Amniotic Fluid Classification and Artificial Intelligence: Challenges and Opportunities
Abstract
:1. Introduction
- Summarizes the use of ML- and DL-based classification and segmentation methods in measuring and classifying AF volume.
- Discusses the factors leading to abnormal AF levels and summarizes the studies focusing on using AI to detect these factors.
- Summarizes the previous studies’ performances in terms of their detection approaches, including their accuracy, dice similarity, etc.
- Presents a comprehensive discussion on the techniques used by the previous studies and the dataset types and sizes.
- Highlights the challenges and future directions in this field of AF detection using AI techniques.
2. Correlation between AF Levels and Gestational Age
3. Methodology
4. Artificial Intelligence-Based Techniques for Diagnosis and Classification of Amniotic Fluid
4.1. Classification
4.2. Segmentation
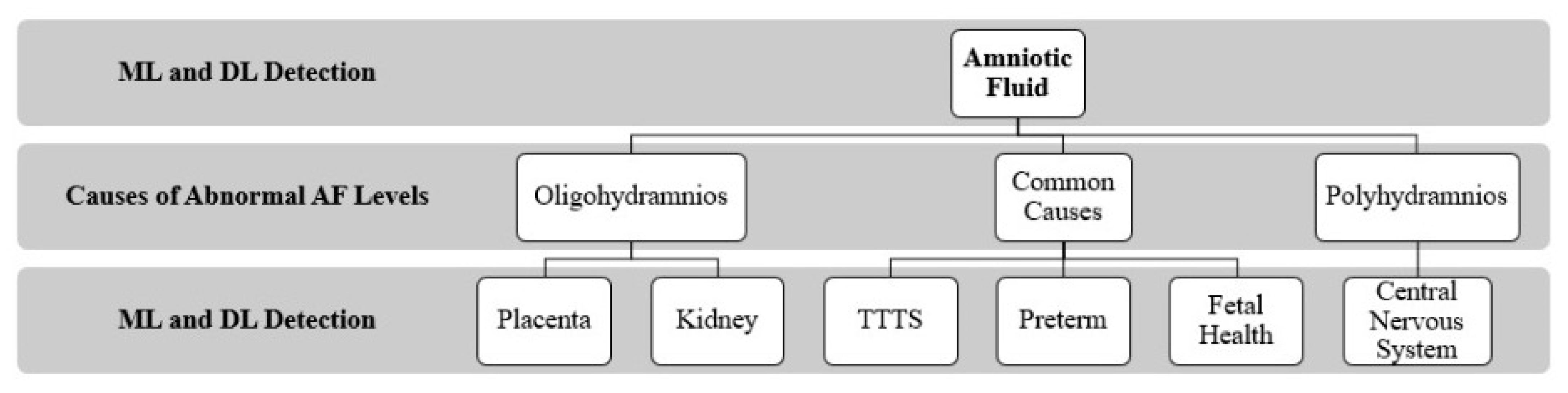
5. Factors/Causes of Abnormal Amniotic Fluid Levels
5.1. Oligohydramnios
5.1.1. Placenta
5.1.2. Kidneys
5.2. Polyhydramnios
Central Nervous System
5.3. Common Factors of Oligohydramnios and Polyhydramnios
5.3.1. TTTS (Twin-to-Twin Transfusion Syndrome)
5.3.2. Preterm
5.3.3. Fetal Health
6. Discussion
7. Challenges and Opportunities
7.1. Limited Dataset Size
7.2. Sonographer Dependency
7.3. Overfitting
7.4. Black Box Nature
7.5. Bias
7.6. Distributed Data
7.7. Privacy and Cyber Security Concerns
8. Future Research
8.1. Image Quality
8.2. Imaging Techniques
8.3. Awareness and Training
9. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beall, M.H.; van den Wijngaard, J.P.H.M.; van Gemert, M.J.C.; Ross, M.G. Amniotic Fluid Water Dynamics. Placenta 2007, 28, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Phelan, J.P.; Ahn, M.O.; Smith, C.V.; Rutherford, S.E.; Anderson, E. Amniotic fluid index measurements during pregnancy. J. Reprod. Med. Obstet. Gynecol. 1987, 32, 601–604. [Google Scholar]
- Magann, E.F.; Chauhan, S.P.; Doherty, D.A.; Lutgendorf, M.A.; Magann, M.I.; Morrison, J.C. A review of idiopathic hydramnios and pregnancy outcomes. Obstet. Gynecol. Surv. 2007, 62, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Kehl, S.; Schelkle, A.; Thomas, A.; Puhl, A.; Meqdad, K.; Tuschy, B.; Berlit, S.; Weiss, C.; Bayer, C.; Heimrich, J.; et al. Single deepest vertical pocket or amniotic fluid index as evaluation test for predicting adverse pregnancy outcome (SAFE trial): A multicenter, open-label, randomized controlled trial. Ultrasound Obstet. Gynecol. 2016, 47, 674–679. [Google Scholar] [CrossRef]
- Cho, H.C.; Sun, S.; Hyun, C.M.; Kwon, J.Y.; Kim, B.; Park, Y.; Seo, J.K. Automated ultrasound assessment of amniotic fluid index using deep learning. Med. Image Anal. 2021, 69, 101951. [Google Scholar] [CrossRef]
- Drukker, L.; Noble, J.A.; Papageorghiou, A.T.P. Introduction to artificial intelligence in ultrasound imaging in obstetrics and gynecology. Ultrasound Obstet. Gynecol. 2020, 56, 498–505. [Google Scholar] [CrossRef]
- Fischer, R.L. Amniotic fluid: Physiology and assessment. Gynecol. Obstet. 1995, 8, 1–9. [Google Scholar] [CrossRef]
- Papageorghiou, A. Fetal physiology. In MRCOG Part One: Your Essential Revision Guide; Fiander, A., Thilaganathan, B., Eds.; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Bakhsh, H.; Alenizy, H.; Alenazi, S.; Alnasser, S.; Alanazi, N.; Alsowinea, M.; Alharbi, L.; Alfaifi, B. Amniotic fluid disorders and the effects on prenatal outcome: A retrospective cohort study. BMC Pregnancy Childbirth 2021, 21, 75. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Sonek, J.; McKenna, D.; Cool, D.; Aydas, B.; Turkoglu, O.; Bjorndahl, T.; Mandal, R.; Wishart, D.; Friedman, P.; et al. Artificial intelligence and amniotic fluid multiomics: Prediction of perinatal outcome in asymptomatic women with short cervix. Ultrasound Obstet. Gynecol. 2019, 54, 110–118. [Google Scholar] [CrossRef]
- Ramya, R.; Srinivasan, K. Classification of Amniotic Fluid Level Using Bi-LSTM with Homomorphic filter and Contrast Enhancement Techniques. Wirel. Pers. Commun. 2021, 124, 1123–1150. [Google Scholar] [CrossRef]
- Ayu, P.D.W.; Hartati, S.; Musdholifah, A.; Nurdiati, D.S. Amniotic Fluids Classification Using Combination of Rules-Based and Random Forest Algorithm. In Communications in Computer and Information Science; Springer: Berlin/Heidelberg, Germany, 2021; Volume 1489, pp. 267–285. [Google Scholar] [CrossRef]
- Ayu, P.D.W.; Hartati, S. Pixel Classification Based on Local Gray Level Rectangle Window Sampling for Amniotic Fluid Segmentation. Int. J. Intell. Eng. Syst. 2021, 14, 420–432. [Google Scholar] [CrossRef]
- Amuthadevi, C.; Subarnan, G.M. Development of fuzzy approach to predict the fetus safety and growth using AFI. J. Supercomput. 2020, 76, 5981–5995. [Google Scholar] [CrossRef]
- Sun, S.; Kwon, J.Y.; Park, Y.; Cho, H.C.; Hyun, C.M.; Seo, J.K. Complementary Network for Accurate Amniotic Fluid Segmentation from Ultrasound Images. IEEE Access 2021, 9, 108223–108235. [Google Scholar] [CrossRef]
- Li, Y.; Xu, R.; Ohya, J.; Iwata, H. Automatic fetal body and amniotic fluid segmentation from fetal ultrasound images by encoder-decoder network with inner layers. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), Jeju, Korea, 11–15 July 2017. [Google Scholar] [CrossRef]
- Ayu, D.W.; Hartati, S.; Musdholifah, A. Amniotic Fluid Segmentation by Pixel Classification in B-Mode Ultrasound Image for Computer Assisted Diagnosis. In Communications in Computer and Information Science; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1100. [Google Scholar] [CrossRef]
- Ayu, P.D.W.; Hartati, S.; Musdholifah, A.; Nurdiati, D.S. Amniotic fluid segmentation based on pixel classification using local window information and distance angle pixel. Appl. Soft Comput. 2021, 107, 107196. [Google Scholar] [CrossRef]
- Looney, P.; Yin, Y.; Collins, S.L.; Nicolaides, K.H.; Plasencia, W.; Molloholli, M.; Natsis, S.; Stevenson, G.N. Fully Automated 3-D Ultrasound Segmentation of the Placenta, Amniotic Fluid, and Fetus for Early Pregnancy Assessment. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2021, 68, 2038–2047. [Google Scholar] [CrossRef]
- Anquez, J.; Angelini, E.D.; Grange, G.; Bloch, I. Automatic segmentation of antenatal 3-D ultrasound images. IEEE Trans. Biomed. Eng. 2013, 60, 1388–1400. [Google Scholar] [CrossRef] [Green Version]
- Friedman, P.; Ogunyemi, D. Oligohydramnios. In Obstetric Imaging: Fetal Diagnosis and Care, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Han, M.; Bao, Y.; Sun, Z.; Wen, S.; Xia, L.; Zhao, J.; Du, J.; Yan, Z. Automatic Segmentation of Human Placenta Images with U-Net. IEEE Access 2019, 7, 180083–180092. [Google Scholar] [CrossRef]
- Yang, X.; Yu, L.; Li, S.; Wang, X.; Wang, N.; Qin, J.; Ni, D.; Heng, P.-A. Towards Automatic Semantic Segmentation in Volumetric Ultrasound. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017, Quebec City, QC, Canada, 11–13 September 2017; pp. 711–719. [Google Scholar]
- Hu, R.; Singla, R.; Yan, R.; Mayer, C.; Rohling, R.N. Automated Placenta Segmentation with a Convolutional Neural Network Weighted by Acoustic Shadow Detection. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6718–6723. [Google Scholar] [CrossRef]
- Zimmer, V.A.; Gomez, A.; Skelton, E.; Ghavami, N.; Wright, R.; Li, L.; Matthew, J.; Hajnal, J.V.; Schnabel, J.A. A Multi-task Approach Using Positional Information for Ultrasound Placenta Segmentation. In Proceedings of the Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis, Lima, Peru, 4–8 October 2020; pp. 264–273. [Google Scholar]
- Hu, Z.; Hu, R.; Yan, R.; Mayer, C.; Rohling, R.N.; Singla, R. Automatic Placenta Abnormality Detection Using Convolutional Neural Networks on Ultrasound Texture. In Proceedings of the Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging, Placental and Preterm Image Analysis, Strasbourg, France, 1 October 2021; pp. 147–156. [Google Scholar]
- Schilpzand, M.; Neff, C.; van Dillen, J.; van Ginneken, B.; Heskes, T.; de Korte, C.; van den Heuvel, T. Automatic Placenta Localization From Ultrasound Imaging in a Resource-Limited Setting Using a Predefined Ultrasound Acquisition Protocol and Deep Learning. Ultrasound Med. Biol. 2022, 48, 663–674. [Google Scholar] [CrossRef]
- Zimmer, V.A.; Gomez, A.; Skelton, E.; Toussaint, N.; Zhang, T.; Khanal, B.; Wright, R.; Noh, Y.; Ho, A.; Matthew, J.; et al. Towards Whole Placenta Segmentation at Late Gestation Using Multi-view Ultrasound Images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Shenzhen, China, 13–17 October 2019; pp. 628–636. [Google Scholar]
- Looney, P.; Stevenson, G.N.; Nicolaides, K.H.; Plasencia, W.; Molloholli, M.; Natsis, S.; Collins, S.L. Automatic 3D ultrasound segmentation of the first trimester placenta using deep learning. In Proceedings of the International Symposium on Biomedical Imaging, Melbourne, VIC, Australia, 18–21 April 2017. [Google Scholar] [CrossRef]
- Looney, P.; Stevenson, G.N.; Nicolaides, K.H.; Plasencia, W.; Molloholli, M.; Natsis, S.; Collins, S.L. Fully automated, real-time 3D ultrasound segmentation to estimate first trimester placental volume using deep learning. JCI Insight 2018, 3, e120178. [Google Scholar] [CrossRef]
- Saavedra, A.C.; Arroyo, J.; Tamayo, L.; Egoavil, M.; Ramos, B.; Castaneda, B. Automatic ultrasound assessment of placenta previa during the third trimester for rural areas. In Proceedings of the 2020 IEEE International Ultrasonics Symposium (IUS), Las Vegas, NV, USA, 7–11 September 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Oguz, B.U.; Wang, J.; Yushkevich, N.; Pouch, A.; Gee, J.; Yushkevich, P.A.; Schwartz, N.; Oguz, I. Combining Deep Learning and Multi-atlas Label Fusion for Automated Placenta Segmentation from 3DUS. In Proceedings of the Data Driven Treatment Response Assessment and Preterm, Perinatal, and Paediatric Image Analysis, Granada, Spain, 16 September 2018; pp. 138–148. [Google Scholar]
- Qi, H.; Collins, S.; Noble, J.A. Knowledge-guided Pretext Learning for Utero-placental Interface Detection. Med. Image Comput. Comput. Interv. MICCAI 2020, 12261, 582–593. [Google Scholar] [CrossRef]
- Romeo, V.; Ricciardi, C.; Cuocolo, R.; Stanzione, A.; Verde, F.; Sarno, L.; Improta, G.; Mainenti, P.P.; D’Armiento, M.; Brunetti, A.; et al. Machine learning analysis of MRI-derived texture features to predict placenta accreta spectrum in patients with placenta previa. Magn. Reson. Imaging 2019, 64, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, C.; Zhang, Z.; Goldstein, J.A.; Gernand, A.D.; Wang, J.Z. PlacentaNet: Automatic Morphological Characterization of Placenta Photos with Deep Learning. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; pp. 487–495. [Google Scholar] [CrossRef]
- Sridar, P.; Kumar, A.; Quinton, A.; Nanan, R.; Kim, J.; Krishnakumar, R. Decision Fusion-Based Fetal Ultrasound Image Plane Classification Using Convolutional Neural Networks. Ultrasound Med. Biol. 2019, 45, 1259–1273. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y. Prediction and Value of Ultrasound Image in Diagnosis of Fetal Central Nervous System Malformation under Deep Learning Algorithm. Adv. Sci. Program. Methods Health Inform. 2021, 2021, 6246274. [Google Scholar] [CrossRef]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Deep Learning Techniques for Automatic Detection of Embryonic Neurodevelopmental Disorders. Diagnostics 2020, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bano, S.; Vasconcelos, F.; Vander Poorten, E.; Vercauteren, T.; Ourselin, S.; Deprest, J.; Stoyanov, D. FetNet: A recurrent convolutional network for occlusion identification in fetoscopic videos. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Bano, S.; Vasconcelos, F.; Tella-Amo, M.; Dwyer, G.; Gruijthuijsen, C.; Vander Poorten, E.; Vercauteren, T.; Ourselin, S.; Deprest, J.; Stoyanov, D. Deep learning-based fetoscopic mosaicking for field-of-view expansion. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.A.; Ourak, M.; Gruijthuijsen, C.; Deprest, J.; Vercauteren, T.; Vander Poorten, E. Deep learning-based monocular placental pose estimation: Towards collaborative robotics in fetoscopy. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1561–1571. [Google Scholar] [CrossRef]
- Casella, A.; Moccia, S.; Frontoni, E.; Paladini, D.; De Momi, E.; Mattos, L.S. Inter-foetus Membrane Segmentation for TTTS Using Adversarial Networks. Ann. Biomed. Eng. 2020, 48, 848–859. [Google Scholar] [CrossRef] [Green Version]
- Sufriyana, H.; Wu, Y.-W.; Su, E.C.-Y. Prognostication for prelabor rupture of membranes and the time of delivery in nationwide insured women: Development, validation, and deployment. medRxiv 2021. [Google Scholar] [CrossRef]
- Gao, C.; Osmundson, S.; Velez Edwards, D.R.; Jackson, G.P.; Malin, B.A.; Chen, Y. Deep learning predicts extreme preterm birth from electronic health records. J. Biomed. Inform. 2019, 100, 103334. [Google Scholar] [CrossRef]
- Lee, K.-S.; Kim, H.Y.; Lee, S.J.; Kwon, S.O.; Na, S.; Hwang, H.S.; Park, M.H.; Ahn, K.H.; Korean Society of Ultrasound in Obstetrics and Gynecology Research Group. Prediction of newborn’s body mass index using nationwide multicenter ultrasound data: A machine-learning study. BMC Pregnancy Childbirth 2021, 21, 172. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Wan, L.; Li, Z.; Qing, L.; Qi, X. Fetal Weight Estimation via Ultrasound Using Machine Learning. IEEE Access 2019, 7, 87783–87791. [Google Scholar] [CrossRef]
- Ghelich Oghli, M.; Shabanzadeh, A.; Moradi, S.; Sirjani, N.; Gerami, R.; Ghaderi, P.; Sanei Taheri, M.; Shiri, I.; Arabi, H.; Zaidi, H. Automatic fetal biometry prediction using a novel deep convolutional network architecture. Phys. Med. 2021, 88, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Y.; Yang, X.; Lei, B.; Liu, L.; Li, S.X.; Ni, D.; Wang, T. Deep Learning in Medical Ultrasound Analysis: A Review. Engineering 2019, 5, 261–275. [Google Scholar] [CrossRef]
- Park, S.H. Artificial intelligence for ultrasonography: Unique opportunities and challenges. Ultrasonography 2021, 40, 3–6. [Google Scholar] [CrossRef]
- Wadden, J.J. Defining the undefinable: The black box problem in healthcare artificial intelligence. J. Med. Ethics 2021. Online First. [Google Scholar] [CrossRef]


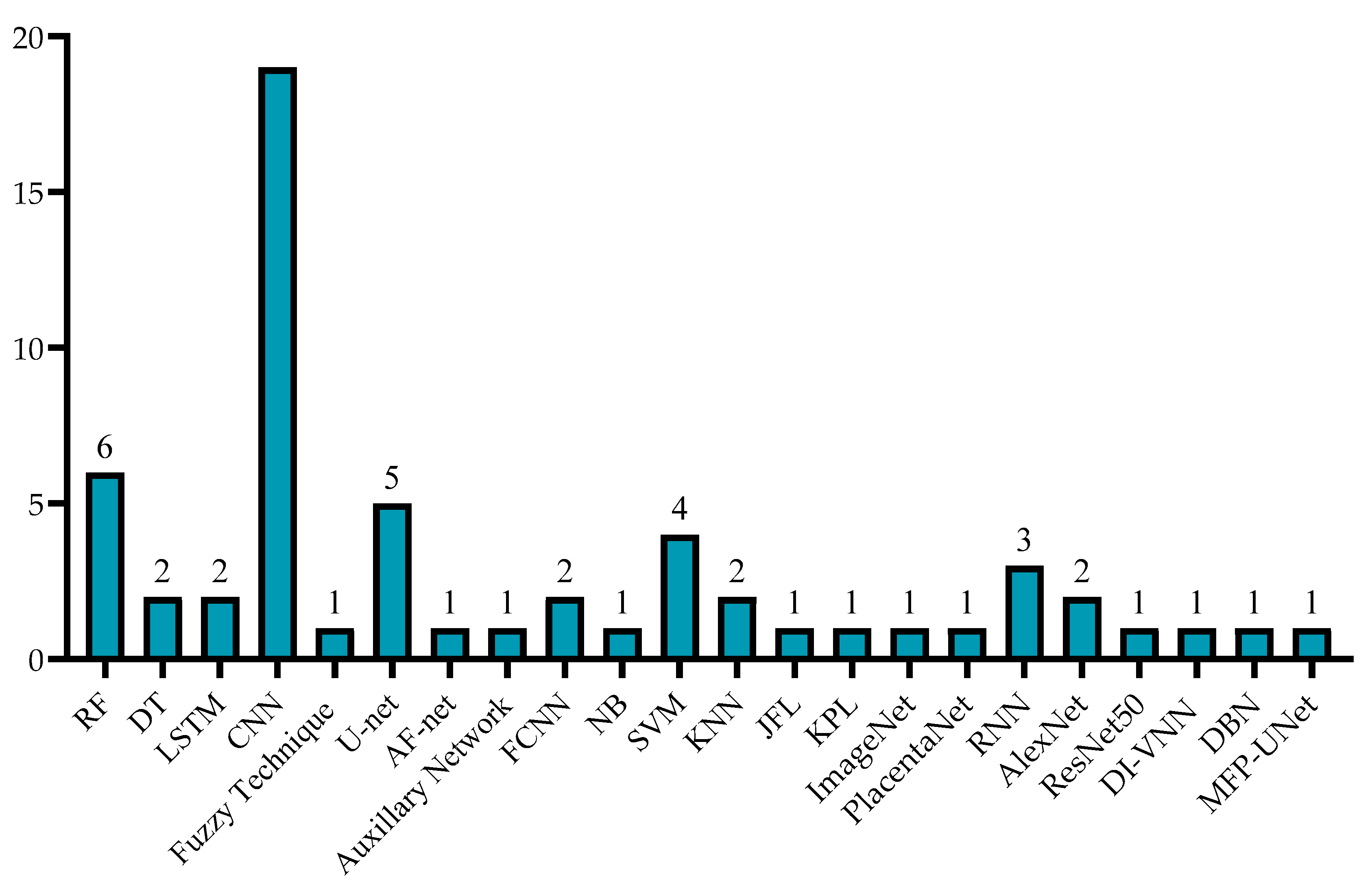
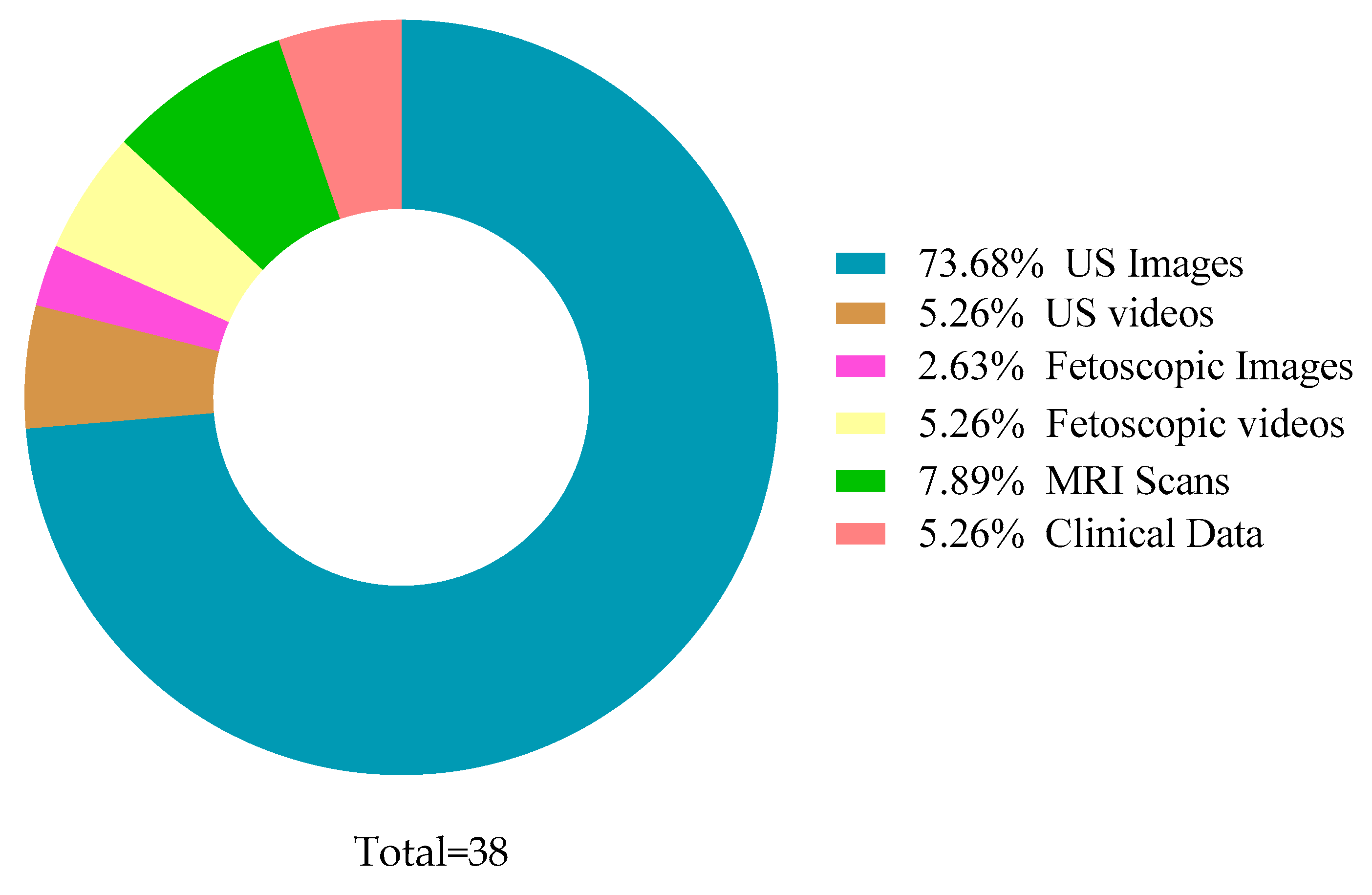

| Domain | Ref. | Year | Data Type | Dataset Size | Binary/ Multi-Class | Augmentation | Methods | MSE | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| ML | [12] | 2021 | US Images | 95 | Multi | No | RF | 0.9052 | |
| [13] | 2021 | US Images | 50 | Multi | No | RF, DT | 0.995 | ||
| DL | [11] | 2021 | US Images | 4000 | Multi | No | HBU-LSTM | 0.5244 | |
| [10] | 2018 | US Images | 26 | Binary | Yes | CNN | 0.95 | ||
| [14] | 2020 | US Images | 50 | Multi | No | Fuzzy Technique | 0.925 |
| Domain | Ref. | Year | Data Type | Dataset Size | Binary/ Multi-Class | Augmentation | Methods | DSC | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| DL | [5] | 2021 | US Images | 435 | Binary | Yes | U-net (CNN) | 0.877 | |
| [15] | 2021 | US Images | 2380 | Multi | Yes | AF-net + Auxiliary Network | 0.8599 | ||
| [19] | 2021 | US Images | 2393 | Binary, Multi | No | FCNN | 0.84 | ||
| [16] | 2017 | US Videos | 601 | Multi | Yes | CNN, Encoder–Decoder Network | 0.93 | ||
| ML | [18] | 2021 | US Images | 55 | Binary | No | RF, DT, NB, SVM, and KNN | RF—0.876 | |
| [17] | 2019 | US Images | 50 | Multi | Yes | RF | 0.5553 | 0.8586 | |
| [20] | 2013 | US Images | 19 | Binary | No | Bayesian Formulation | Overlap Measure—0.89 |
| Condition | Ref. | Type | Year | Data Type | Dataset Size | Domain | Methods | Performance Measure |
|---|---|---|---|---|---|---|---|---|
| Oligo | [29] | Placenta | 2017 | US Images | 2893 | DL | CNN, Random Walker | DSC-0.73 |
| [22] | Placenta | 2019 | MRI Images | 1110 | DL | CNN (U-net) | Acc-0.98 | |
| [30] | Placenta | 2018 | US Images | 1200 | DL | CNN | DSC-0.81 | |
| [25] | Placenta | 2020 | US Images (3D) | 1054 | DL | U-net | DSC-0.87 | |
| [24] | Placenta | 2019 | US Images | 1364 | DL | CNN | DSC-0.92 | |
| [26] | Placenta | 2021 | US Images | 321 (patients) | DL | CNN | Acc-0.81 | |
| [27] | Placenta | 2021 | US Images | 6576 | DL | U-net | Acc-0.84 | |
| [31] | Placenta | 2020 | US Images | 11,014 | DL | U-net | Sen-0.75 Spe-0.92 | |
| [32] | Placenta | 2018 | US Images | 47 (patients) | DL | JLF + CNN | DSC-0.863 | |
| [33] | Placenta | 2020 | US Volumes | 101 | DL | KPL, ImageNet | ODS-0.605 | |
| [34] | Placenta | 2019 | MRI Images | 64 (patients) | ML | KNN | Acc-0.981 | |
| [35] | Placenta | 2019 | Photographic Images | 1003 | DL | CNN (PlacentaNet) | Acc-0.9751 | |
| [23] | Placenta | 2017 | US Images (3D) | 104 (patients) | DL | FCNN, RNN | DSC-0.882 | |
| [28] | Placenta | 2019 | US Images (3D) | 127 | DL | CNN | DSC-0.8 | |
| [36] | Kidney | 2019 | US Images | 4074 | DL | CNN (AlexNet) | Acc-0.9705 | |
| Poly | [38] | Neuro | 2020 | MRI Images | 227 | DL | SVM + CNN (AlexNet, ResNet50) | Acc-0.886 |
| [37] | Neuro | 2021 | US Images | 63 | DL | CNN | p < 0.05 | |
| Both | [42] | TTTS | 2019 | US Videos | 900 | DL | CNN | DSC-0.9191 |
| [39] | TTTS | 2020 | Fetoscopic Videos | 138,780 | DL | CNN and LSTM | Precision-0.96 | |
| [40] | TTTS | 2020 | Fetoscopic Videos | 2400 | DL | CNN | - | |
| [41] | TTTS | 2020 | Fetoscopic Images | 30,000 | DL | CNN | Acc-0.87 | |
| [43] | Preterm | 2021 | Clinical Data | 219,272 (patients) | DL | DI-VNN | Sen-0.494 Spe-0.816 | |
| [44] | Preterm | 2019 | Clinical Data | 25,689 (patients) | DL | RNN | Sen-0.819 AUC-0.777 | |
| [45] | Health | 2021 | US Images | 3159 (patients) | ML | RF | MSE-0.02161 | |
| [46] | Health | 2019 | US Images | 7875 (patients) | ML | SVM, DBN | MAPE-0.0609 | |
| [47] | Health | 2021 | US Images | 1334 | DL | CNN (MFP-U-net) | DSC-0.98 |
| Ref. | Limitations | Solutions |
|---|---|---|
| [18] | Limited features to differentiate between actual AF and reflected waves. Insufficient uneven window shapes for relevant results. | Considering AF coordinates. Adding data for uneven and rectangular window shapes. |
| [12] | As there is no accurate measuring number for echogenicity, only doctor’s insight applied in observing the gray texture in US images. | - |
| [11] | Fetal weight ignored for results. | Present findings considering fetal weight. |
| [20] | Not entirely automated—Images manually labeled. Advance dataset with all different forms of US images is required. | Automize labelling of US images. |
| [13] | Uncertain aspects, such as angle and direction of the transducer, are ignored. Decreased AF appearance in overweight women due to US beam. | Considering maternal position during Amniotic Fluid Volume (AFV) measurement. |
| [5] | Errors in final finding due to secondary path in complementation procedure. US Images with noise disturbance. | Distinction between both paths. Unified image settings. |
| [15] | Moderate accuracy. Specific situation. | More broad case with additional clinical data and adequate labels. |
| [16] | Average accuracy. | - |
| [17] | Clinical data and US images exhibit moderate accuracy. | Omics data analysis to provide understanding to patients and help with clinical management. |
| [10] | Various measurements scattered. | First trimester screening tool combining different measures and characteristics. |
| [19] | Manual refinement of models. | Automate procedure. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, I.U.; Aslam, N.; Anis, F.M.; Mirza, S.; AlOwayed, A.; Aljuaid, R.M.; Bakr, R.M. Amniotic Fluid Classification and Artificial Intelligence: Challenges and Opportunities. Sensors 2022, 22, 4570. https://doi.org/10.3390/s22124570
Khan IU, Aslam N, Anis FM, Mirza S, AlOwayed A, Aljuaid RM, Bakr RM. Amniotic Fluid Classification and Artificial Intelligence: Challenges and Opportunities. Sensors. 2022; 22(12):4570. https://doi.org/10.3390/s22124570
Chicago/Turabian StyleKhan, Irfan Ullah, Nida Aslam, Fatima M. Anis, Samiha Mirza, Alanoud AlOwayed, Reef M. Aljuaid, and Razan M. Bakr. 2022. "Amniotic Fluid Classification and Artificial Intelligence: Challenges and Opportunities" Sensors 22, no. 12: 4570. https://doi.org/10.3390/s22124570
APA StyleKhan, I. U., Aslam, N., Anis, F. M., Mirza, S., AlOwayed, A., Aljuaid, R. M., & Bakr, R. M. (2022). Amniotic Fluid Classification and Artificial Intelligence: Challenges and Opportunities. Sensors, 22(12), 4570. https://doi.org/10.3390/s22124570






