | 0.9390 | 0.9680 | 0.7358 | - | ||
| Shah et al. [6] | 2019 | 0.9470 | 0.9724 | 0.7760 | - | ||
| Deep learning based methods | Original image | U-Net [10] | 2018 | 0.9531 | 0.9820 | 0.7537 | 0.8142 |
| Guo et al. [51] | 2020 | 0.9691 | 0.9839 | 0.8149 | 0.8222 | ||
| AG-UNet [52] | 2020 | 0.9558 | 0.9810 | 0.7854 | 0.8216 | ||
| FANet [53] | 2021 | 0.8189 | 0.9826 | - | 0.8183 | ||
| Tong et al. [54] | 2021 | 0.9684 | 0.9870 | 0.8117 | 0.8174 | ||
| Only pre-processing | SCS-Net [47] | 2021 | 0.9697 | 0.9838 | 0.8289 | 0.8189 | |
| PLRS-Net [33] | 2022 | 0.9682 | 0.9817 | 0.8269 | - | ||
| Patch+ Pre-processing | RVSeg-Net [55] | 2020 | 0.9681 | 0.9845 | 0.8107 | - | |
| R2U-Net [46] | 2018 | 0.9556 | 0.9813 | 0.7792 | 0.8171 | ||
| UNet++ [13] | 2018 | 0.9710 | 0.9861 | 0.8120 | 0.8302 | ||
| DUNet [56] | 2019 | 0.9566 | 0.9800 | 0.7963 | 0.8237 | ||
| CA-Net [9] | 2020 | 0.9605 | 0.9788 | 0.7727 | 0.7733 | ||
| Huang et al. [57] | 2021 | 0.9701 | 0.9849 | 0.8011 | - | ||
| Ours | 2021 | 0.9718 | 0.9836 | 0.8410 | 0.8318 | ||
| Type | Methods | Year | Acc | Sp | Se | Dice | |
|---|---|---|---|---|---|---|---|
| Deep learning based methods | Original image | U-Net [10] | 2018 | 0.9690 | 0.9842 | 0.8270 | 0.8373 |
| Tong et al. [54] | 2021 | 0.9805 | 0.9927 | 0.8072 | 0.8270 | ||
| Only pre-processing | SCS-Net [47] | 2021 | 0.9736 | 0.9839 | 0.8207 | - | |
| PLRS-Net [33] | 2022 | 0.9715 | 0.9803 | 0.8635 | - | ||
| Patch+ Pre-processing | UNet++ [13] | 2018 | 0.9753 | 0.9843 | 0.8646 | 0.8393 | |
| R2U-Net [46] | 2018 | 0.9712 | 0.9862 | 0.8298 | 0.8475 | ||
| Iter-Net [58] | 2020 | 0.9782 | 0.9919 | 0.7715 | 0.8146 | ||
| CA-Net [9] | 2020 | 0.9703 | 0.9705 | 0.8685 | 0.8397 | ||
| Ours | 2022 | 0.9773 | 0.9841 | 0.8938 | 0.8557 | ||
| Type | Methods | Year | Acc | Sp | Se | Dice | |
|---|---|---|---|---|---|---|---|
| Non-learning based methods | Azzopardi et al. [48] | 2015 | 0.9387 | 0.9587 | 0.7585 | - | |
| Deep learning based methods | Original image | U-Net [10] | 2018 | 0.9578 | 0.9701 | 0.8288 | 0.7783 |
| AG-UNet [52] | 2020 | 0.9752 | 0.9870 | 0.8110 | 0.8116 | ||
| FANet [53] | 2021 | 0.7722 | 0.9830 | - | 0.8108 | ||
| Tong et al. [54] | 2021 | 0.9739 | 0.9868 | 0.8340 | 0.7911 | ||
| Only pre-processing | SCS-Net [47] | 2021 | 0.9744 | 0.9839 | 0.8365 | - | |
| PLRS-Net [33] | 2022 | 0.9731 | 0.9839 | 0.8301 | - | ||
| Patch+ Pre-processing | RVSeg-Net [55] | 2020 | 0.9726 | 0.9836 | 0.8069 | - | |
| UNet++ [13] | 2018 | 0.9760 | 0.9810 | 0.8184 | 0.8139 | ||
| R2U-Net [46] | 2018 | 0.9634 | 0.9820 | 0.7756 | 0.7928 | ||
| DUNet [56] | 2019 | 0.9752 | 0.9610 | 0.8155 | 0.7883 | ||
| CA-Net [9] | 2020 | 0.9645 | 0.9749 | 0.8120 | 0.7409 | ||
| Ours | 2022 | 0.9762 | 0.9858 | 0.8437 | 0.8164 | ||
| Model | BaseBine | BaseBine + SCI | BaseBine + SCI + MT | MTPA_Unet |
|---|---|---|---|---|
| params | 12.2 M | 13.0 M | 15.9 M | 11.8 M |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Liang, J.; Cheng, T.; Lin, X.; Zhang, Y.; Dong, J. MTPA_Unet: Multi-Scale Transformer-Position Attention Retinal Vessel Segmentation Network Joint Transformer and CNN. Sensors 2022, 22, 4592. https://doi.org/10.3390/s22124592
Jiang Y, Liang J, Cheng T, Lin X, Zhang Y, Dong J. MTPA_Unet: Multi-Scale Transformer-Position Attention Retinal Vessel Segmentation Network Joint Transformer and CNN. Sensors. 2022; 22(12):4592. https://doi.org/10.3390/s22124592
Chicago/Turabian StyleJiang, Yun, Jing Liang, Tongtong Cheng, Xin Lin, Yuan Zhang, and Jinkun Dong. 2022. "MTPA_Unet: Multi-Scale Transformer-Position Attention Retinal Vessel Segmentation Network Joint Transformer and CNN" Sensors 22, no. 12: 4592. https://doi.org/10.3390/s22124592
APA StyleJiang, Y., Liang, J., Cheng, T., Lin, X., Zhang, Y., & Dong, J. (2022). MTPA_Unet: Multi-Scale Transformer-Position Attention Retinal Vessel Segmentation Network Joint Transformer and CNN. Sensors, 22(12), 4592. https://doi.org/10.3390/s22124592






