Steered Molecular Dynamics of Lipid Membrane Indentation by Carbon and Silicon-Carbide Nanotubes—The Impact of Indenting Angle Uncertainty
Abstract
:1. Introduction
2. Simulations’ Details
3. Results and Discussion
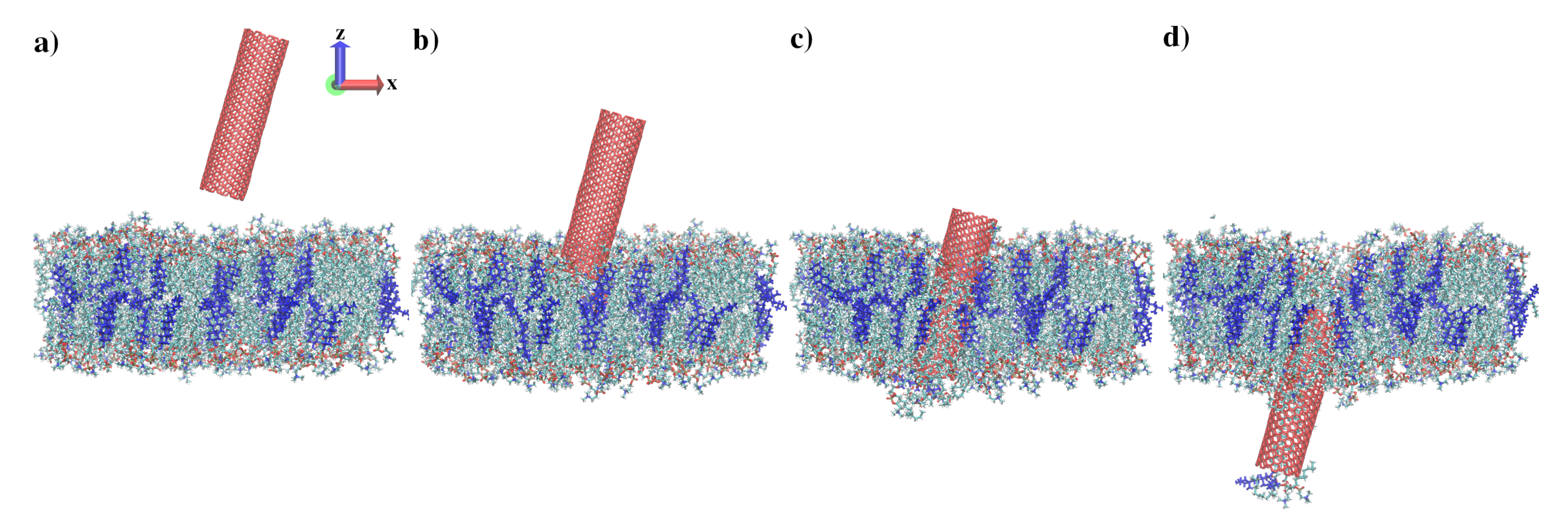
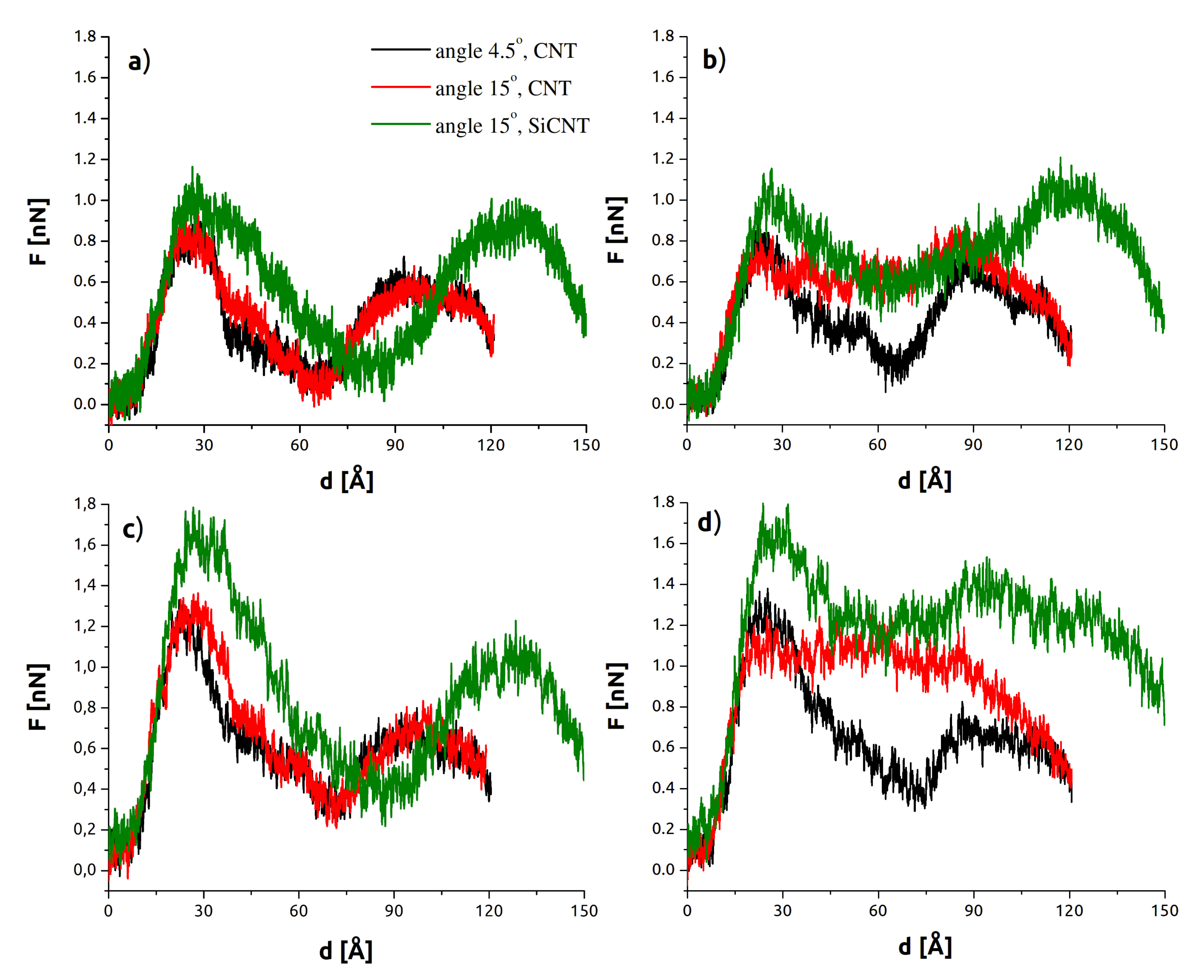
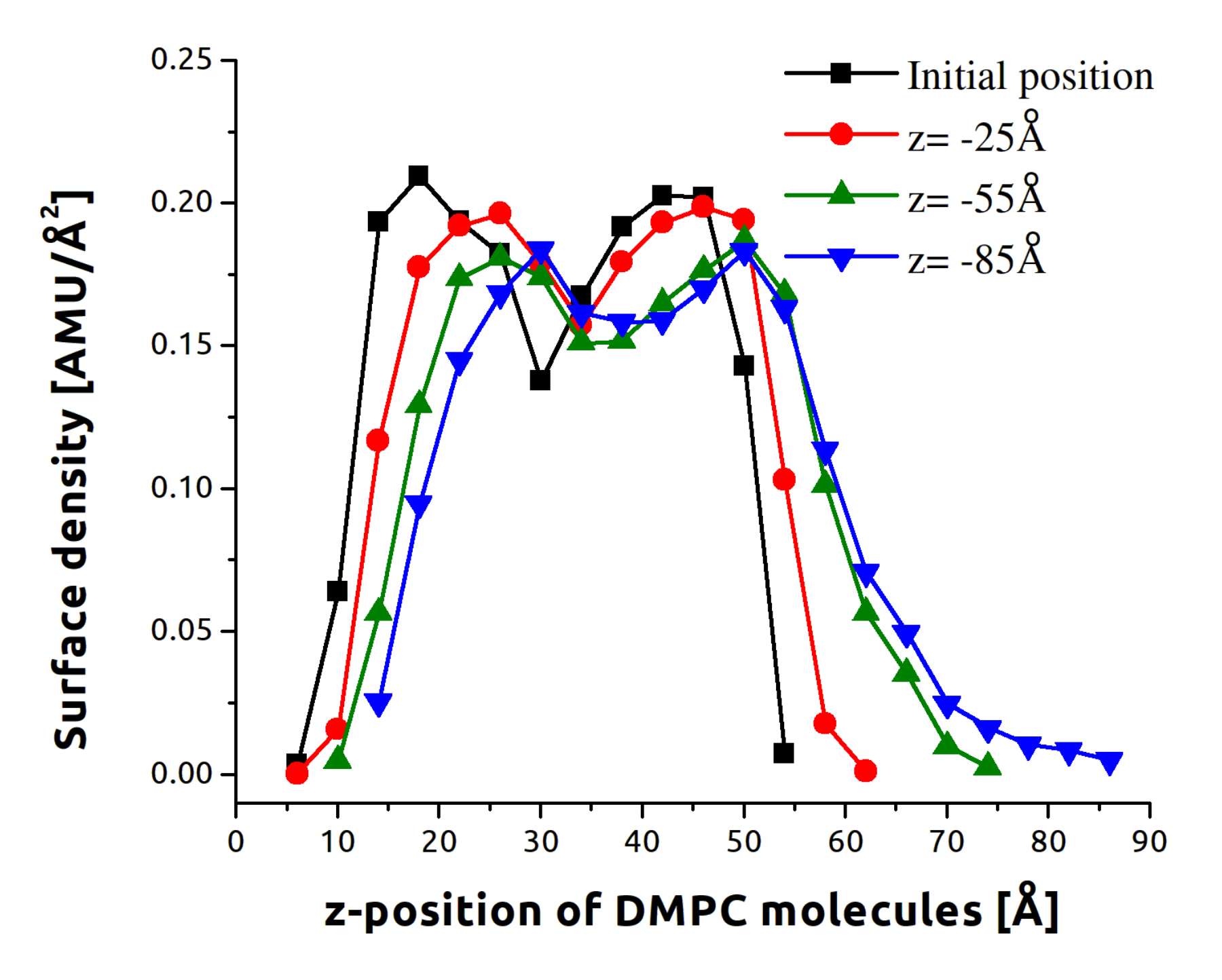
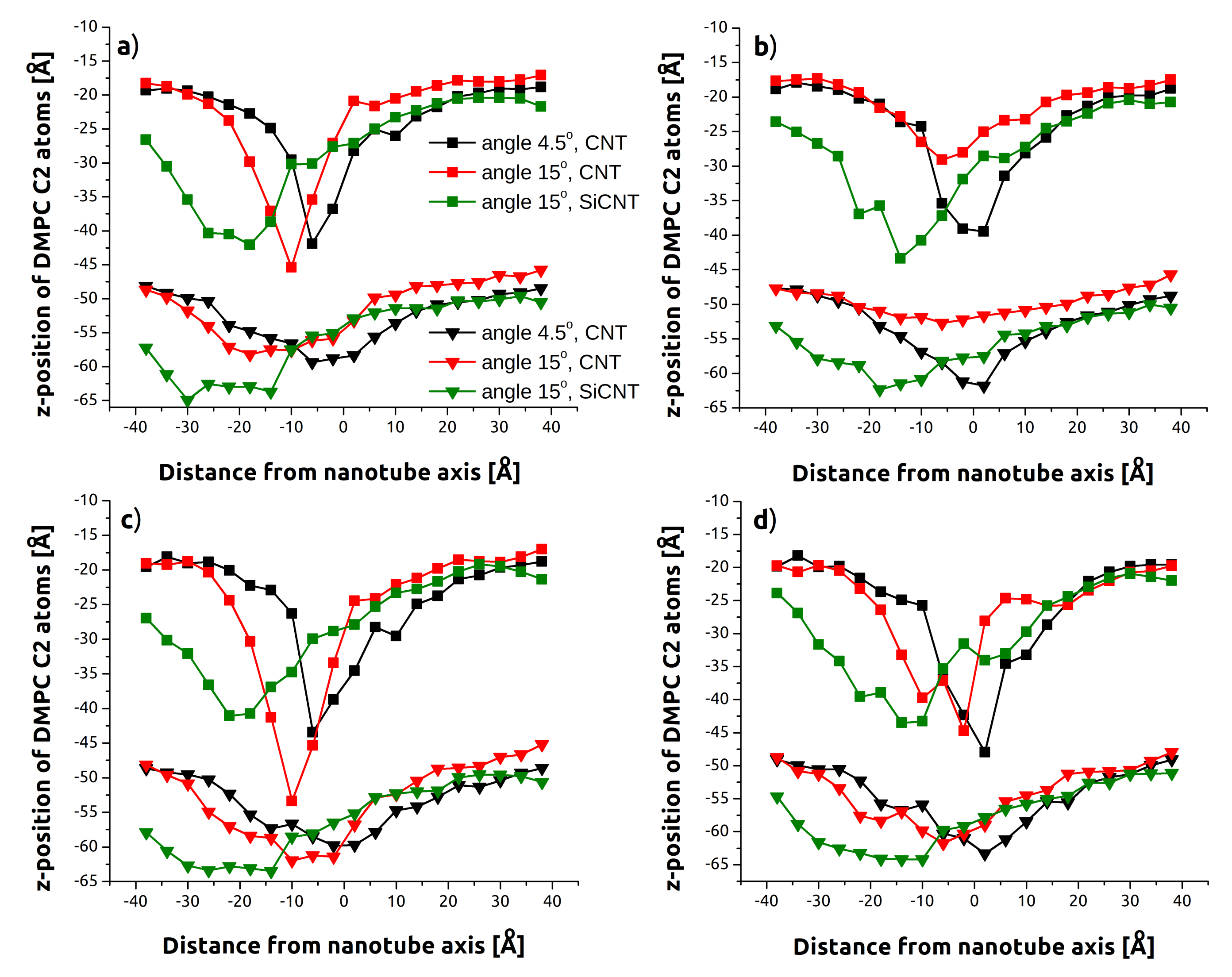
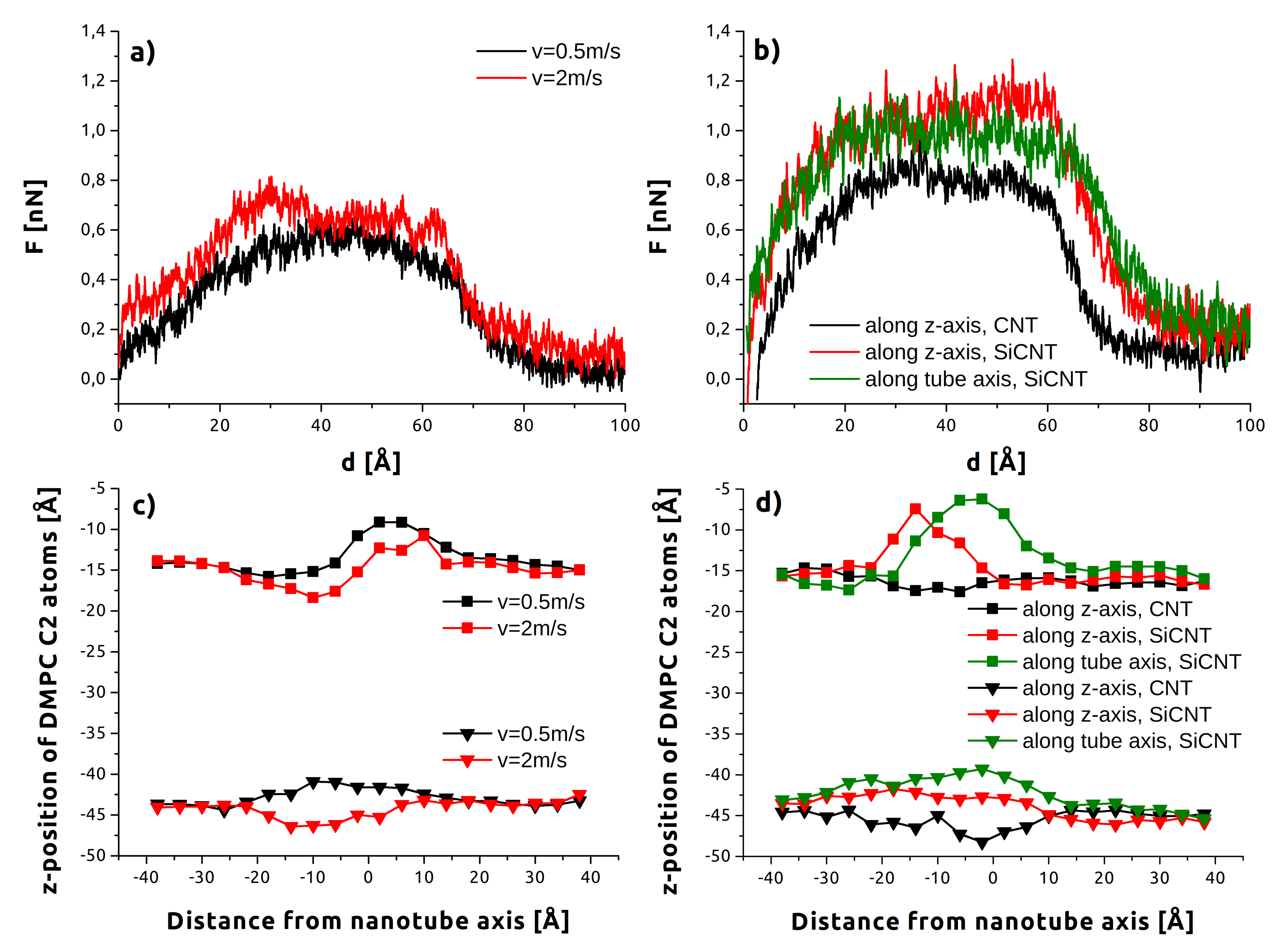
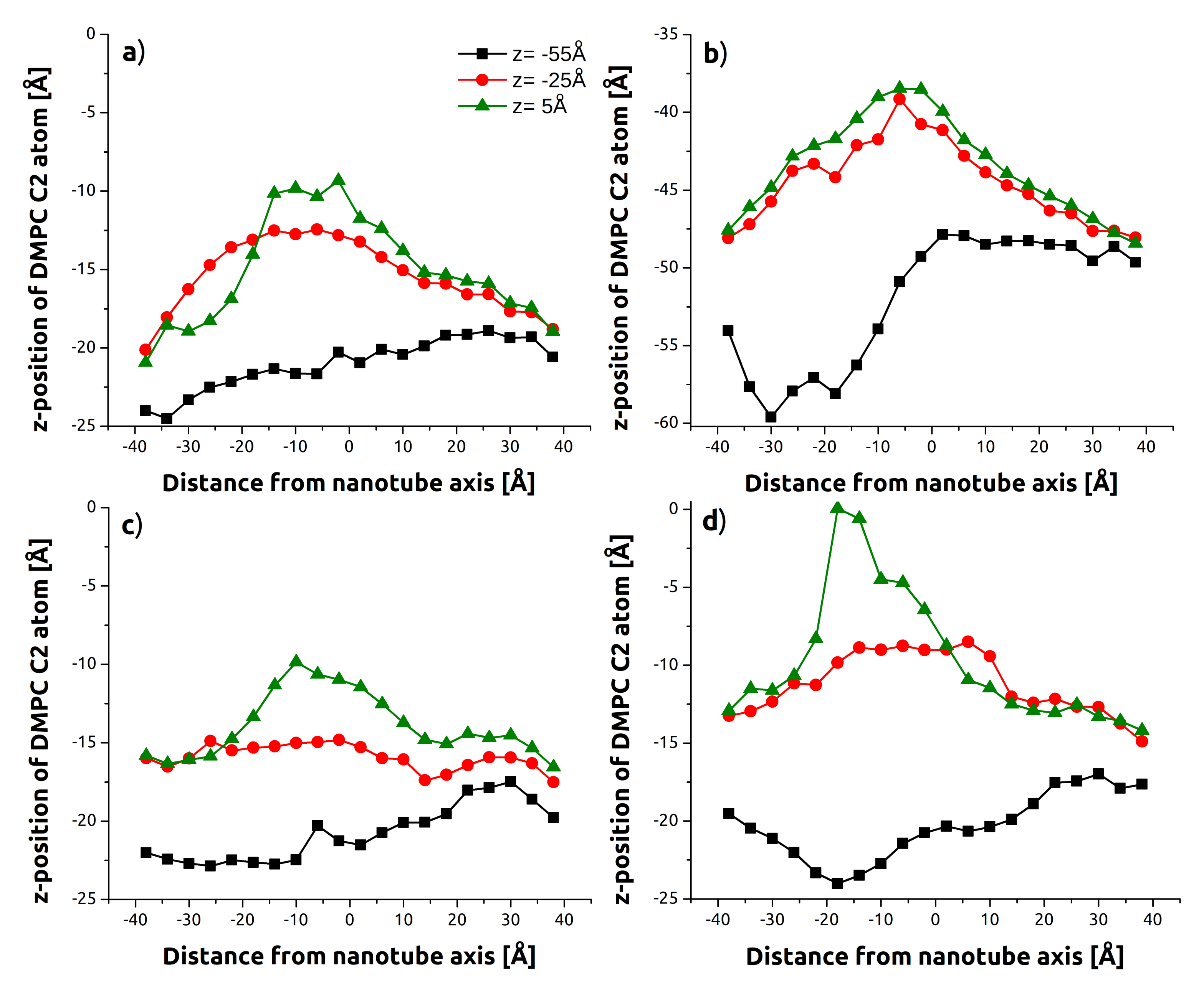
3.1. Indentation Process
3.2. Withdrawal Process
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Golnabi, H. Carbon nanotube research developments in terms of published papers and patents, synthesis and production. Sci. Iran. 2012, 19, 2012–2022. [Google Scholar] [CrossRef] [Green Version]
- Rao, R.; Pint, C.L.; Islam, A.E.; Weatherup, R.S.; Hofmann, S.; Meshot, E.R.; Wu, F.; Zhou, C.; Dee, N.; Amama, P.B.; et al. Carbon Nanotubes and Related Nanomaterials: Critical Advances and Challenges for Synthesis toward Mainstream Commercial Applications. ACS Nano 2018, 12, 11756–11784. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Pham-Huy, L.A.; Dramou, P.; Xiao, D.; Zuo, P.; Pham-Huy, C. Carbon Nanotubes: Applications in Pharmacy and Medicine. BioMed Res. Int. 2013, 2013, e578290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liuyang, Z.; Bingqian, X.; Wang, X. Cholesterol Extraction from Cell Membrane by Graphene Nanosheets: A Computational Study. J. Phys. Chem. B 2016, 120, 957–964. [Google Scholar] [CrossRef]
- Raphey, V.R.; Henna, T.K.; Nivitha, K.P.; Mufeedha, P.; Sabu, C.; Pramod, K. Advanced biomedical applications of carbon nanotube. Mater. Sci. Eng. C 2019, 100, 616–630. [Google Scholar] [CrossRef]
- Raczynski, P.; Gorny, K.; Beldowski, P.; Yuvan, S.; Dendzik, Z. Application of Graphene as a Nanoindenter Interacting with Phospholipid Membranes-Computer Simulation Study. J. Phys. Chem. B 2020, 124, 6592–6602. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, R.; Ashrafizadeh, M.; Pardakhty, A.; Uzieliene, I.; Denkovskij, J.; Bernotiene, E.; Janssen, L.; Lorite, G.S.; Saarakkala, S.; Mobasheri, A. Nanotechnological Strategies for Osteoarthritis Diagnosis, Monitoring, Clinical Management, and Regenerative Medicine: Recent Advances and Future Opportunities. Curr. Rheumatol. Rep. 2020, 22, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacchetti, C.; Liu-Bryan, R.; Magrini, A.; Rosato, N.; Bottini, N.; Bottini, M. Polyethylene-Glycol-Modified Single-Walled Carbon Nanotubes for Intra-Articular Delivery to Chondrocytes. ACS Nano 2014, 8, 12280–12291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vardharajula, S.; Ali, S.Z.; Tiwari, P.M.; Eroğlu, E.; Vig, K.; Dennis, V.A.; Singh, S.R. Functionalized carbon nanotubes: Biomedical applications. Int. J. Nanomed. 2012, 7, 5361–5374. [Google Scholar] [CrossRef] [Green Version]
- Maiti, D.; Tong, X.; Mou, X.; Yang, K. Carbon-Based Nanomaterials for Biomedical Applications: A Recent Study. Front. Pharmacol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Yaron, P.N.; Holt, B.D.; Short, P.A.; Lösche, M.; Islam, M.F.; Dahl, K.N. Single wall carbon nanotubes enter cells by endocytosis and not membrane penetration. J. Nanobiotechnol. 2011, 9, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vakarelski, I.U.; Brown, S.C.; Higashitani, K.; Moudgil, B.M. Penetration of Living Cell Membranes with Fortified Carbon Nanotube Tips. Langmuir 2007, 23, 10893–10896. [Google Scholar] [CrossRef]
- Chen, X.; Kis, A.; Zettl, A.; Bertozzi, C.R. A cell nanoinjector based on carbon nanotubes. Proc. Natl. Acad. Sci. USA 2007, 104, 8218–8222. [Google Scholar] [CrossRef] [Green Version]
- Zakaria, A.; Picaud, F.; Guillaume, Y.C.; Gharbi, T.; Micheau, O.; Herlem, G. Enhanced DR5 binding capacity of nanovectorized TRAIL compared to its cytotoxic version by affinity chromatography and molecular docking studies. J. Mol. Recognit. 2016, 29, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Boukari, K.; Caoduro, C.; Kacem, R.; Skandrani, N.; Borg, C.; Boulahdour, H.; Gharbi, T.; Delage-Mourroux, R.; Hervouet, E.; Pudlo, M.; et al. Nanovectorization of DNA Through Cells Using Protamine Complexation. J. Membr. Biol. 2016, 249, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Caoduro, C.; Kacem, R.; Boukari, K.; Picaud, F.; Brachais, C.H.; Monchaud, D.; Borg, C.; Boulahdour, H.; Gharbi, T.; Delage-Mourroux, R.; et al. Carbon nanotube—Protamine hybrid: Evaluation of DNA cell penetration. Carbon 2016, 96, 742–752. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Mori-Yamamoto, N.; Mori-Shuto, S.; Mori, K.; Kawashiri, N.; Minami, R. Basic Studies of Carbon Nanotube/DNA- protamine Complex Membranes for Guided Bone Regeneration. J. Oral Tissue Eng. 2020, 18, 63–70. [Google Scholar] [CrossRef]
- Sireesha, M.; Babu, V.J.; Kiran, A.S.K.; Ramakrishna, S. A review on carbon nanotubes in biosensor devices and their applications in medicine. Nanocomposites 2018, 4, 36–57. [Google Scholar] [CrossRef]
- Mousavi, S.Z.; Amjad-Iranagh, S.; Nademi, Y.; Modarress, H. Carbon Nanotube-Encapsulated Drug Penetration Through the Cell Membrane: An Investigation Based on Steered Molecular Dynamics Simulation. J. Membr. Biol. 2013, 246, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Raczynski, P.; Gorny, K.; Pabiszczak, M.; Gburski, Z. Nanoindentation of biomembrane by carbon nanotubes–MD simulation. Comput. Mater. Sci. 2013, 70, 13–18. [Google Scholar] [CrossRef]
- Solares, S.D.; Matsuda, Y.; Goddard, W.A. Influence of the Carbon Nanotube Probe Tilt Angle on the Effective Probe Stiffness and Image Quality in Tapping-Mode Atomic Force Microscopy. J. Phys. Chem. B 2005, 109, 16658–16664. [Google Scholar] [CrossRef] [Green Version]
- Wilson, N.; Macpherson, J. Carbon nanotube tips for atomic force microscopy. Nat. Nanotechnol. 2009, 4, 483–491. [Google Scholar] [CrossRef]
- Yum, K.; Wang, N.; Yu, M.F. Nanoneedle: A multifunctional tool for biological studies in living cells. Nanoscale 2010, 2, 363–372. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Kale, L.; Skeel, R.; Bhandarkar, M.; Brunner, R.; Gursoy, A.; Krawetz, N.; Phillips, J.; Shinozaki, A.; Varadarajan, K.; Schulten, K. NAMD2: Greater Scalability for Parallel Molecular Dynamics. J. Comput. Phys. 1999, 151, 283–312. [Google Scholar] [CrossRef]
- Kraszewski, S.; Picaud, F.; Elhechmi, I.; Gharbi, T.; Ramseyer, C. How long a functionalized carbon nanotube can passively penetrate a lipid membrane. Carbon 2012, 50, 5301–5308. [Google Scholar] [CrossRef]
- Duverger, E.; Balme, S.; Bechelany, M.; Miele, P.; Picaud, F. Natural payload delivery of the doxorubicin anticancer drug from boron nitride oxide nanosheets. Appl. Surf. Sci. 2019, 475, 666–675. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M.; Tuszynski, J.A. Membrane Biophysics; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Alberts, B. Molecular Biology of the Cell; Taylor and Francis: Abingdon, UK, 2015. [Google Scholar]
- Raczynski, P.; Gorny, K.; Raczynska, V.; Pabiszczak, M.; Dendzik, Z.; Gburski, Z. On the impact of nanotube diameter on biomembrane indentation—Computer simulations study. Biochim. Biophys. Acta-Biomembr. 2018, 1860, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Raczynski, P.; Gorny, K.; Dendzik, Z.; Samios, J.; Gburski, Z. Modeling the Impact of Silicon-Carbide Nanotube on the Phospholipid Bilayer Membrane: Study of Nanoindentation and Removal Processes via Molecular Dynamics Simulation. J. Phys. Chem. C 2019, 123, 18726–18733. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Brünger, A.; Brooks, C.L., III; Karplus, M. Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 1984, 105, 495–500. [Google Scholar] [CrossRef]
- MacKerell, A.D.J.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VDM: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–28. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613. [Google Scholar] [CrossRef]
- Wallace, E.J.; Sansom, M.S.P. Blocking of carbon nanotube based nanoinjectors by lipids: A simulation study. Nano Lett. 2008, 8, 2751–2756. [Google Scholar] [CrossRef]
- Pogodin, S.; Baulin, V. Can a Carbon Nanotube Pierce through a Phospholipid Bilayer? ACS Nano 2010, 4, 5293–5300. [Google Scholar] [CrossRef] [Green Version]
- Gangupomu, V.; Capaldi, F. Interactions of Carbon Nanotube with Lipid Bilayer Membranes. J. Nanomater. 2011, 2011, 830436. [Google Scholar] [CrossRef] [Green Version]


| Angle | CNT or SiCNT | Along Tube Axis | Along z-Axis |
|---|---|---|---|
| low rate | |||
| 4.5° | CNT | 2.9 (1.9) | 3.8 (1.5) |
| 9° | CNT | 3.1 (1.5) | 3.9 (1.4) |
| 15° | CNT | 3.9 (2.2) | 4.1 (2.4) |
| 15° | SiCNT | 7.5 (2.4) | 10.0 (3.2) |
| high rate | |||
| 4.5° | CNT | 2.4 (1.6) | 2.8 (2.2) |
| 9° | CNT | 2.6 (1.8) | 3.1 (1.4) |
| 15° | CNT | 2.7 (2.4) | 4.4 (2.2) |
| 15° | SiCNT | 4.9 (2.4) | 8.0 (2.6) |
| CNT or SiCNT | Along Tube Axis | Along z-Axis |
|---|---|---|
| low rate | ||
| CNT | 1.9 (1.2) | 1.2 (1.0) |
| SiCNT | 4.8 (2.0) | 4.3 (1.2) |
| high rate | ||
| CNT | 1.0 (0.7) | 0.4 (0.5) |
| SiCNT | 1.4 (1.1) | 1.9 (0.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raczyński, P.; Górny, K.; Bełdowski, P.; Yuvan, S.; Marciniak, B.; Dendzik, Z. Steered Molecular Dynamics of Lipid Membrane Indentation by Carbon and Silicon-Carbide Nanotubes—The Impact of Indenting Angle Uncertainty. Sensors 2021, 21, 7011. https://doi.org/10.3390/s21217011
Raczyński P, Górny K, Bełdowski P, Yuvan S, Marciniak B, Dendzik Z. Steered Molecular Dynamics of Lipid Membrane Indentation by Carbon and Silicon-Carbide Nanotubes—The Impact of Indenting Angle Uncertainty. Sensors. 2021; 21(21):7011. https://doi.org/10.3390/s21217011
Chicago/Turabian StyleRaczyński, Przemysław, Krzysztof Górny, Piotr Bełdowski, Steven Yuvan, Beata Marciniak, and Zbigniew Dendzik. 2021. "Steered Molecular Dynamics of Lipid Membrane Indentation by Carbon and Silicon-Carbide Nanotubes—The Impact of Indenting Angle Uncertainty" Sensors 21, no. 21: 7011. https://doi.org/10.3390/s21217011






