Optimization of Deep Architectures for EEG Signal Classification: An AutoML Approach Using Evolutionary Algorithms
Abstract
1. Introduction
- 1.
- We propose a fully-configurable optimization framework for deep learning architectures. The proposal is not only aimed to optimize hyperparameters, but it can also be setup to modify the architecture, including or removing layers from the initial solutions, covering the inclusion of regularization parameters to reduce the generalization error.
- 2.
- Architecture optimization is performed in a multi-objective way. That means that different and conflicting objectives are taken into account during the optimization process.
- 3.
- It is based on multi-objective optimization. Thus, the result is a pool of non-dominated solutions that provide a trade-off among objectives. This allows for selecting the most appropriate solution by moving through the Pareto front.
- 4.
- The proposed framework uses both CPUs and GPUs to speed up the execution.
2. Materials and Methods
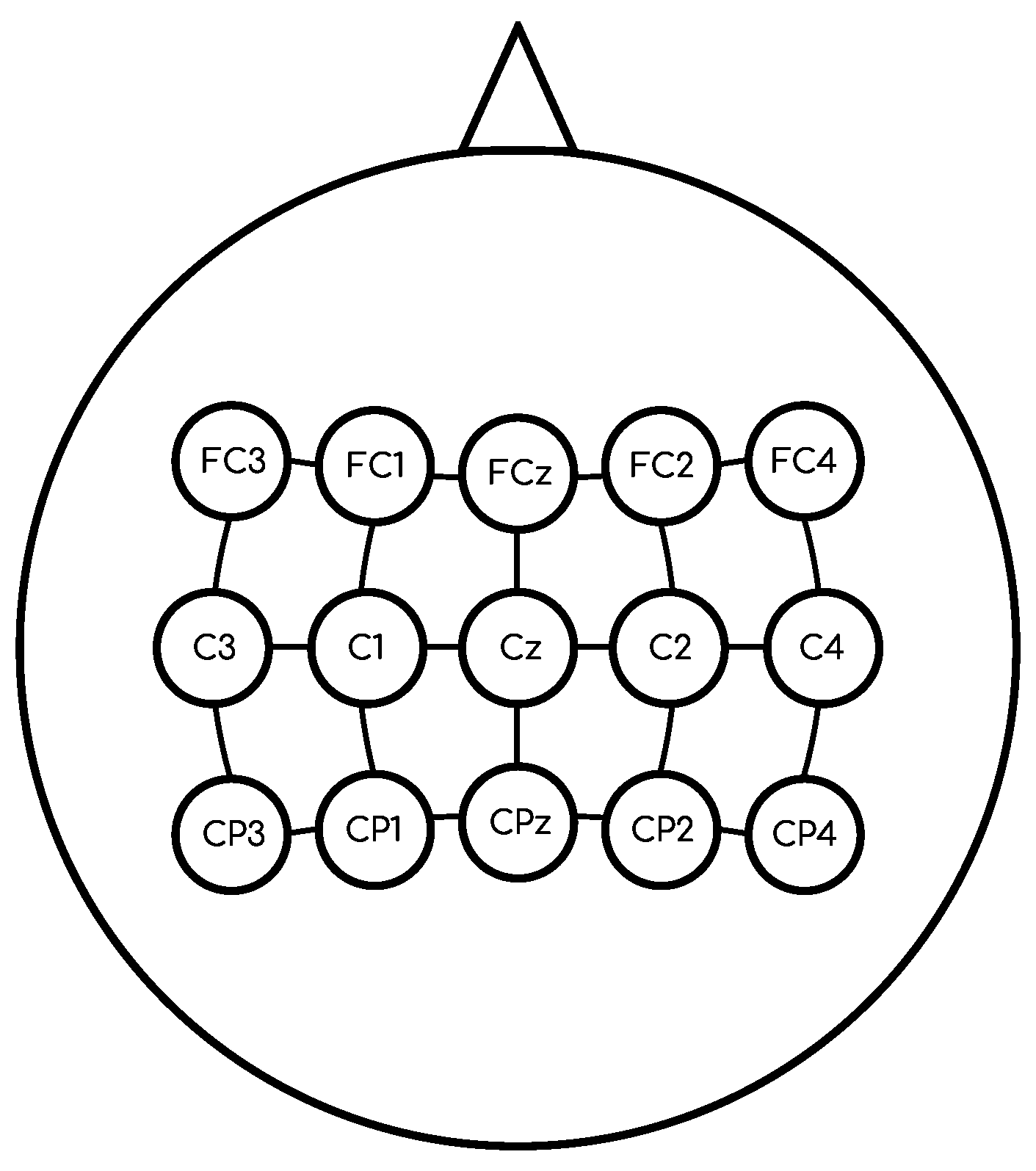
2.1. Data Description
2.2. Deep Neural Networks
- Sigmoid: is a logistic function, where the input values are transformed into output values within a range of . The function definition is given by Equation (2).
- Scaled Exponential Linear Unit (SELU): ensures a slope larger than one for positive inputs. If the input value z is positive then the output value is multiplied by a coefficient . Otherwise, when the input value z is equal or less than 0, then the coefficient multiplies the exponential of the input value z minus the coefficient, and, finally, the result is multiplied by a coefficient . The function definition is given by Equation (3).
- Hyperbolic tangent (TanH): is a useful activation function with a boundary range of , which allows for efficient training. However, its main drawback occurs in the backpropagation process, due to the vanishing gradient problem that limits the adjustment of the weight value. Equation (4) provides the function definition.
- Rectifier Linear Unit (ReLU): a commonly used function, where, if the input value z is equal to or less than 0, then z is converted to 0. In the case of a positive input z, the value is not changed. The function definition is given by Equation (5).
- Leaky ReLU (LReLU): similar to ReLU. The difference occurs when the input value z is equal to or less than 0, then z is multiplied by a coefficient which is usually within the range . Equation (6) provides the function definition.
- Exponential Linear Unit (ELU): compared to its predecessors, such as ReLU and LReLU, this function decreases the vanishing gradient effect using the exponential operation . If the input value z is negative, then is multiplied by a coefficient in the common range of . The function definition is given by Equation (7).
Convolutional Neural Networks
- Convolution 2D: exploits spatial correlations in the data. This layer can be composed of one or more filters, where each one is sliding across a 2D input array and performing a dot product between the filter and the input array for each position.
- Depthwise Convolution 2D: aims to learn spatial patterns from each temporal filter allowing for feature extraction from specific frequencies of the spatial filters. This layer performs an independent spatial convolution on each input channel and, thus, produces a set of output tensors (2D) that are finally stacked together.
- Separable Convolution 2D: aims to reduce the number of parameters to fit. This layer basically decomposes the convolution into two independent operations: the first one performs a depthwise convolution across each input channel, while the second one performs a pointwise convolution that projects the output channels from the depthwise convolution into a new channel space.
- Average Pooling 2D: aims to reduce the representation of the spatial size of each channel. This layer takes the input data array and calculates the average value from all values of each input channel. This way, it generates a smaller tensor than the corresponding input data.
- Batch Normalization [41]: aims to achieve fixed distributions of the input data and address the internal covariate shift problem. This layer performs the calculation of the mean and variance of the input data.
2.3. Overfitting
2.4. Multi-Objective Optimization
2.4.1. NSGA-II Algorithm
| Algorithm 1: Pseudo-code of the Multi-objective Optimization Procedure for Deep Convolutional Architectures using NSGA-II [48,49]. |
 |
2.4.2. Convolutional Neural Networks for EEG Data Classification
2.5. EEGNet: CNN for EEG Classification
2.6. Performance Evaluation
- Pareto front [50]: multi-objective optimization problems consider several objectives at the same time. Therefore, it is usually not possible to obtain an optimal solution that satisfies all the conditions. Instead, the optimization process provides a set of non-dominated solutions with different trade-offs among the objectives: the Pareto optimal solutions.
- Kappa Index [51]: is a statistic measure that is used for multi-class classification problems or imbalanced class problems. The Kappa Index k is defined as:where is the observed agreement and is the expected agreement. The Kappa Index value is always less than or equal to 1.
- CNN Parameters [25]: They are weights changing during the training process that are also known as trainable parameters.
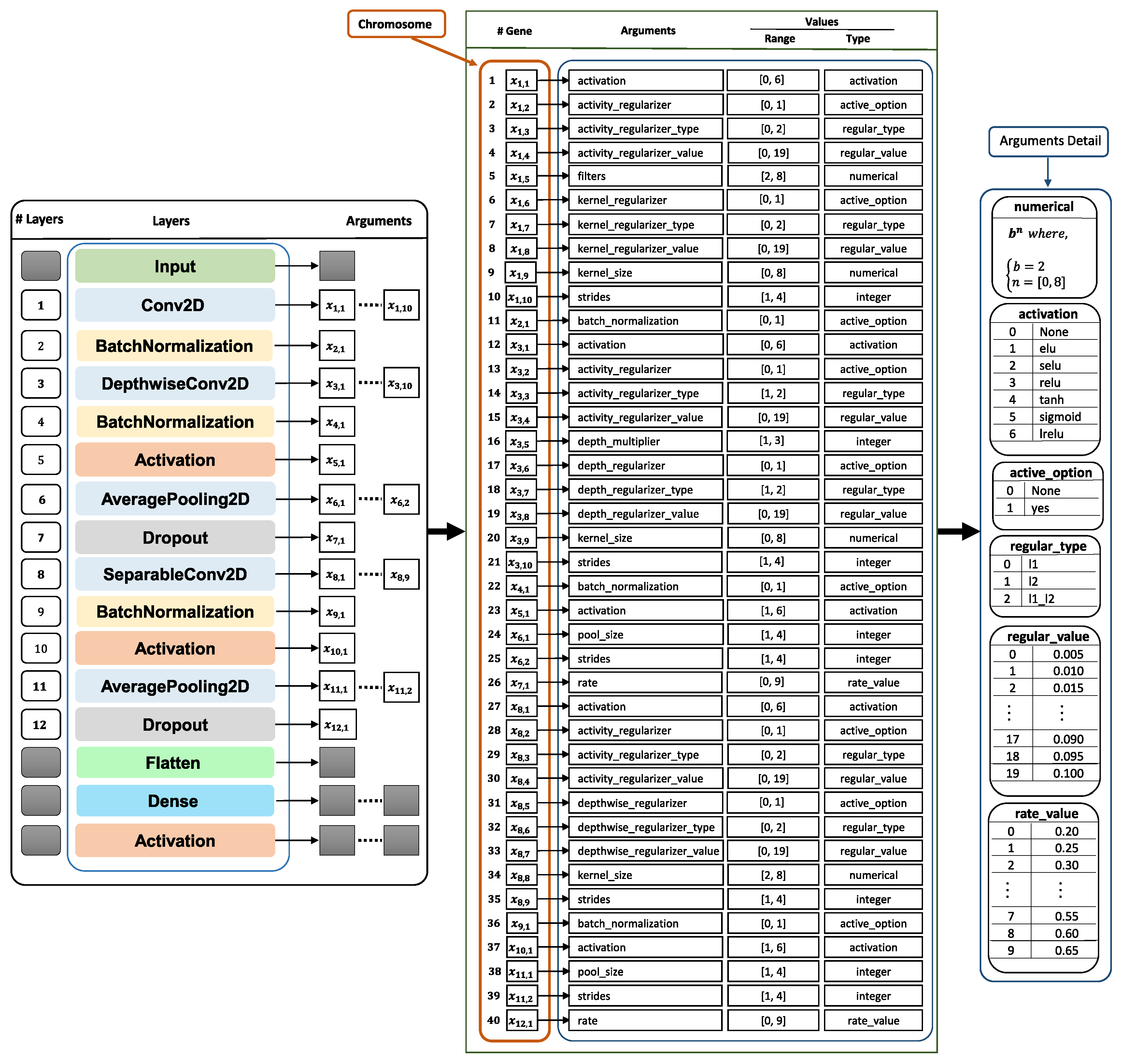
2.7. Proposed Optimization Framework and Application to EEG Signal Classification
Optimization Process and CPU-GPU Workload Distribution
3. Results
3.1. Data Split for Performance Evaluation
3.2. Experimental Results
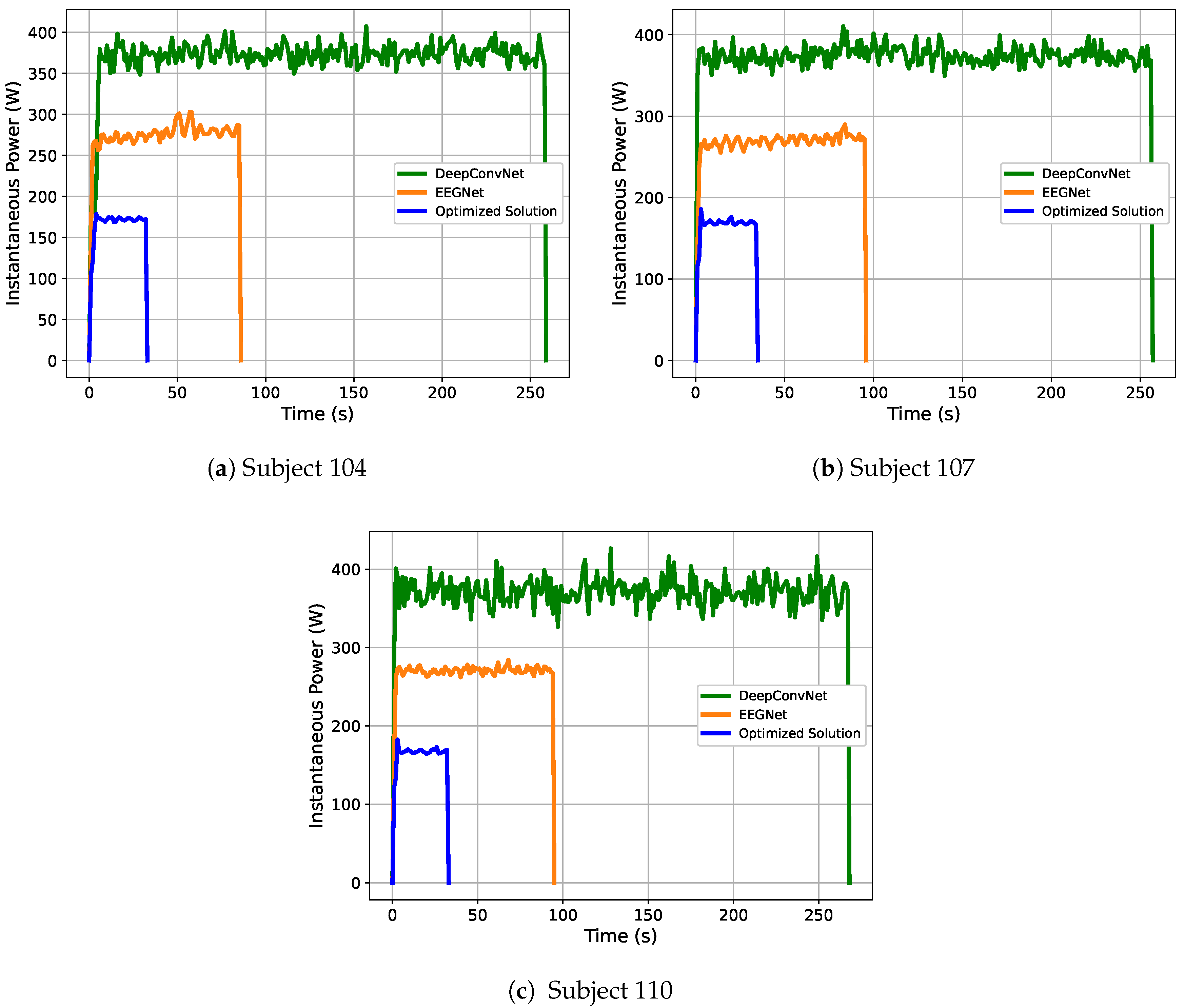
Power Efficiency of the Optimized Solutions
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ben-Nun, T.; Hoefler, T. Demystifying Parallel and Distributed Deep Learning: An in-depth Concurrency Analysis. ACM Comput. Surv. 2019, 52, 1–43. [Google Scholar] [CrossRef]
- Baxevanis, A.D.; Bader, G.D.; Wishart, D.S. Bioinformatics; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; McCarthy, D.; Robinson, M.; Smyth, G.K. edgeR: Differential Expression Analysis of Digital Gene Expression Data User’s Guide. Bioconductor User’s Guide. 2014. Available online: http://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (accessed on 17 September 2008).
- Min, S.; Lee, B.; Yoon, S. Deep Learning in Bioinformatics. Briefings Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Górriz, J.M.; Ramírez, J.; Ortíz, A. Artificial intelligence within the interplay between natural and artificial computation: Advances in data science, trends and applications. Neurocomputing 2020, 410, 237–270. [Google Scholar] [CrossRef]
- León, J.; Escobar, J.J.; Ortiz, A.; Ortega, J.; González, J.; Martín-Smith, P.; Gan, J.Q.; Damas, M. Deep learning for EEG-based Motor Imagery classification: Accuracy-cost trade-off. PLoS ONE 2020, 15, e0234178. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Chugh, N. Signal processing techniques for motor imagery brain computer interface: A review. Array 2019, 1, 100003. [Google Scholar] [CrossRef]
- Hotson, G.; McMullen, D.P.; Fifer, M.S.; Johannes, M.S.; Katyal, K.D.; Para, M.P.; Armiger, R.; Anderson, W.S.; Thakor, N.V.; Wester, B.A.; et al. Individual Finger Control of the Modular Prosthetic Limb using High-Density Electrocorticography in a Human Subject. J. Neural Eng. 2016, 13, 026017. [Google Scholar] [CrossRef]
- Berger, H. Über das Elektrenkephalogramm des Menschen. XIV. In Archiv für Psychiatrie und Nervenkrankheiten; Springer: Cham, Switzerland, 1938. [Google Scholar]
- Hill, N.J.; Lal, T.N.; Schroder, M.; Hinterberger, T.; Wilhelm, B.; Nijboer, F.; Mochty, U.; Widman, G.; Elger, C.; Scholkopf, B.; et al. Classifying EEG and ECoG signals without subject training for fast BCI implementation: Comparison of nonparalyzed and completely paralyzed subjects. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Stokes, M.G.; Wolff, M.J.; Spaak, E. Decoding Rich Spatial Information with High Temporal Resolution. Trends Cogn. Sci. 2015, 19, 636–638. [Google Scholar] [CrossRef]
- Coyle, S.M.; Ward, T.E.; Markham, C.M. Brain–computer interface using a simplified functional near-infrared spectroscopy system. J. Neural Eng. 2007, 4, 219. [Google Scholar] [CrossRef]
- Baig, M.Z.; Aslam, N.; Shum, H.P. Filtering techniques for channel selection in motor imagery EEG applications: A survey. Artif. Intell. Rev. 2020, 53, 1207–1232. [Google Scholar] [CrossRef]
- Martinez-Murcia, F.J.; Ortiz, A.; Gorriz, J.M.; Ramirez, J.; Lopez-Abarejo, P.J.; Lopez-Zamora, M.; Luque, J.L. EEG Connectivity Analysis Using Denoising Autoencoders for the Detection of Dyslexia. Int. J. Neural Syst. 2020, 30, 2050037. [Google Scholar] [CrossRef]
- Ortiz, A.; Martinez-Murcia, F.J.; Luque, J.L.; Giménez, A.; Morales-Ortega, R.; Ortega, J. Dyslexia Diagnosis by EEG Temporal and Spectral Descriptors: An Anomaly Detection Approach. Int. J. Neural Syst. 2020, 30, 2050029. [Google Scholar] [CrossRef]
- Duin, R.P. Classifiers in almost empty spaces. In Proceedings of the 15th International Conference on Pattern Recognition, Barcelona, Spain, 3–8 September 2000; Volume 2, pp. 1–7. [Google Scholar]
- Raudys, S.J.; Jain, A.K. Small sample size effects in statistical pattern recognition: Recommendations for practitioners. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 252–264. [Google Scholar] [CrossRef]
- Lotte, F.; Congedo, M.; Lécuyer, A.; Lamarche, F.; Arnaldi, B. A review of classification algorithms for EEG-based brain–computer interfaces. J. Neural Eng. 2007, 4, R1. [Google Scholar] [CrossRef] [PubMed]
- Martín-Smith, P.; Ortega, J.; Asensio-Cubero, J.; Gan, J.Q.; Ortiz, A. A supervised filter method for multi-objective feature selection in EEG classification based on multi-resolution analysis for BCI. Neurocomputing 2017, 250, 45–56. [Google Scholar] [CrossRef]
- Kimovski, D.; Ortega, J.; Ortiz, A.; Banos, R. Parallel alternatives for evolutionary multi-objective optimization in unsupervised feature selection. Expert Syst. Appl. 2015, 42, 4239–4252. [Google Scholar] [CrossRef]
- Corralejo, R.; Hornero, R.; Alvarez, D. Feature selection using a genetic algorithm in a motor imagery-based Brain Computer Interface. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 7703–7706. [Google Scholar]
- Ortega, J.; Asensio-Cubero, J.; Gan, J.Q.; Ortiz, A. Classification of Motor Imagery Tasks for BCI with Multiresolution Analysis and Multiobjective Feature Selection. Biomed. Eng. Online 2016, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Abootalebi, V.; Moradi, M.H.; Khalilzadeh, M.A. A new approach for EEG feature extraction in P300-based lie detection. Comput. Methods Programs Biomed. 2009, 94, 48–57. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolutional Neural Network for EEG-based Brain—Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Qiao, R.; Qing, C.; Zhang, T.; Xing, X.; Xu, X. A novel deep-learning based framework for multi-subject emotion recognition. In Proceedings of the 2017 4th International Conference on Information, Cybernetics and Computational Social Systems (ICCSS), Dalian, China, 24–26 July 2017; pp. 181–185. [Google Scholar]
- Orr, G.B.; Müller, K.R. Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Domhan, T.; Springenberg, J.T.; Hutter, F. Speeding up automatic hyperparameter optimization of deep neural networks by extrapolation of learning curves. In Proceedings of the Twenty-Fourth International Joint Conference on Artificial Intelligence, Buenos Aires, Argentina, 25 July–1 August 2015. [Google Scholar]
- Young, S.R.; Rose, D.C.; Karnowski, T.P.; Lim, S.H.; Patton, R.M. Optimizing Deep Learning Hyper-Parameters through an Evolutionary Algorithm. In Proceedings of the Workshop on Machine Learning in High-Performance Computing Environments, Austin, TX, USA, 15 November 2015; pp. 1–5. [Google Scholar]
- Loshchilov, I.; Hutter, F. CMA-ES for Hyperparameter Optimization of Deep Neural Networks. arXiv 2016, arXiv:1604.07269. [Google Scholar]
- Such, F.P.; Madhavan, V.; Conti, E.; Lehman, J.; Stanley, K.O.; Clune, J. Deep neuroevolution: Genetic algorithms are a competitive alternative for training deep neural networks for reinforcement learning. arXiv 2017, arXiv:1712.06567. [Google Scholar]
- Galván, E.; Mooney, P. Neuroevolution in deep neural networks: Current trends and future challenges. arXiv 2020, arXiv:2006.05415. [Google Scholar]
- Xie, L.; Yuille, A. Genetic CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 1388–1397. [Google Scholar] [CrossRef]
- Sun, Y.; Xue, B.; Zhang, M.; Yen, G.G. Evolving deep convolutional neural networks for image classification. IEEE Trans. Evol. Comput. 2019, 24, 394–407. [Google Scholar] [CrossRef]
- Bilbao, I.; Bilbao, J. Overfitting problem and the over-training in the era of data: Particularly for Artificial Neural Networks. In Proceedings of the 2017 Eighth International Conference on Intelligent Computing and Information Systems (ICICIS), Cairo, Egypt, 5–7 December 2017; pp. 173–177. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Asensio-Cubero, J.; Gan, J.; Palaniappan, R. Multiresolution analysis over simple graphs for brain computer interfaces. J. Neural Eng. 2013, 10, 046014. [Google Scholar] [CrossRef]
- Shrestha, A.; Mahmood, A. Review of Deep Learning Algorithms and Architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; The MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Hawkins, D.M. The Problem of Overfitting. J. Chem. Inf. Comput. Sci. 2004, 44, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Amari, S.; Murata, N.; Muller, K.R.; Finke, M.; Yang, H.H. Asymptotic statistical theory of overtraining and cross-validation. IEEE Trans. Neural Netw. 1997, 8, 985–996. [Google Scholar] [CrossRef]
- Zhang, C.; Bengio, S.; Hardt, M.; Recht, B.; Vinyals, O. Understanding deep learning requires rethinking generalization. arXiv 2016, arXiv:1611.03530. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Prechelt, L. Early Stopping - But When? In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 1998; pp. 55–69. [Google Scholar]
- Cui, Y.; Geng, Z.; Zhu, Q.; Han, Y. Review: Multi-objective optimization methods and application in energy saving. Energy 2017, 125, 681–704. [Google Scholar] [CrossRef]
- Deb, K.; Pratap, A.; Agarwal, S.; Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002, 6, 182–197. [Google Scholar] [CrossRef]
- Deb, K.; Agrawal, S.; Pratap, A.; Meyarivan, T. A Fast Elitist Non-dominated Sorting Genetic Algorithm for Multi-objective Optimization: NSGA-II. In Parallel Problem Solving from Nature PPSN VI; Schoenauer, M., Deb, K., Rudolph, G., Yao, X., Lutton, E., Merelo, J.J., Schwefel, H.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 849–858. [Google Scholar]
- Rachmawati, L.; Srinivasan, D. Multiobjective Evolutionary Algorithm With Controllable Focus on the Knees of the Pareto Front. IEEE Trans. Evol. Comput. 2009, 13, 810–824. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Zhang, X.; Tian, Y.; Jin, Y. A Knee Point-Driven Evolutionary Algorithm for Many-Objective Optimization. IEEE Trans. Evol. Comput. 2014, 19, 761–776. [Google Scholar] [CrossRef]
- Stonebraker, M. PostgreSQL: The World’s Most Advanced Open Source Relational Database; O’Reilly Media, Inc.: Sebastopol, CA, USA, 1996. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]






| Subject | Left Hand | Right Hand | Feet | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | |
| 104 | 60 | 56 | 66 | 65 | 52 | 58 | 178 | 179 |
| 107 | 56 | 58 | 57 | 58 | 65 | 63 | 178 | 179 |
| 110 | 58 | 59 | 60 | 60 | 60 | 60 | 178 | 179 |
| Parameter | Value |
|---|---|
| Chromosome length (genes) | 40 |
| Population size | 100 |
| Number of generations | 100 |
| Mutation probability | 0.05 |
| Crossover probability | 0.5 |
| Maximum CNN training epochs | 500 |
| Layers | Parameters | Subject | ||
|---|---|---|---|---|
| 104 | 107 | 110 | ||
| Conv2D | activation | selu | sigmoid | selu |
| activity_regularizer | None | None | None | |
| activity_regularizer_type | None | None | None | |
| activity_regularizer_value | None | None | None | |
| filters | 4 | 4 | 4 | |
| kernel_regularizer | None | Yes | None | |
| kernel_regularizer_type | None | l1 | None | |
| kernel_regularizer_value | None | 0.065 | None | |
| kernel_size | (1, 1) | (1, 2) | (1, 1) | |
| strides | 1 | 1 | 3 | |
| BatchNormalization | batch_normalization | Yes | Yes | Yes |
| DepthwiseConv2D | activation | relu | tanh | None |
| activity_regularizer | None | None | None | |
| activity_regularizer_type | None | None | None | |
| activity_regularizer_value | None | None | None | |
| depth_multiplier | 1 | 1 | 1 | |
| depthwise_regularizer | Yes | None | None | |
| depthwise_regularizer_type | l2 | None | None | |
| depthwise_regularizer_value | 0.065 | None | None | |
| kernel_size | 1 | 1 | 1 | |
| strides | 3 | 4 | 2 | |
| BatchNormalization | batch_normalization | Yes | Yes | Yes |
| Activation | activation | elu | tanh | relu |
| AveragePooling2D | pool_size | (1, 4) | (1, 1) | (1, 3) |
| strides | 3 | 1 | 3 | |
| Dropout | rate | 0.50 | 0.40 | 0.40 |
| SeparableConv2D | activation | relu | relu | tanh |
| activity_regularizer | Yes | Yes | None | |
| activity_regularizer_type | l2 | l2 | None | |
| activity_regularizer_value | 0.040 | 0.025 | None | |
| depthwise_regularizer | Yes | Yes | Yes | |
| depthwise_regularizer_type | l2 | l2 | l2 | |
| depthwise_regularizer_value | 0.040 | 0.070 | 0.045 | |
| kernel_size | (1, 4) | (1, 16) | (1, 8) | |
| strides | 4 | 4 | 2 | |
| BatchNormalization | batch_normalization | Yes | Yes | Yes |
| Activation | activation | selu | relu | relu |
| AveragePooling2D | pool_size | (1, 4) | (1, 3) | (1, 4) |
| strides | 2 | 2 | 3 | |
| Dropout | rate | 0.55 | 0.40 | 0.25 |
| Subject | Accuracy | Kappa Index | p-Values | ||
|---|---|---|---|---|---|
| Average | Std. Dev. | Average | Std. Dev. | ||
| DeepConvNet [26] | |||||
| 104 | 0.58 | 0.03 | 0.38 | 0.04 | |
| 107 | 0.66 | 0.04 | 0.49 | 0.06 | |
| 110 | 0.48 | 0.02 | 0.22 | 0.03 | |
| EEGNet (baseline) [26] | |||||
| 104 | 0.63 | 0.05 | 0.44 | 0.08 | |
| 107 | 0.63 | 0.06 | 0.44 | 0.08 | |
| 110 | 0.72 | 0.05 | 0.58 | 0.08 | |
| Optimized Solution | |||||
| 104 | 0.75 | 0.01 | 0.63 | 0.01 | |
| 107 | 0.88 | 0.02 | 0.82 | 0.02 | |
| 110 | 0.83 | 0.01 | 0.74 | 0.01 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aquino-Brítez, D.; Ortiz, A.; Ortega, J.; León, J.; Formoso, M.; Gan, J.Q.; Escobar, J.J. Optimization of Deep Architectures for EEG Signal Classification: An AutoML Approach Using Evolutionary Algorithms. Sensors 2021, 21, 2096. https://doi.org/10.3390/s21062096
Aquino-Brítez D, Ortiz A, Ortega J, León J, Formoso M, Gan JQ, Escobar JJ. Optimization of Deep Architectures for EEG Signal Classification: An AutoML Approach Using Evolutionary Algorithms. Sensors. 2021; 21(6):2096. https://doi.org/10.3390/s21062096
Chicago/Turabian StyleAquino-Brítez, Diego, Andrés Ortiz, Julio Ortega, Javier León, Marco Formoso, John Q. Gan, and Juan José Escobar. 2021. "Optimization of Deep Architectures for EEG Signal Classification: An AutoML Approach Using Evolutionary Algorithms" Sensors 21, no. 6: 2096. https://doi.org/10.3390/s21062096
APA StyleAquino-Brítez, D., Ortiz, A., Ortega, J., León, J., Formoso, M., Gan, J. Q., & Escobar, J. J. (2021). Optimization of Deep Architectures for EEG Signal Classification: An AutoML Approach Using Evolutionary Algorithms. Sensors, 21(6), 2096. https://doi.org/10.3390/s21062096







