Explainable Transformer-Based Deep Learning Model for the Detection of Malaria Parasites from Blood Cell Images
Abstract
:1. Introduction
- (1)
- A multiheaded attention transformer-based model was implemented for the detection of malaria parasites for the first time.
- (2)
- The gradient-weighted class activation map (Grad-CAM) technique was applied to interpret and visualize the trained model.
- (3)
- Original and modified datasets of malaria parasites were used for experimental analysis.
- (4)
- The proposed model for malaria parasite detection was compared with SOTA models.
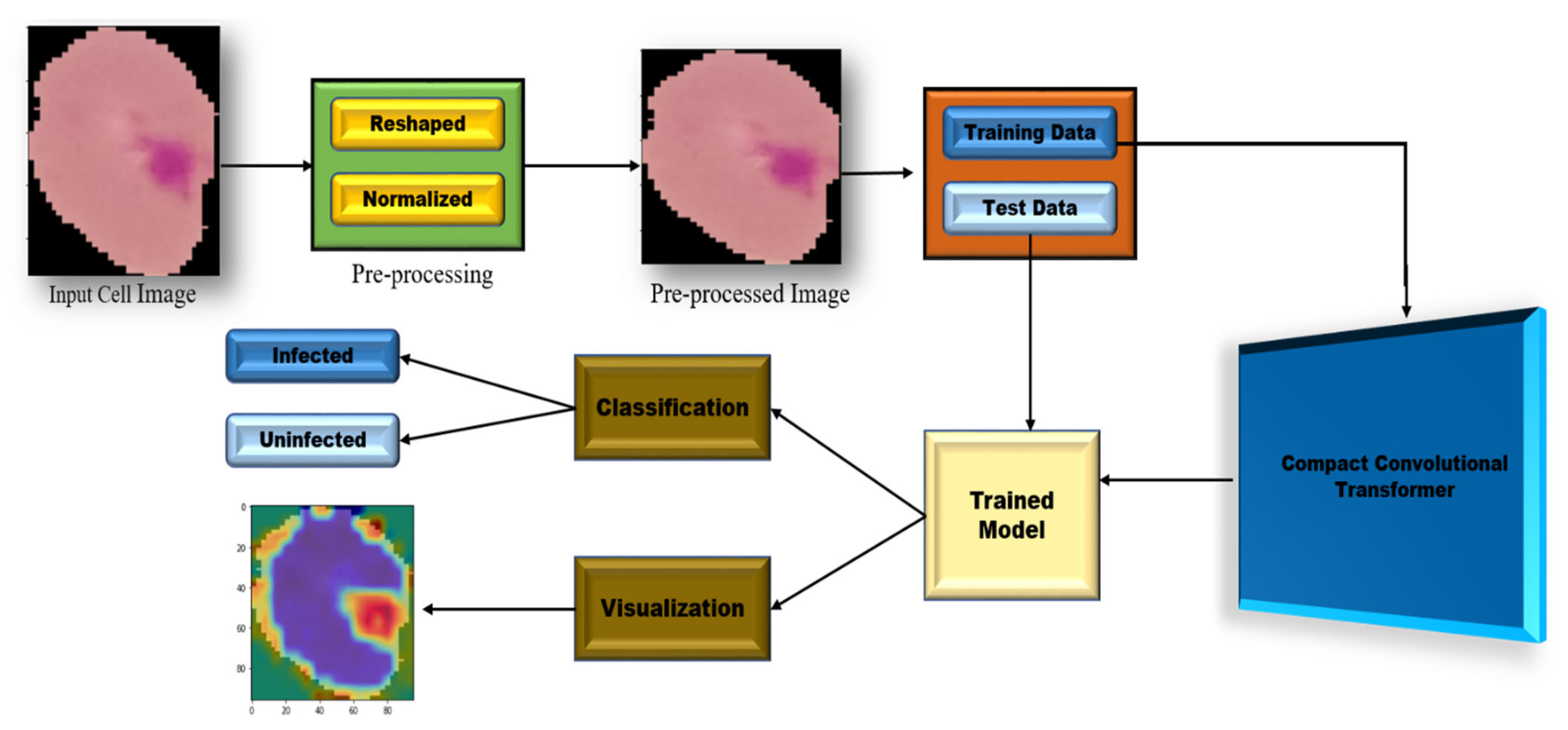
2. Proposed Methodology
2.1. Dataset Description
2.2. Preprocessing (Resize)
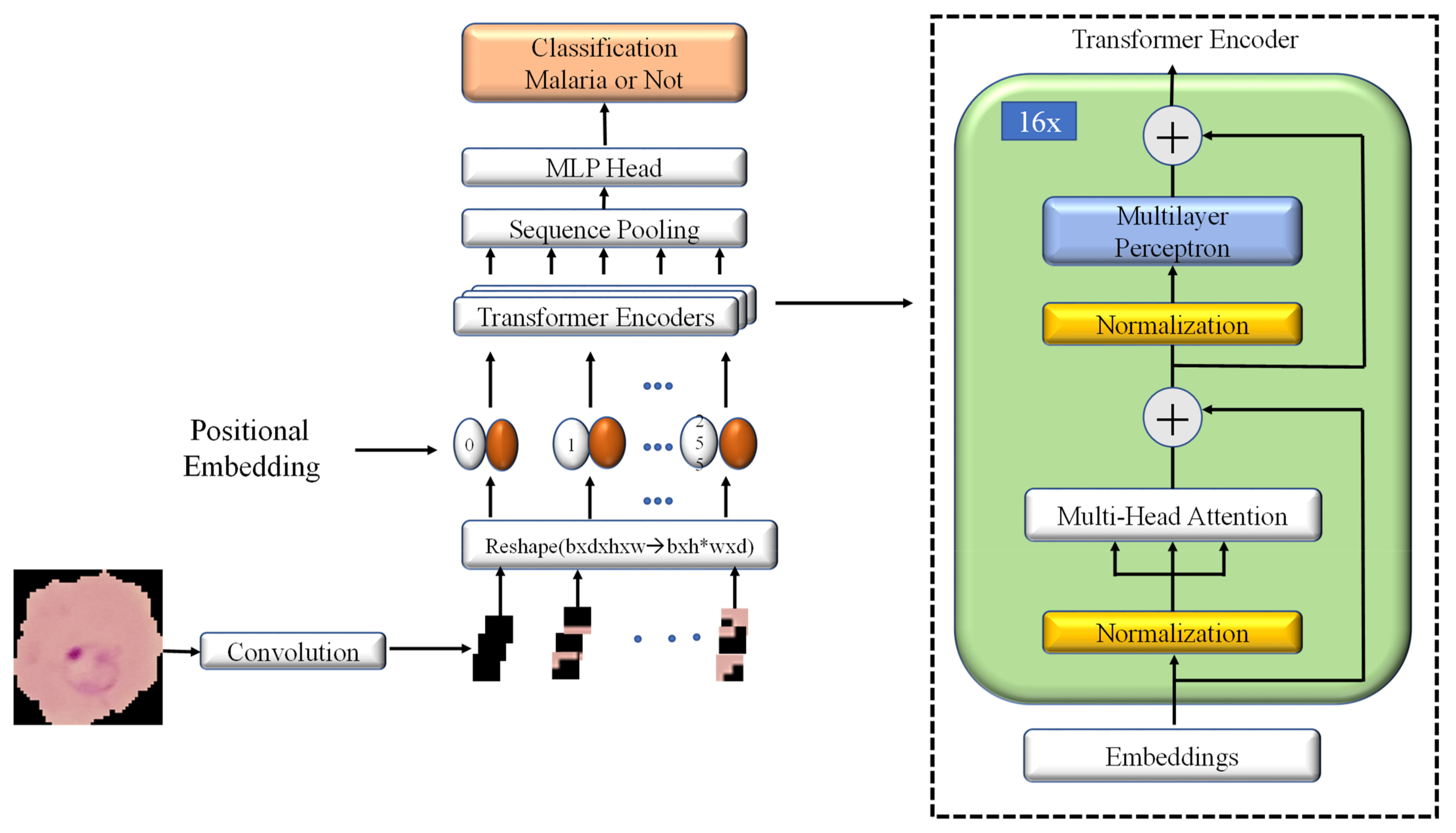
2.3. Model Architecture
2.3.1. Convolutional Block
2.3.2. Multiheaded Attention Mechanism
2.3.3. Transformer Encoder
2.3.4. Sequence Pooling
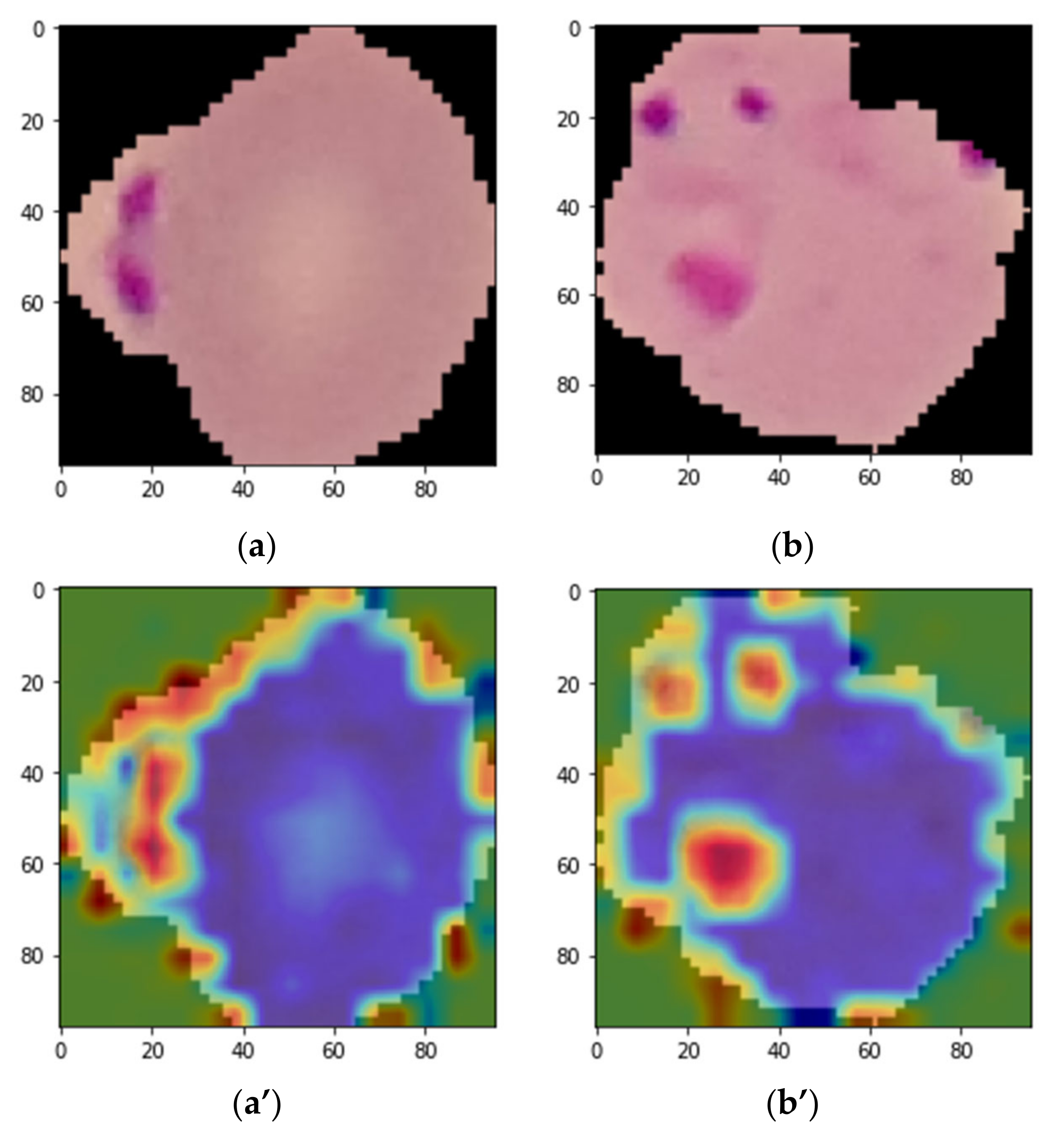
3. Grad-CAM Visualization
4. Result Analysis
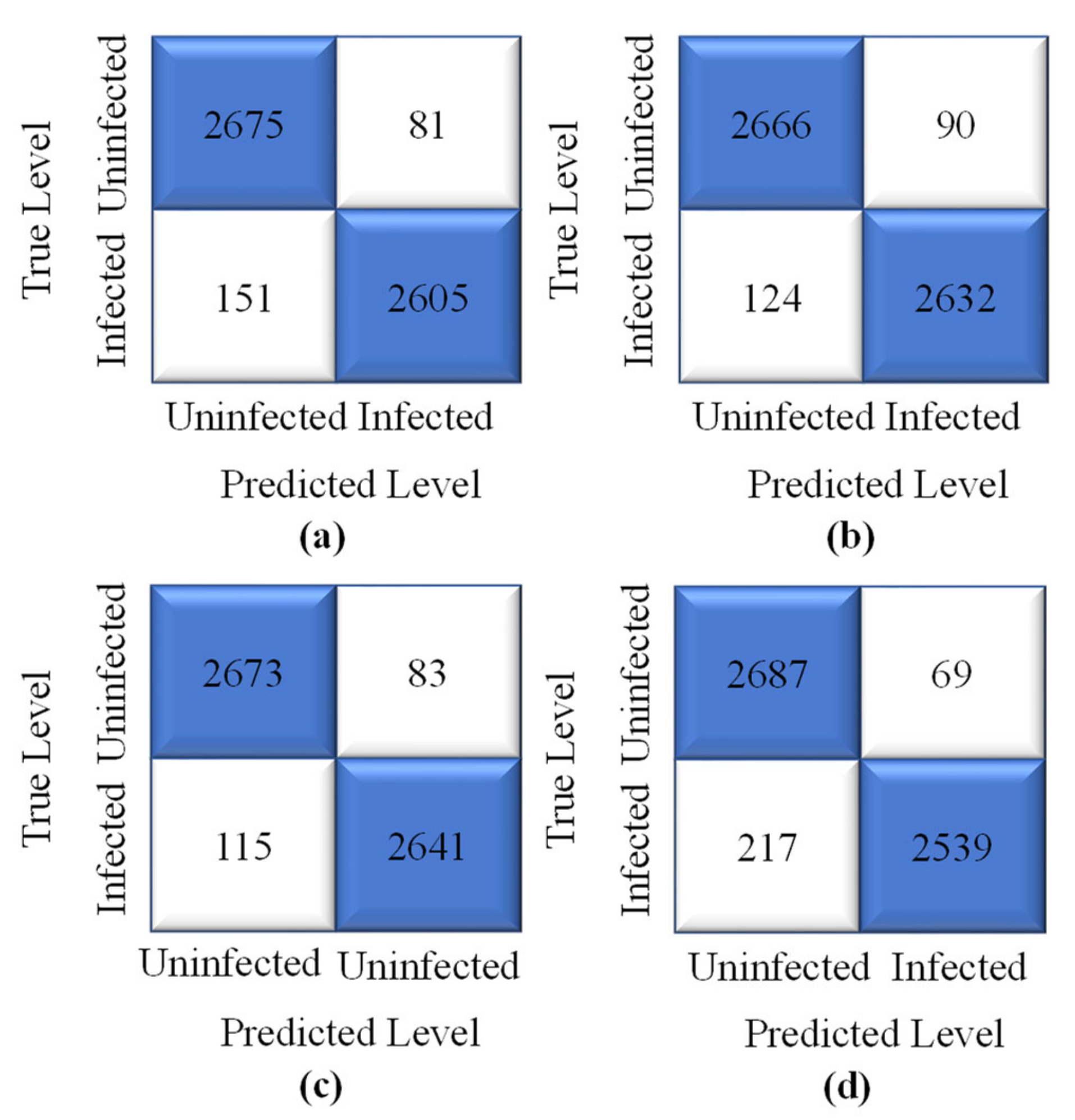
4.1. Performance Evaluation Procedure
4.2. Results Obtained with Original Dataset
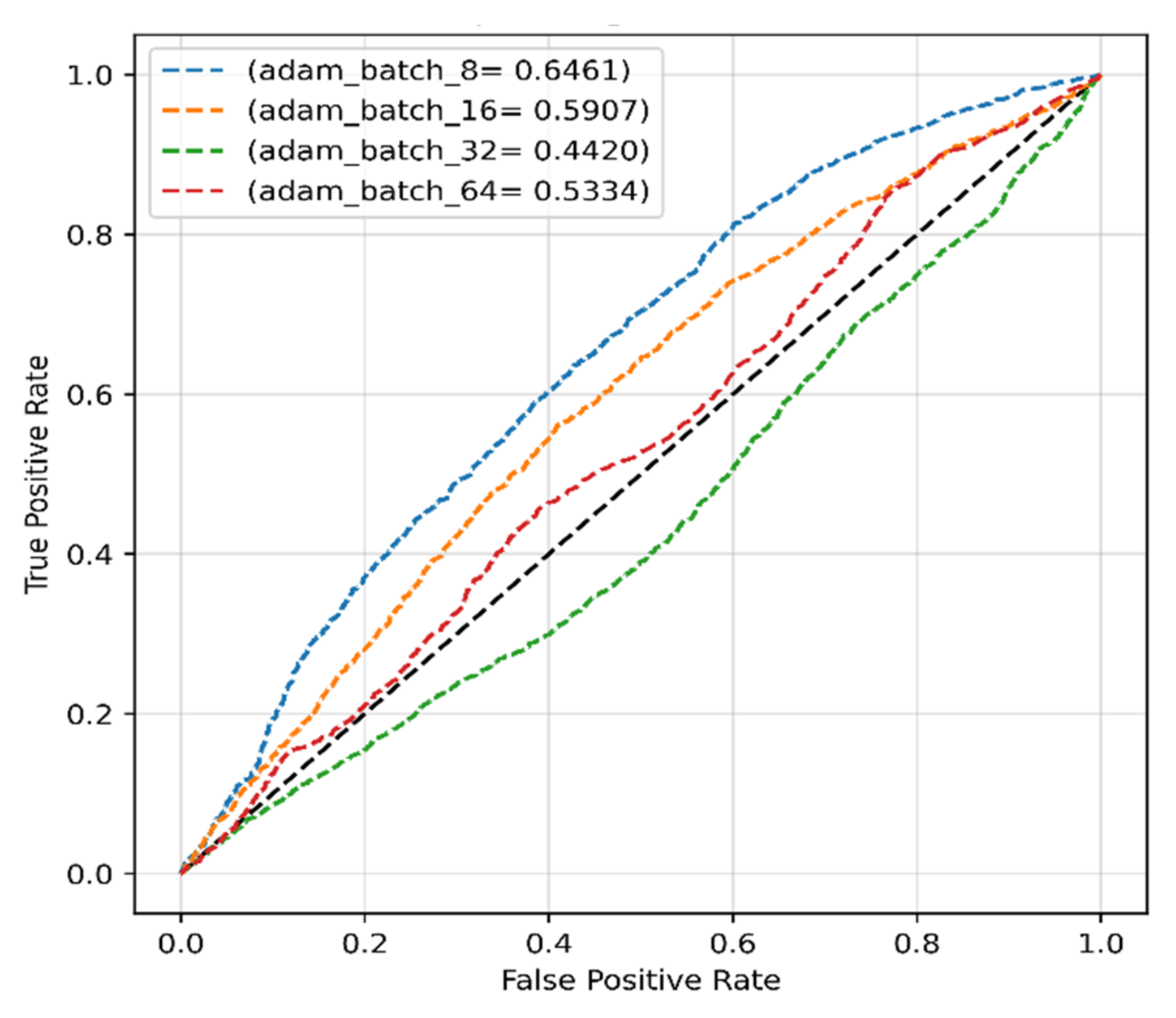
4.2.1. Adam Optimizer for Original Dataset
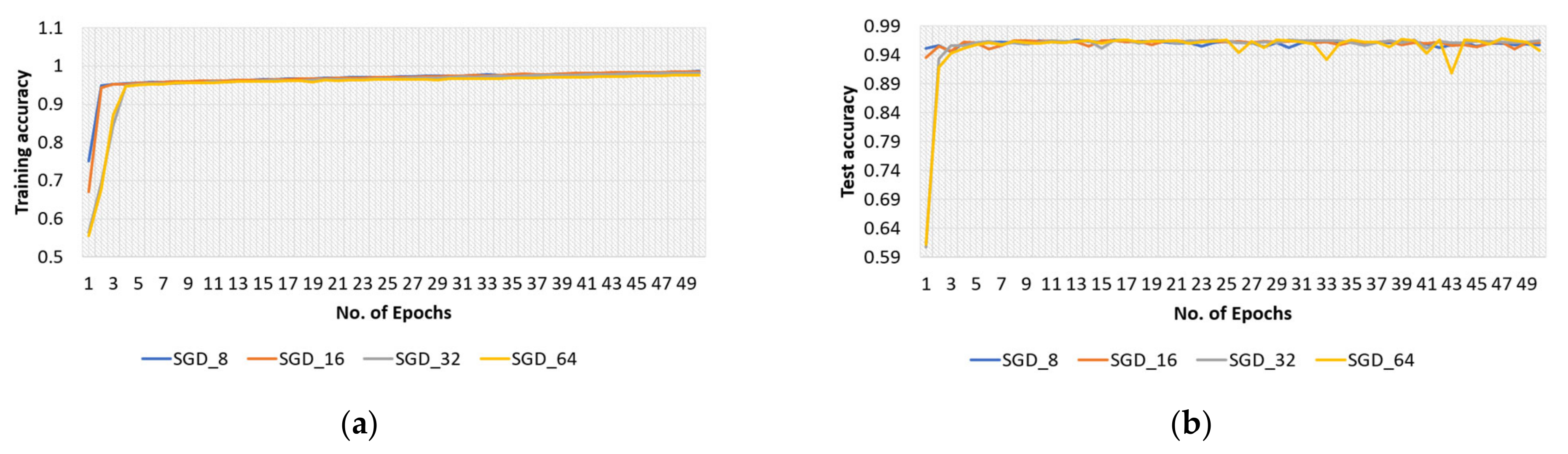
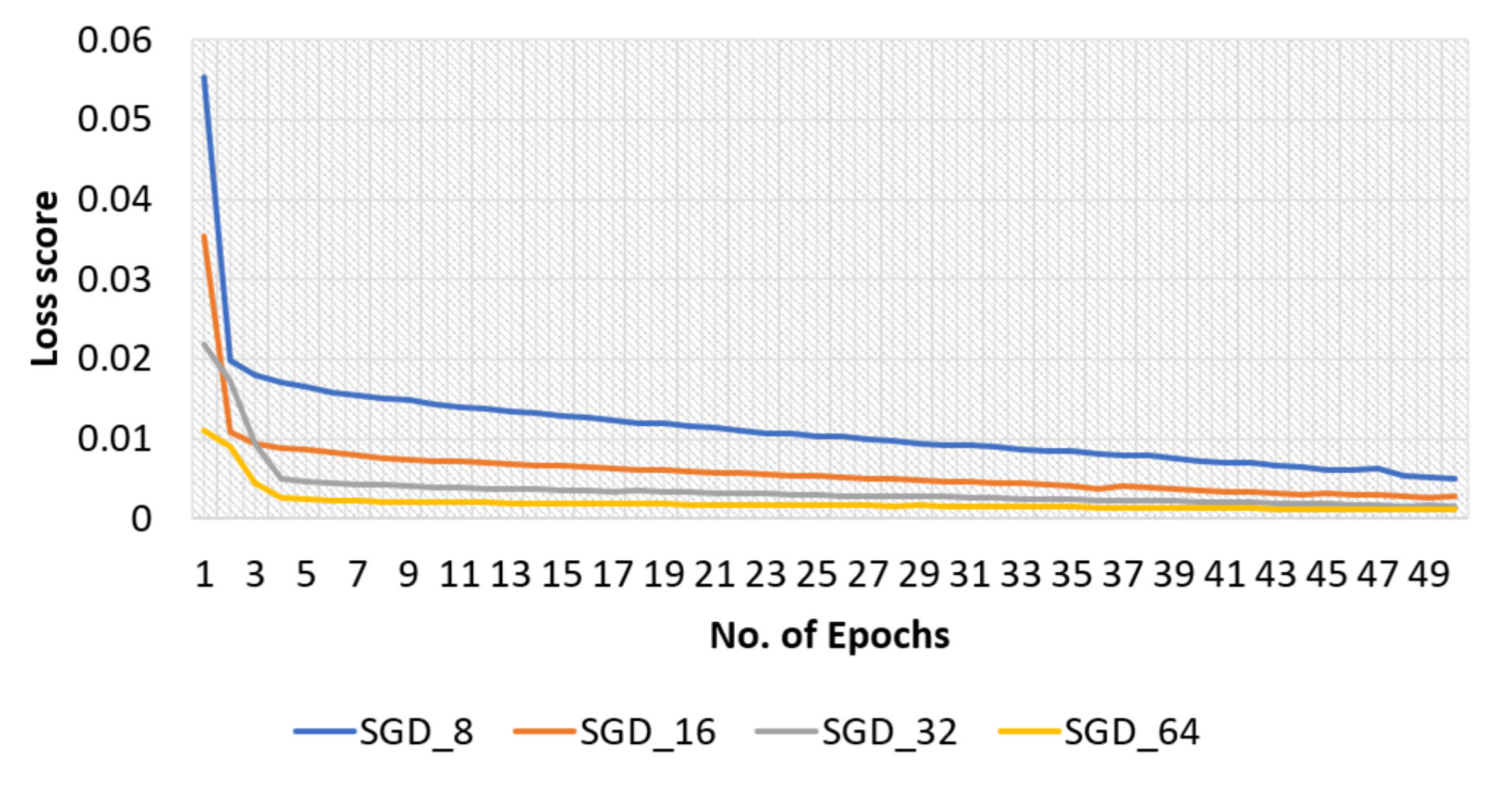
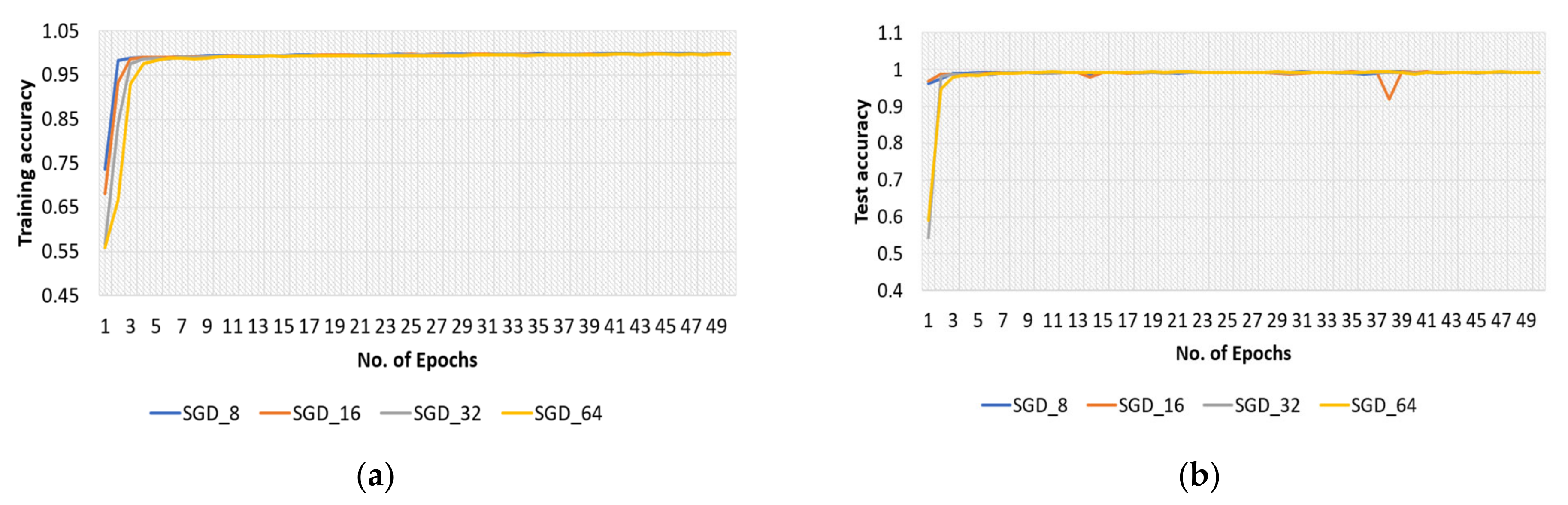
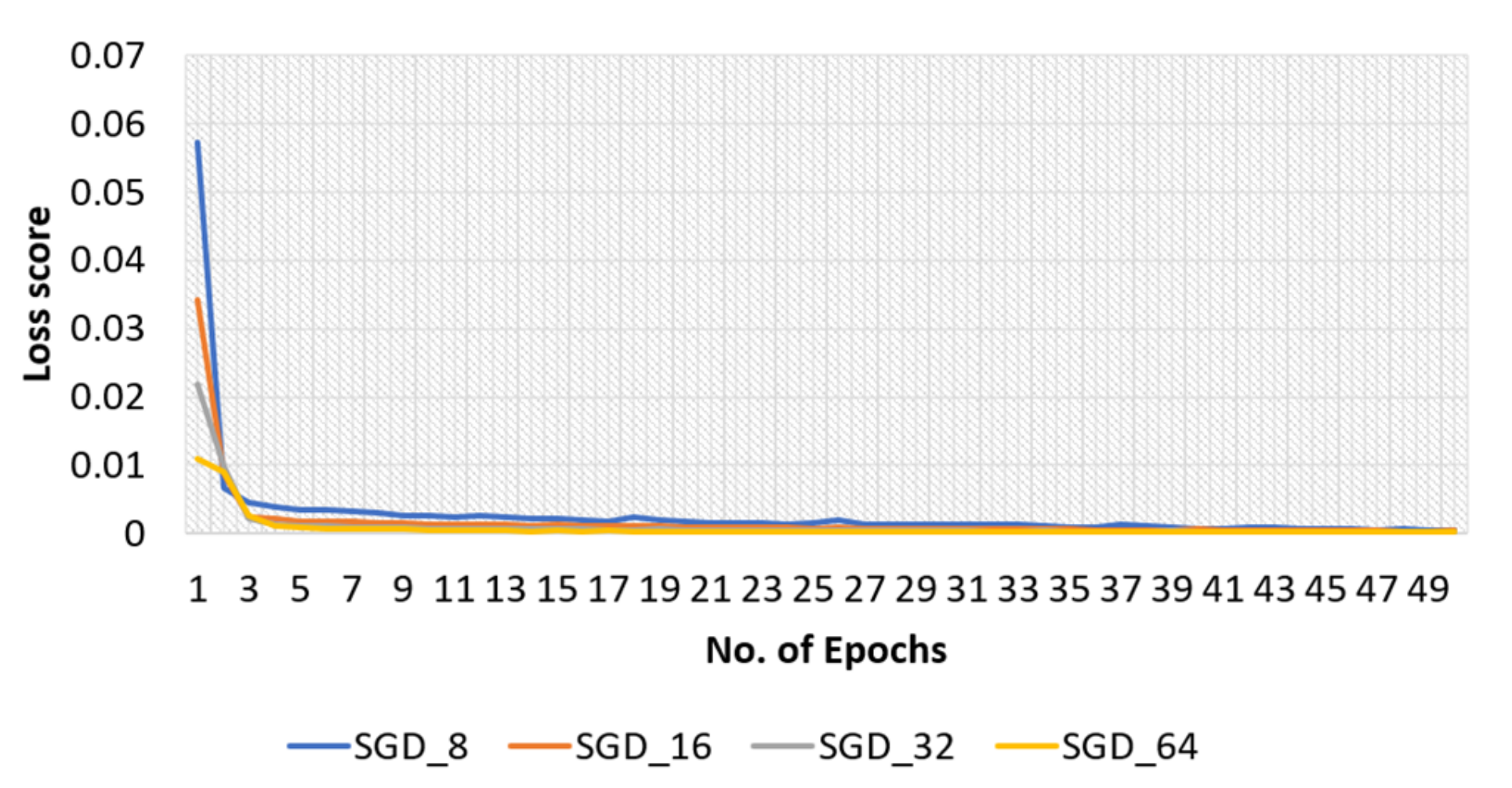
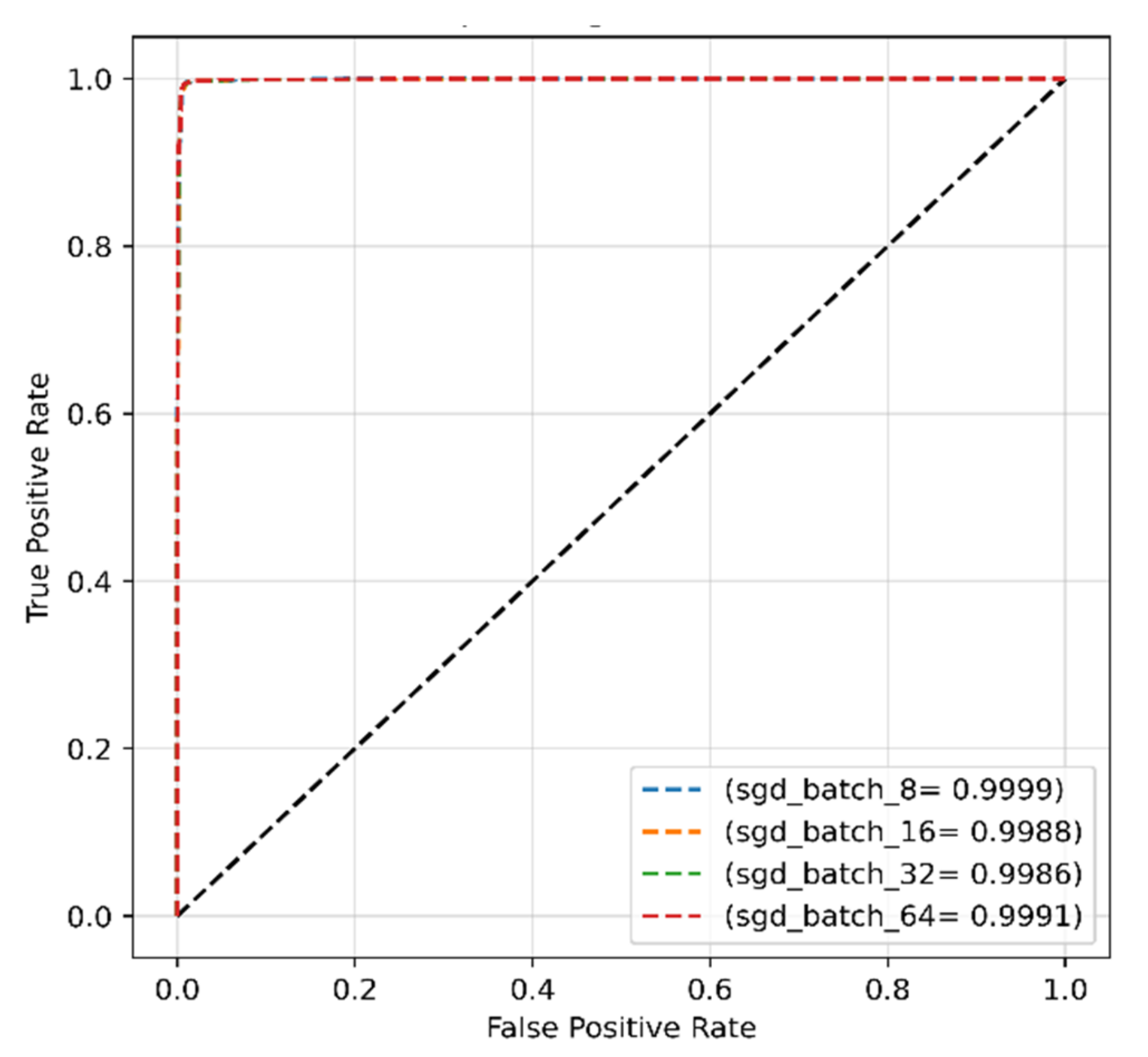
4.2.2. SGD Optimizer for Original Dataset
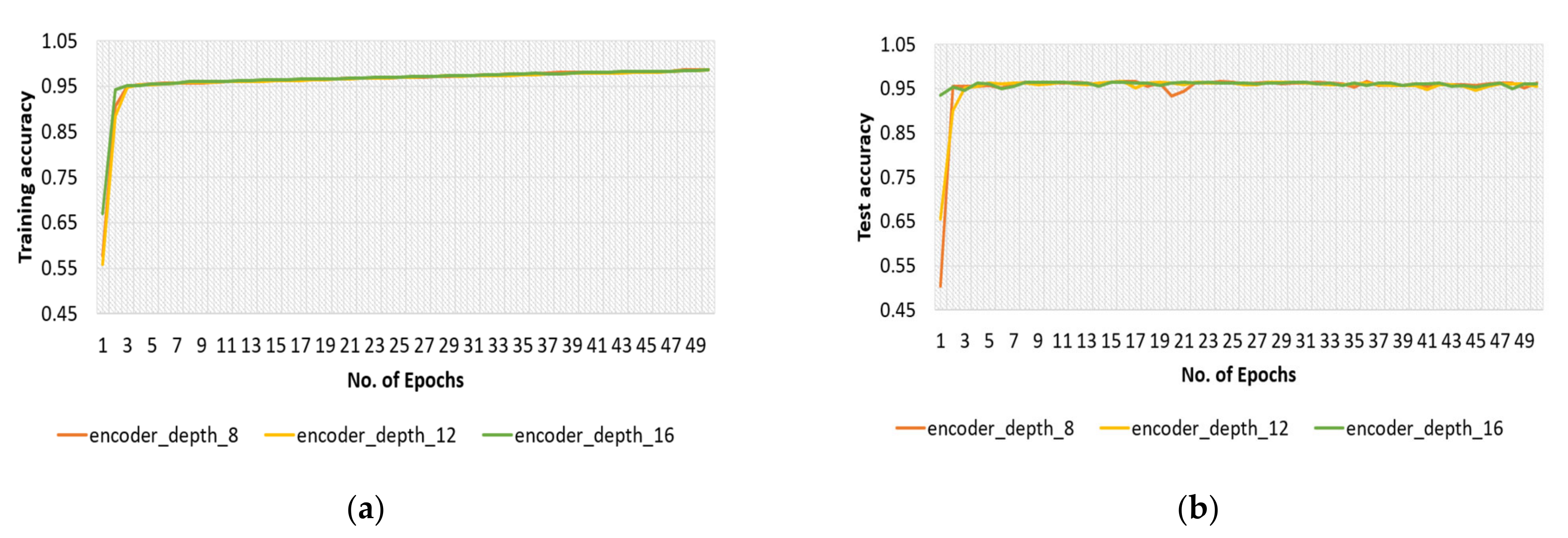
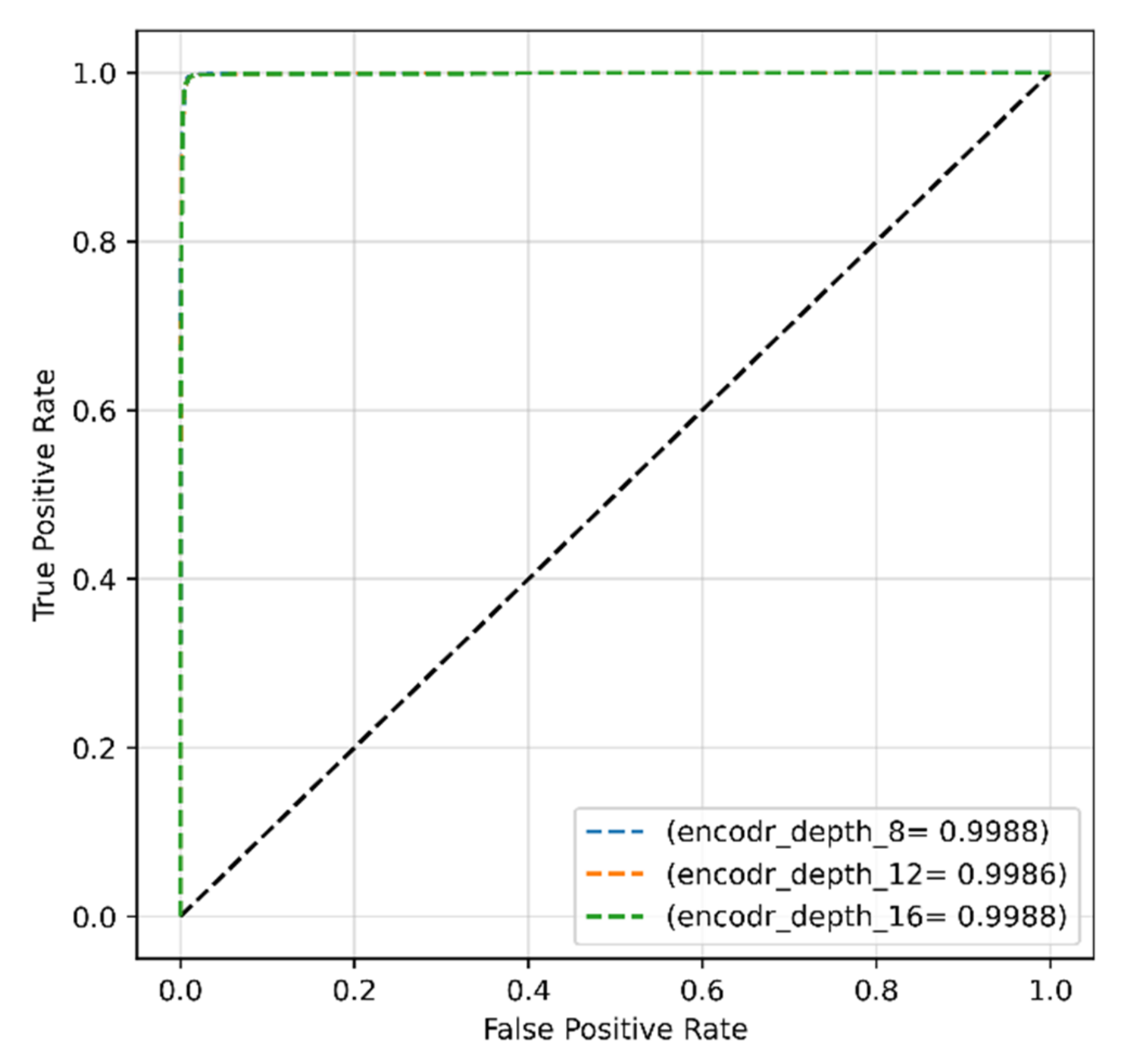
4.2.3. Encoder Depth for Original Dataset
4.3. Results Obtained with Modified Dataset
4.3.1. Adam Optimizer for Modified Dataset
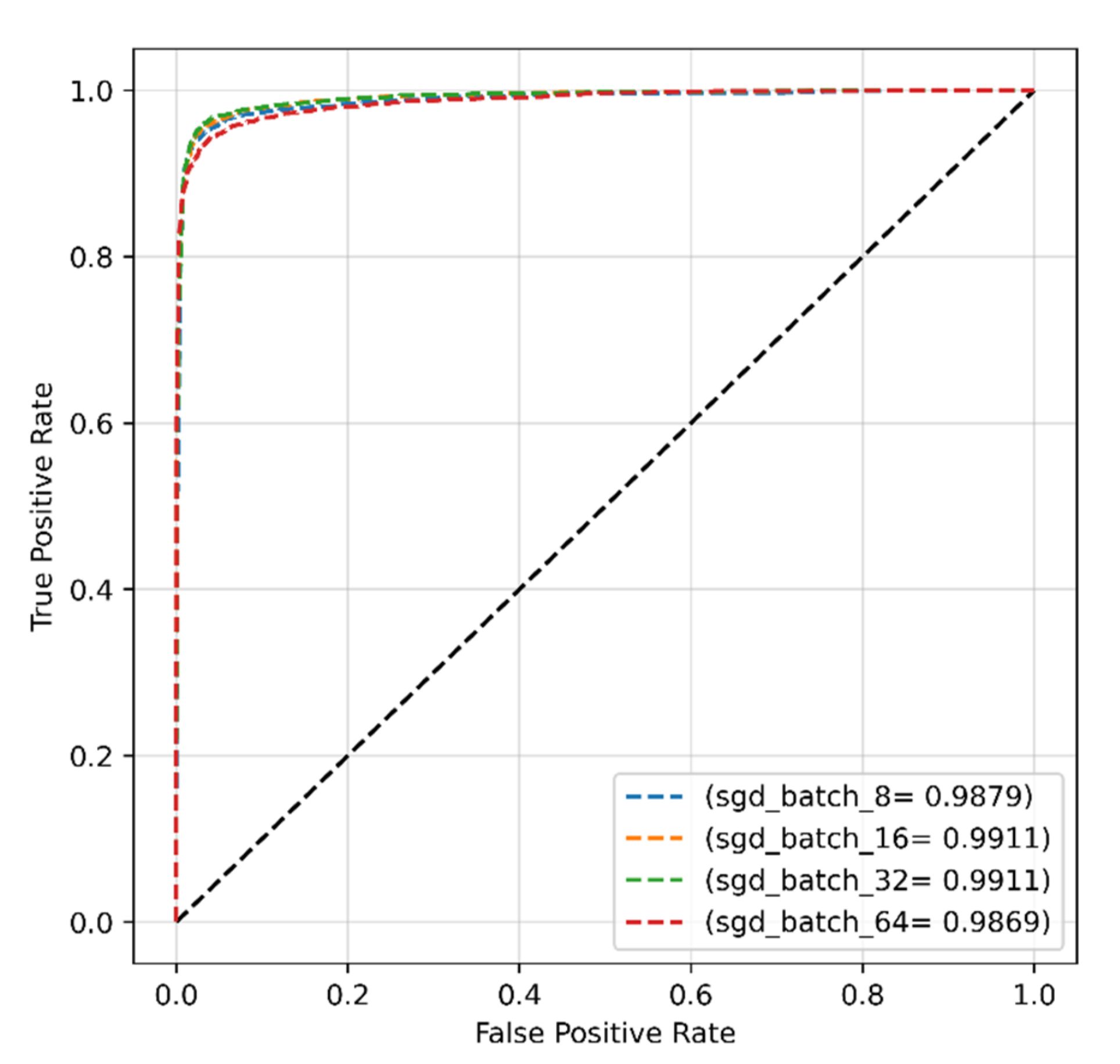
4.3.2. SGD Optimizer for Modified Dataset
4.3.3. Encoder Depth for Modified Dataset
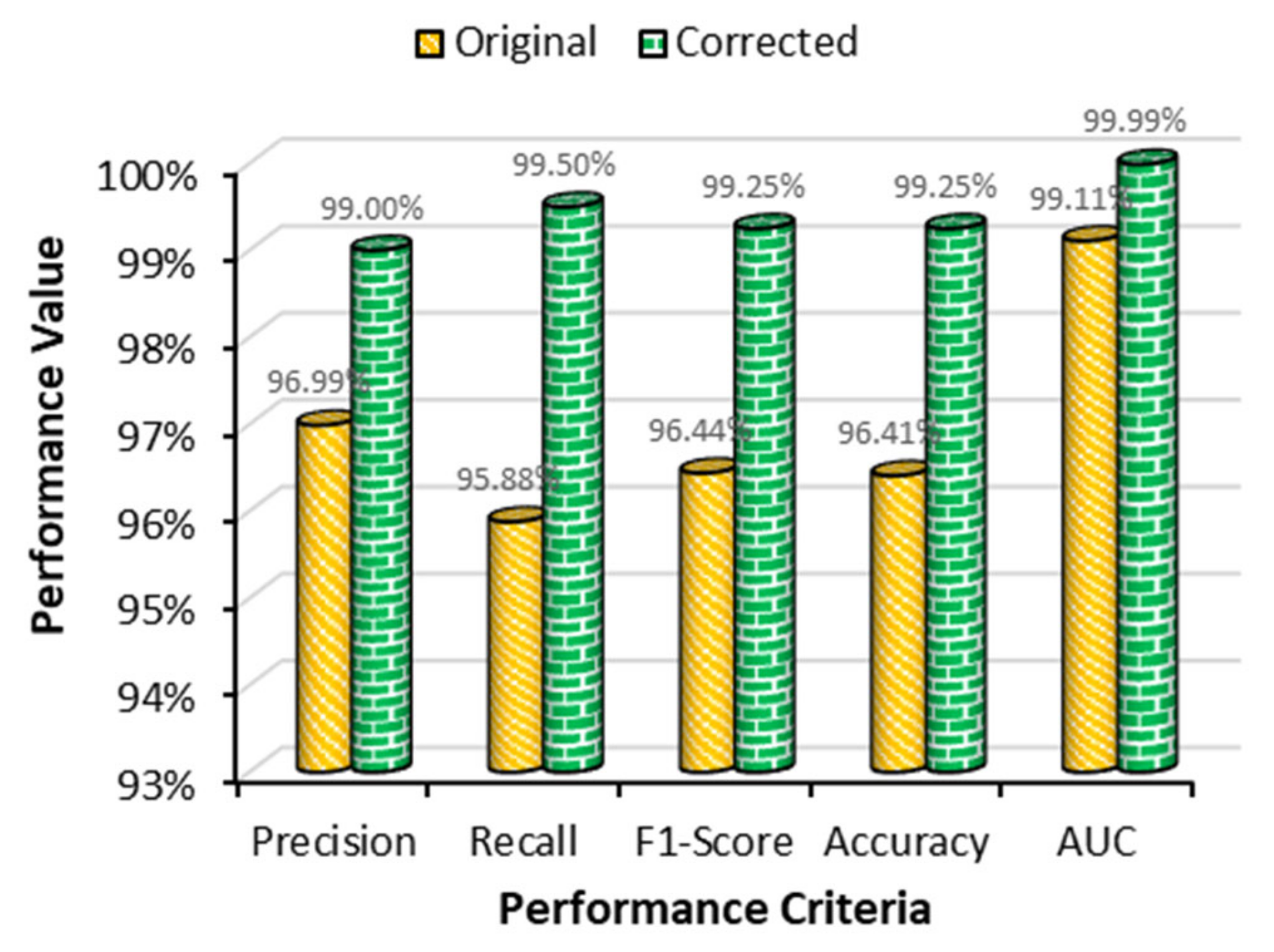
4.4. Performance Comparison between Two Datasets
4.5. Performance Comparison with Previous Works
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Malaria Microscopy Quality Assurance Manual-Version 2; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Caraballo, H.; King, K. Emergency Department Management of Mosquito-Borne Illness: Malaria, Dengue, and West Nile Virus. Emerg. Med. Pract. 2014. Available online: https://europepmc.org/article/med/25207355 (accessed on 10 January 2022).
- World Health Organization. Malaria, “Fact Sheet. No,”; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Wang, H.; Naghavi, M.; Allen, C.; Naghavi, M.; Bhutta, Z.; Carter, A.R.; Casey, D.C.; Charlson, F.J.; Chen, A.; Coates, M.M.; et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef] [Green Version]
- Rajaraman, S.; Antani, S.K.; Poostchi, M.; Silamut, K.; Hossain, M.A.; Maude, R.J.; Jaeger, S.; Thoma, G.R. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ 2018, 6, e4568. [Google Scholar] [CrossRef] [PubMed]
- Bibin, D.; Nair, M.S.; Punitha, P. Malaria parasite detection from peripheral blood smear images using deep belief networks. IEEE Access 2017, 5, 9099–9108. [Google Scholar] [CrossRef]
- Pandit, P.; Anand, A. Artificial neural networks for detection of malaria in RBCs. arXiv 2016, arXiv:1608.06627. [Google Scholar]
- Jain, N.; Chauhan, A.; Tripathi, P.; Moosa, S.B.; Aggarwal, P.; Oznacar, B. Cell image analysis for malaria detection using deep convolutional network. Intell. Decis. Technol. 2020, 14, 55–65. [Google Scholar] [CrossRef]
- Alqudah, A.; Alqudah, A.M.; Qazan, S. Lightweight Deep Learning for Malaria Parasite Detection Using Cell-Image of Blood Smear Images. Rev. d’Intell. Artif. 2020, 34, 571–576. [Google Scholar] [CrossRef]
- Sriporn, K.; Tsai, C.-F.; Tsai, C.-E.; Wang, P. Analyzing Malaria Disease Using Effective Deep Learning Approach. Diagnostics 2020, 10, 744. [Google Scholar] [CrossRef]
- Fuhad, K.M.F.; Tuba, J.F.; Sarker, M.R.A.; Momen, S.; Mohammed, N.; Rahman, T. Deep learning based automatic malaria parasite detection from blood smear and its smartphone based application. Diagnostics 2020, 10, 329. [Google Scholar] [CrossRef]
- Masud, M.; Alhumyani, H.; Alshamrani, S.S.; Cheikhrouhou, O.; Ibrahim, S.; Muhammad, G.; Hossain, M.S.; Shorfuzzaman, M. Leveraging deep learning techniques for malaria parasite detection using mobile application. Wirel. Commun. Mob. Comput. 2020, 2020, 8895429. [Google Scholar] [CrossRef]
- Maqsood, A.; Farid, M.S.; Khan, M.H.; Grzegorzek, M. Deep Malaria Parasite Detection in Thin Blood Smear Microscopic Images. Appl. Sci. 2021, 11, 2284. [Google Scholar] [CrossRef]
- Umer, M.; Sadiq, S.; Ahmad, M.; Ullah, S.; Choi, G.S.; Mehmood, A. A novel stacked CNN for malarial parasite detection in thin blood smear images. IEEE Access 2020, 8, 93782–93792. [Google Scholar] [CrossRef]
- Hung, J.; Carpenter, A. Applying faster R-CNN for object detection on malaria images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 56–61. [Google Scholar]
- Pattanaik, P.A.; Mittal, M.; Khan, M.Z. Unsupervised deep learning cad scheme for the detection of malaria in blood smear microscopic images. IEEE Access 2020, 8, 94936–94946. [Google Scholar] [CrossRef]
- Olugboja, A.; Wang, Z. Malaria parasite detection using different machine learning classifier. In Proceedings of the 2017 International Conference on Machine Learning and Cybernetics (ICMLC), Ningbo, China, 9–12 July 2017; Volume 1, pp. 246–250. [Google Scholar]
- Gopakumar, G.P.; Swetha, M.; Siva, G.S.; Subrahmanyam, G.R.K.S. Convolutional neural network-based malaria diagnosis from focus stack of blood smear images acquired using custom-built slide scanner. J. Biophotonics 2018, 11, e201700003. [Google Scholar] [CrossRef]
- Khan, A.; Gupta, K.D.; Venugopal, D.; Kumar, N. Cidmp: Completely interpretable detection of malaria parasite in red blood cells using lower-dimensional feature space. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar]
- Fatima, T.; Farid, M.S. Automatic detection of Plasmodium parasites from microscopic blood images. J. Parasit. Dis. 2020, 44, 69–78. [Google Scholar] [CrossRef]
- Makhzani, A.; Shlens, J.; Jaitly, N.; Goodfellow, I.; Frey, B. Adversarial autoencoders. arXiv 2015, arXiv:1511.05644. [Google Scholar]
- Kohonen, T. The self-organizing map. Proc. IEEE 1990, 78, 1464–1480. [Google Scholar] [CrossRef]
- Mohanty, I.; Pattanaik, P.A.; Swarnkar, T. Automatic detection of malaria parasites using unsupervised techniques. In Proceedings of the International Conference on ISMAC in Computational Vision and Bio-Engineering, Palladam, India, 16–17 May 2018; pp. 41–49. [Google Scholar]
- El-Sawy, A.; Hazem, E.-B.; Loey, M. CNN for handwritten arabic digits recognition based on LeNet-5. In Proceedings of the International Conference on Advanced Intelligent Systems and Informatics, Cairo, Egypt, 24–26 November 2016; pp. 566–575. [Google Scholar]
- Zhong, Z.; Jin, L.; Xie, Z. High performance offline handwritten chinese character recognition using googlenet and directional feature maps. In Proceedings of the 2015 13th International Conference on Document Analysis and Recognition (ICDAR), Tunis, Tunisia, 23–26 August 2015; pp. 846–850. [Google Scholar]
- Dong, Y.; Jiang, Z.; Shen, H.; Pan, W.D.; Williams, L.A.; Reddy, V.V.; Benjamin, W.H.; Bryan, A.W. Evaluations of deep convolutional neural networks for automatic identification of malaria infected cells. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical\Health Informatics (BHI), Orlando, FL, USA, 16–19 February 2017; pp. 101–104. [Google Scholar]
- Anggraini, D.; Nugroho, A.S.; Pratama, C.; Rozi, I.E.; Iskandar, A.A.; Hartono, R.N. Automated status identification of microscopic images obtained from malaria thin blood smears. In Proceedings of the 2011 International Conference on Electrical Engineering and Informatics, Bandung, Indonesia, 17–19 July 2011; pp. 1–6. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. October 2020. Available online: https://arxiv.org/abs/2010.11929v2 (accessed on 11 January 2022).
- Corrected Malaria Data—Google Drive. 2019. Available online: https://drive.google.com/drive/folders/10TXXa6B_D4AKuBV085tX7UudH1hINBRJ?usp=sharing (accessed on 10 January 2022).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual Recognition. Proc. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef] [Green Version]
- Hassani, A.; Walton, S.; Shah, N.; Abuduweili, A.; Li, J.; Shi, H. Escaping the Big Data Paradigm with Compact Transformers. 2021. Available online: http://arxiv.org/abs/2104.05704 (accessed on 21 February 2022).
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Red Hook, NY, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Poostchi, M.; Silamut, K.; Maude, R.J.; Jaeger, S.; Thoma, G. Image analysis and machine learning for detecting malaria. Transl. Res. 2018, 194, 36–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menditto, A.; Patriarca, M.; Magnusson, B. Understanding the meaning of accuracy, trueness and precision. Accredit. Qual. Assur. 2007, 12, 45–47. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness and Correlation. October 2020. Available online: http://arxiv.org/abs/2010.16061 (accessed on 10 January 2022).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Ketkar, N. Stochastic gradient descent. In Deep Learning with Python; Springer: Berlin/Heidelberg, Germany, 2017; pp. 113–132. [Google Scholar]
- Shah, D.; Kawale, K.; Shah, M.; Randive, S.; Mapari, R. Malaria Parasite Detection Using Deep Learning: (Beneficial to humankind). In Proceedings of the 2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 13–15 May 2020; pp. 984–988. [Google Scholar]
- Mondal, A.K.; Bhattacharjee, A.; Singla, P.; Prathosh, A.P. xViTCOS: Explainable Vision Transformer Based COVID-19 Screening Using Radiography. IEEE J. Transl. Eng. Health Med. 2021, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
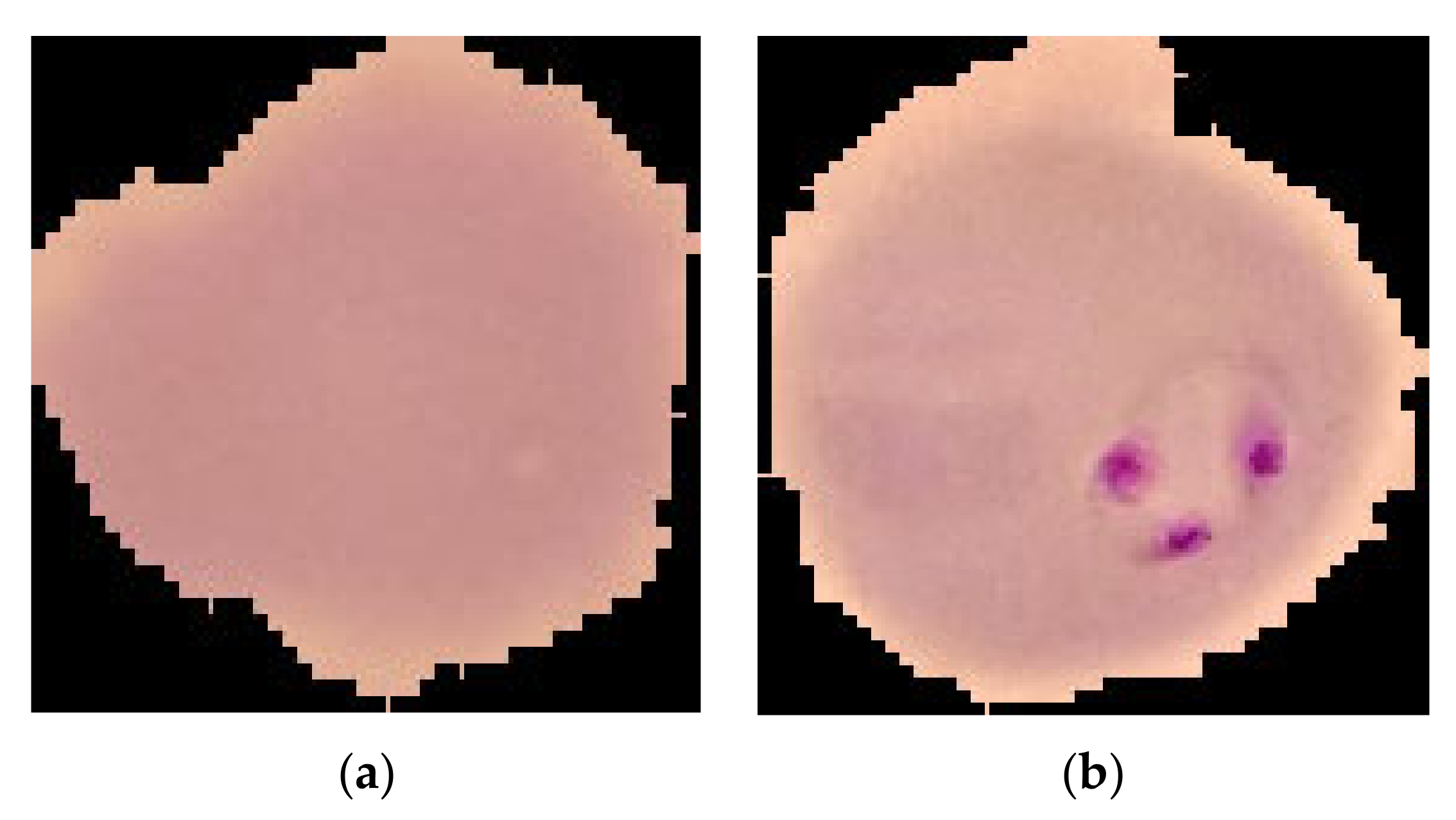




| Dataset | Number of Healthy Images | Number of Infected Images | Total | Total Training Samples (80%) | Total Testing Samples (20%) |
|---|---|---|---|---|---|
| Original dataset [5] | 13,779 | 13,779 | 27,558 | 22,046 | 5512 |
| Modified dataset [11] | 13,029 | 13,132 | 26,161 | 20,928 | 5233 |
| Batch Size | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|
| 8 | 52.10 | 62.03 | 56.64 | 60.11 |
| 16 | 63.17 | 56.05 | 59.40 | 56.82 |
| 32 | 100 | 50 | 66.67 | 50 |
| 64 | 99.96 | 49.99 | 66.65 | 49.98 |
| Batch Size | Precision (%) | Recall (%) | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| 8 | 97.06 | 94.66 | 95.84 | 95.79 |
| 16 | 96.73 | 95.56 | 96.14 | 96.12 |
| 32 | 96.99 | 95.88 | 96.44 | 96.41 |
| 64 | 97.50 | 92.53 | 94.95 | 94.81 |
| Batch Size | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|
| 8 | 58.37 | 53.58 | 55.87 | 54.08 |
| 16 | 48.12 | 59.69 | 53.28 | 57.98 |
| 32 | 100 | 49.80 | 66.49 | 49.80 |
| 64 | 100 | 49.80 | 66.49 | 49.80 |
| Batch Size | Precision (%) | Recall (%) | F1-Score (%) | Accuracy (%) |
|---|---|---|---|---|
| 8 | 99.08 | 99.42 | 99.25 | 99.25 |
| 16 | 99.12 | 99.16 | 99.14 | 99.14 |
| 32 | 99.19 | 99.27 | 99.23 | 99.24 |
| 64 | 99.00 | 99.50 | 99.25 | 99.25 |
| Reference No | Model Used | Optimizer | Learning Rate | Batch Size | Precision (%) | Recall (%) | AUC (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| [39] | Custom CNN | Adam | - | - | - | - | - | 95 |
| [20] | Image processing | - | - | - | 94.66 | - | - | 91.80 |
| [19] | Random forest | - | - | - | 82.00 | 86.00 | - | - |
| [5] | CNN | SGD | 0.0005 | - | 94.70 | 95.90 | 99.90 | - |
| [16] | Neural network | - | - | - | 93.90 | - | - | 83.10 |
| Proposed work (original dataset) | Transformer | SGD | 0.001 | 32 | 96.99 | 95.88 | 99.11 | 96.41 |
| [9] | CNN | Adam | 0.001 | 128 | 98.79 | - | - | 98.85 |
| [11] | Custom CNN | SGD | 0.01 | 32 | 98.92 | 99.52 | - | 99.23 |
| Proposed work (modified dataset) | Transformer | SGD | 0.001 | 32 | 99.00 | 99.50 | 99.99 | 99.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.R.; Nahiduzzaman, M.; Goni, M.O.F.; Sayeed, A.; Anower, M.S.; Ahsan, M.; Haider, J. Explainable Transformer-Based Deep Learning Model for the Detection of Malaria Parasites from Blood Cell Images. Sensors 2022, 22, 4358. https://doi.org/10.3390/s22124358
Islam MR, Nahiduzzaman M, Goni MOF, Sayeed A, Anower MS, Ahsan M, Haider J. Explainable Transformer-Based Deep Learning Model for the Detection of Malaria Parasites from Blood Cell Images. Sensors. 2022; 22(12):4358. https://doi.org/10.3390/s22124358
Chicago/Turabian StyleIslam, Md. Robiul, Md. Nahiduzzaman, Md. Omaer Faruq Goni, Abu Sayeed, Md. Shamim Anower, Mominul Ahsan, and Julfikar Haider. 2022. "Explainable Transformer-Based Deep Learning Model for the Detection of Malaria Parasites from Blood Cell Images" Sensors 22, no. 12: 4358. https://doi.org/10.3390/s22124358
APA StyleIslam, M. R., Nahiduzzaman, M., Goni, M. O. F., Sayeed, A., Anower, M. S., Ahsan, M., & Haider, J. (2022). Explainable Transformer-Based Deep Learning Model for the Detection of Malaria Parasites from Blood Cell Images. Sensors, 22(12), 4358. https://doi.org/10.3390/s22124358










