Compensation Strategies for Bioelectric Signal Changes in Chronic Selective Nerve Cuff Recordings: A Simulation Study
Abstract
1. Introduction
2. Methods
2.1. Model Construction
2.2. Simulation of Chronic Factors
2.2.1. Growth of Encapsulation Tissue
2.2.2. Rotation of the Nerve Cuff Electrode
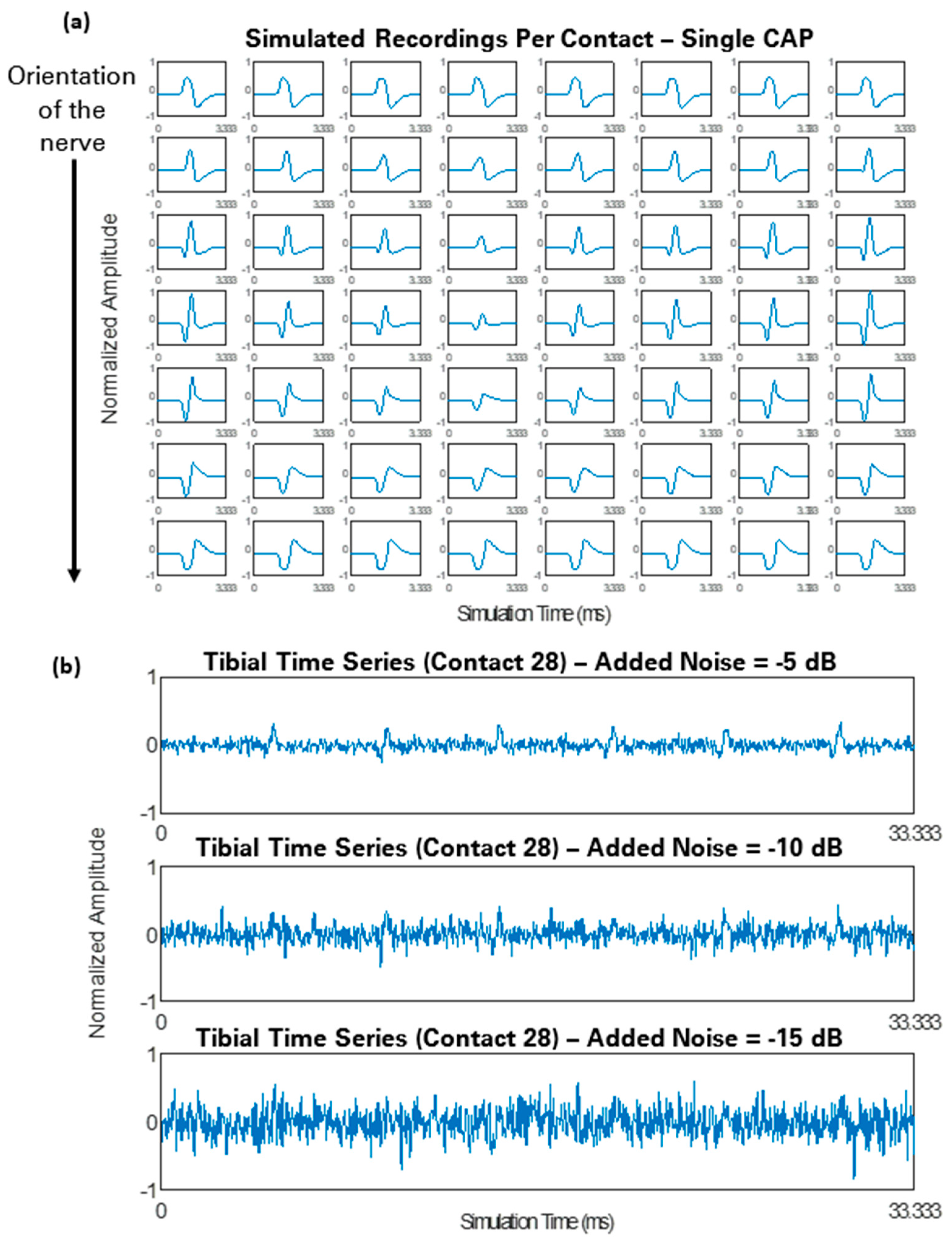
2.3. Simulated Recordings
2.4. CAP Classification
2.4.1. Datasets
2.4.2. Convolutional Neural Network
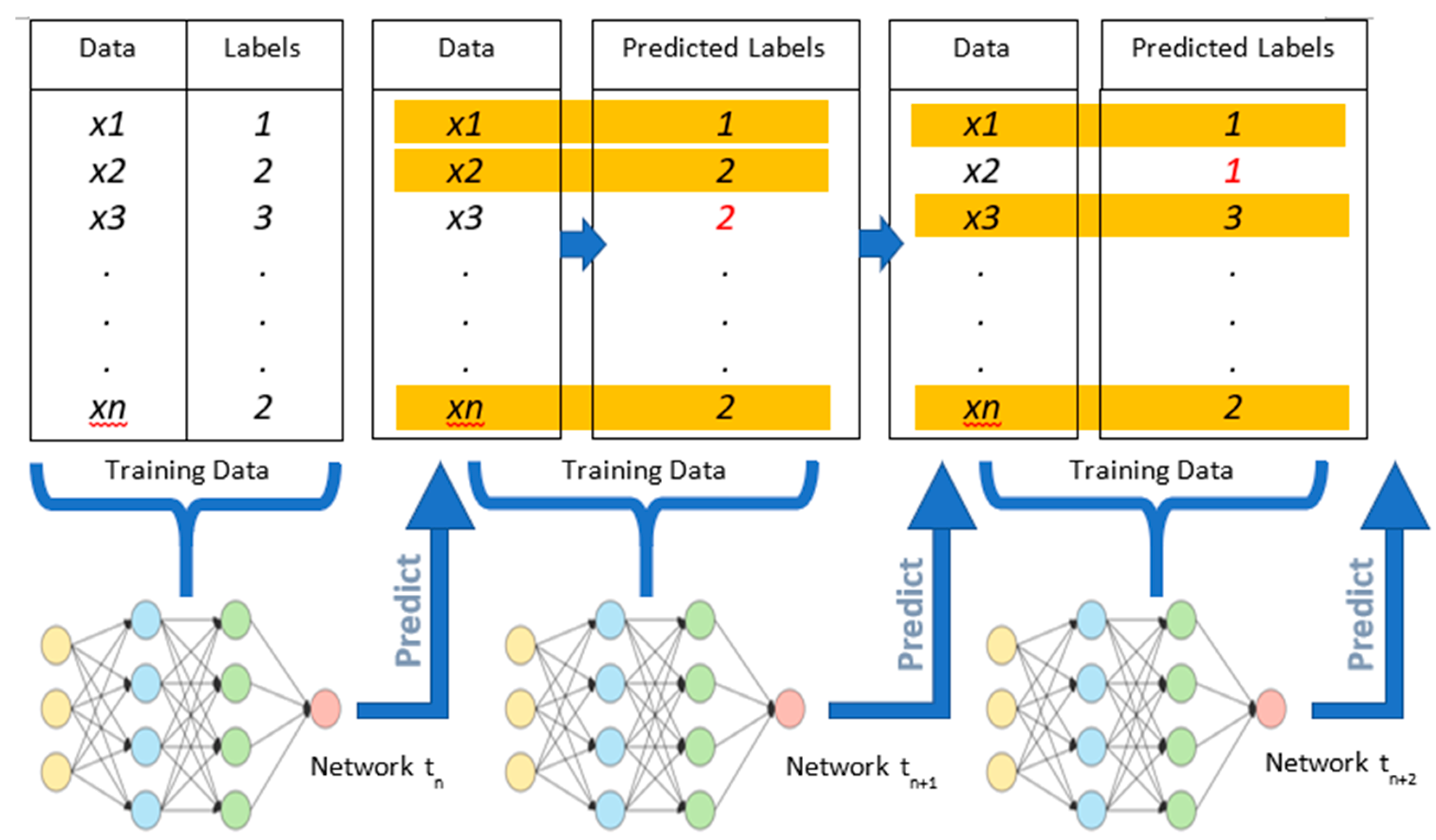
2.5. Classifier Update Strategies
2.5.1. Baseline Calibration
2.5.2. Periodic Recalibration
2.5.3. Self-Learning Approach
2.5.4. Further Analysis of the Self-Learning Approach
Training Frequency
Initial Performance Level
Evaluation
3. Results
3.1. Simulated Recordings
3.2. Classification
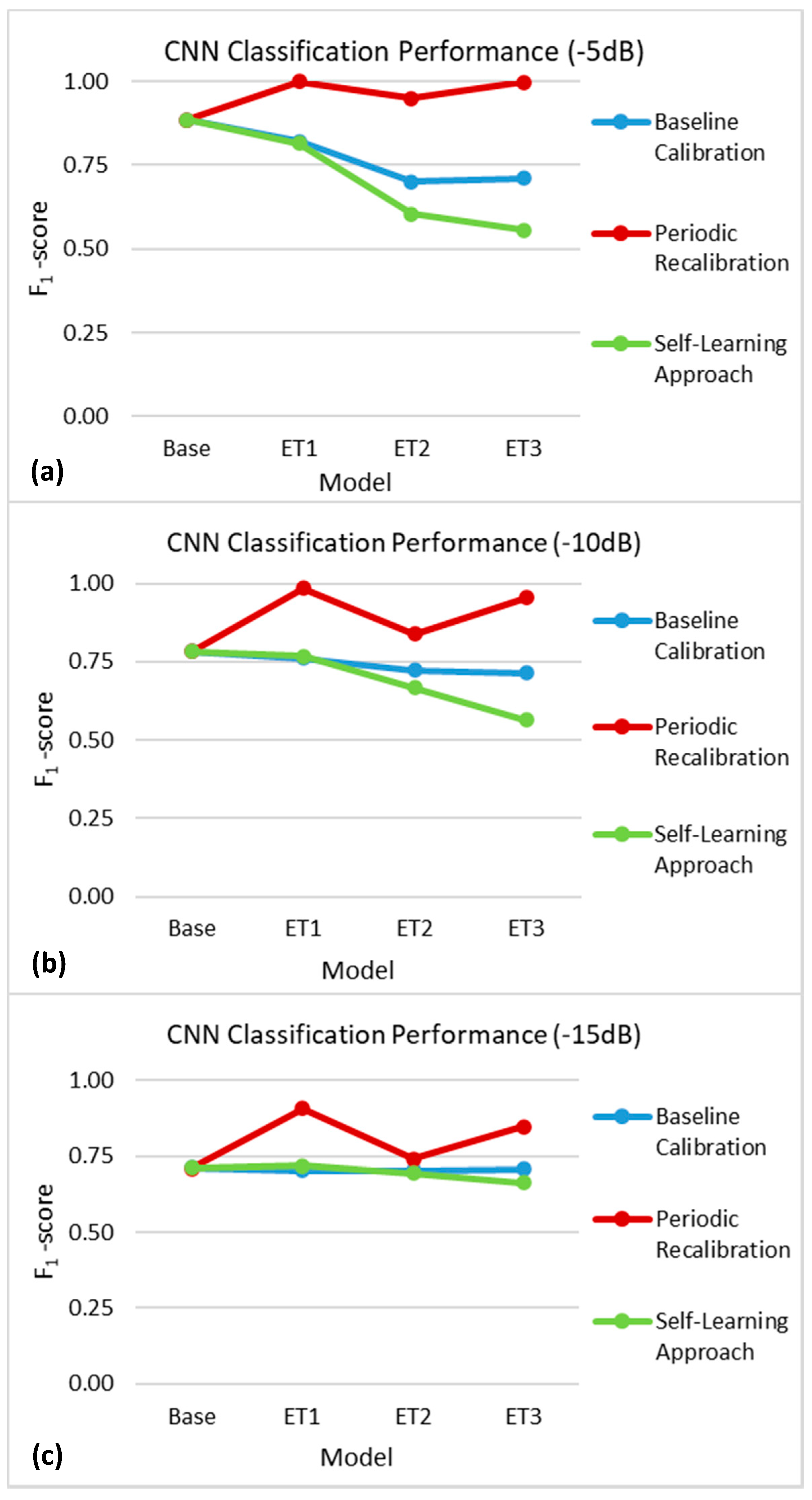
3.2.1. Growth of Encapsulation Tissue
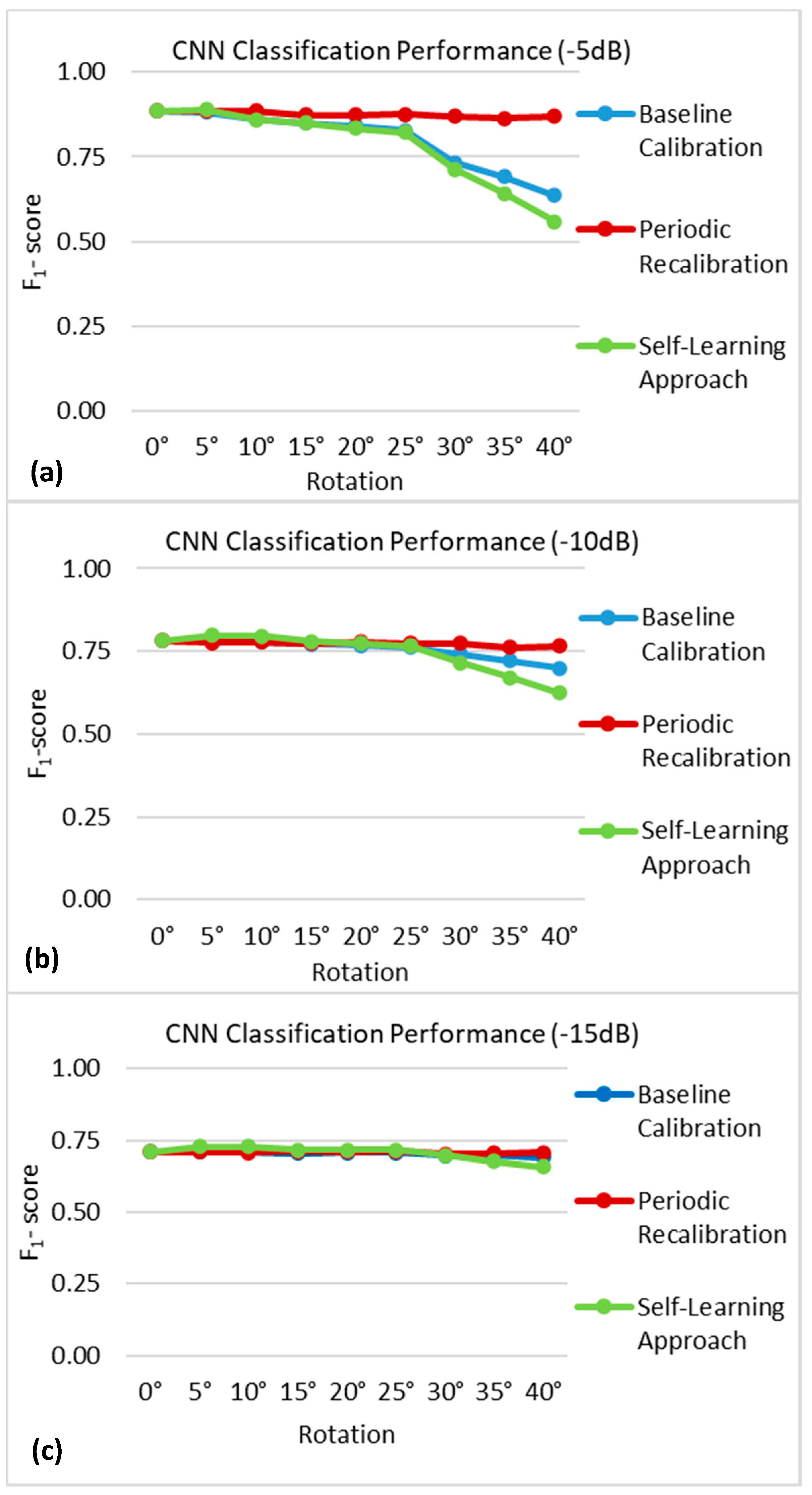
3.2.2. Rotation of the Nerve Cuff Electrode
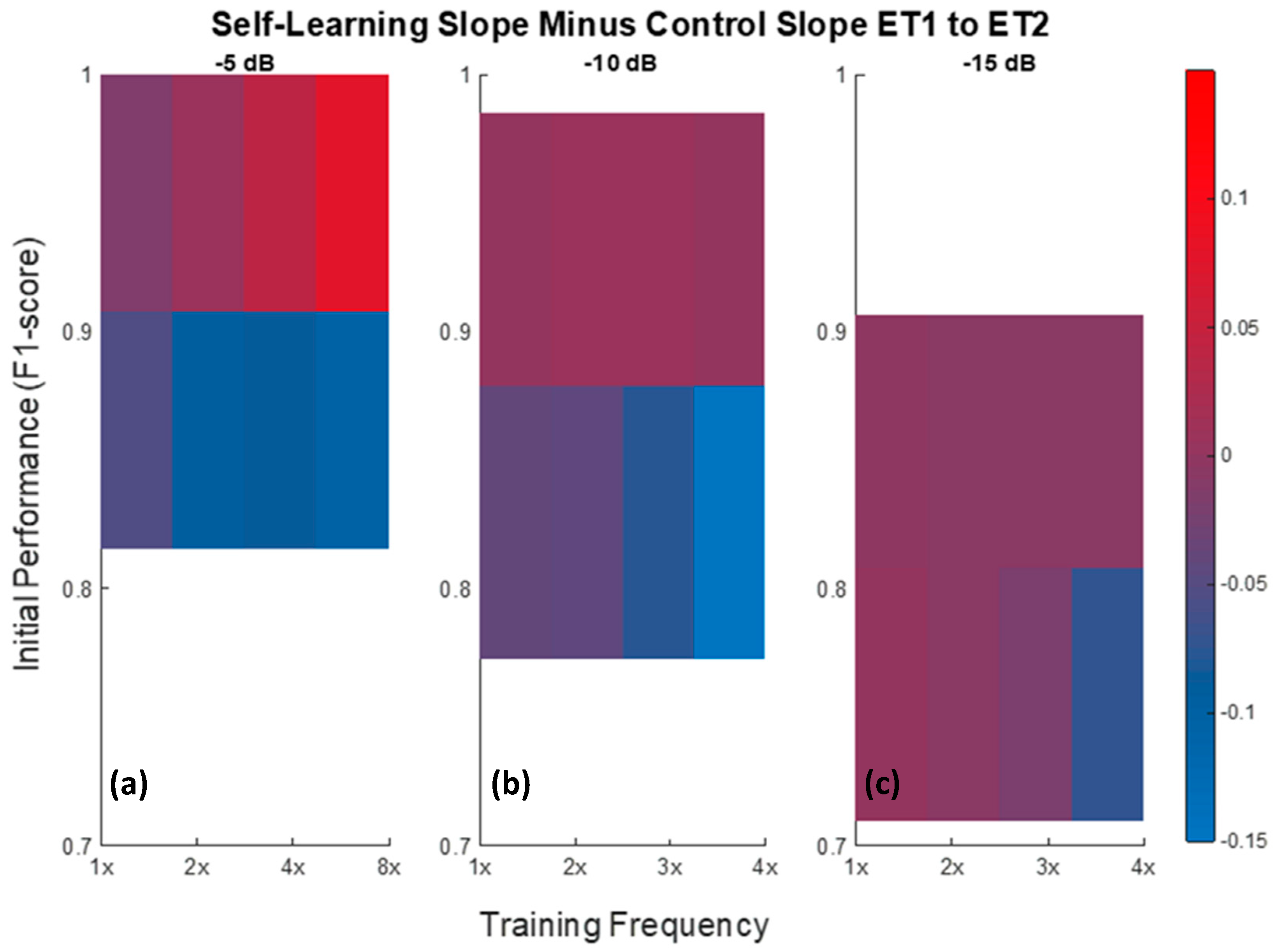
3.2.3. Influence of Training Frequency and Initial Performance on Self-Learning Approach Performance
4. Discussion
4.1. Baseline Calibration
4.2. Periodic Recalibration
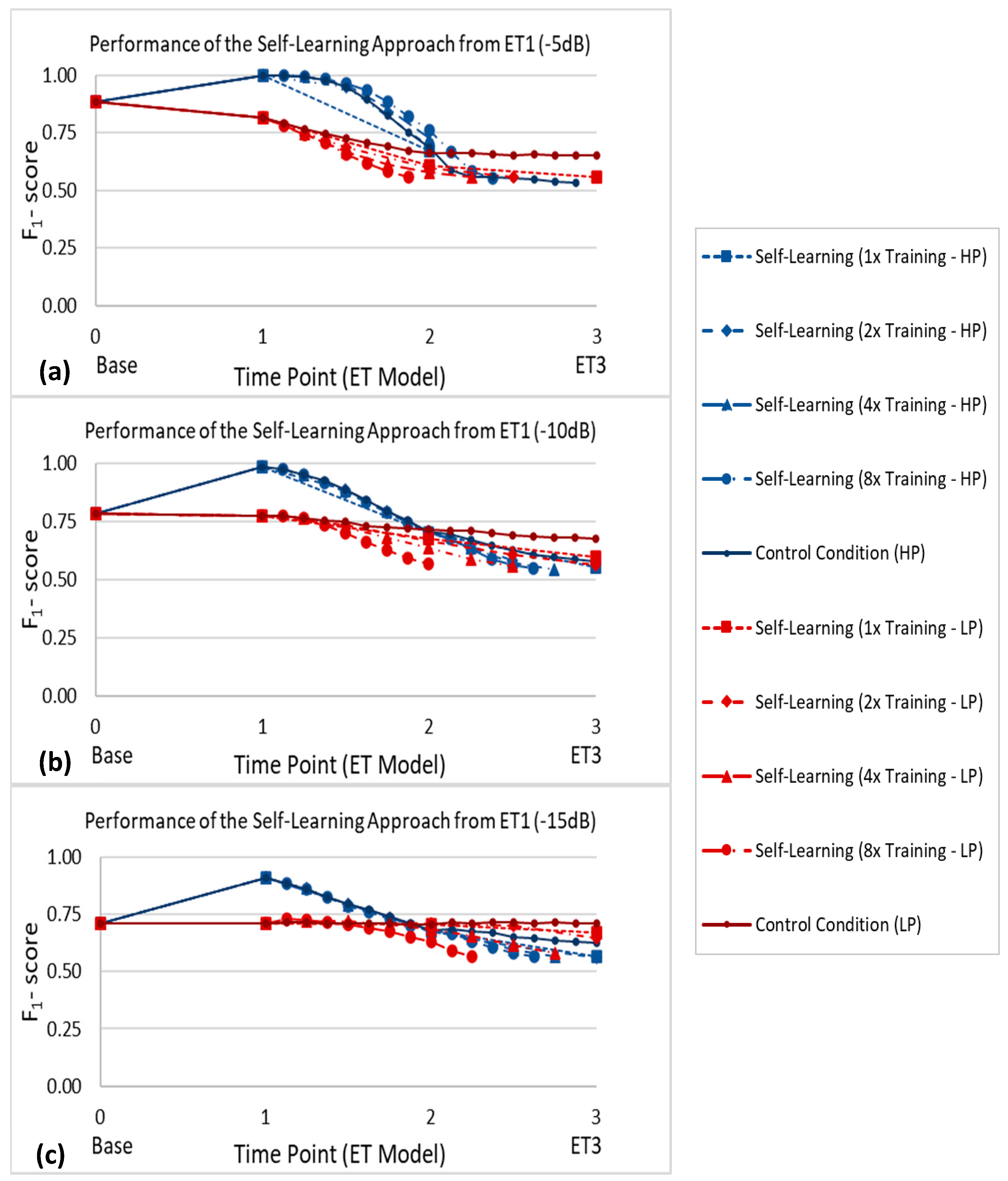
4.3. Self-Learning Approach
4.4. Influence of Training Frequency and Initial Performance on Self-Learning Approach Performance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhillon, G.S.; Lawrence, S.M.; Hutchinson, D.T.; Horch, K.W. Residual function in peripheral nerve stumps of amputees: Implications for neural control of artificial limbs. J. Hand Surg. 2004, 29, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, G.S.; Krüger, T.B.; Sandhu, J.S.; Horch, K.W. Effects of short-term training on sensory and motor function in severed nerves of long-term human amputees. J. Neurophysiol. 2005, 93, 2625–2633. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.S.; Wark, H.A.C.; Hutchinson, D.T.; Warren, D.J.; O’Neill, K.; Scheinblum, T.; Clark, G.A.; Normann, R.A.; Greger, B. Restoring motor control and sensory feedback in people with upper extremity amputations using arrays of 96 microelectrodes implanted in the median and ulnar nerves. J. Neural Eng. 2016, 13. [Google Scholar] [CrossRef]
- Micera, S.; Citi, L.; Rigosa, J.; Carpaneto, J.; Raspopovic, S.; di Pino, G.; Rossini, L.; Yoshida, K.; Denaro, L.; Dario, P.; et al. Decoding information from neural signals recorded using intraneural electrodes: Toward the development of a neurocontrolled hand prosthesis. Proc. IEEE 2010, 98, 407–417. [Google Scholar] [CrossRef]
- Inmann, A.; Haugland, M. Functional evaluation of natural sensory feedback incorporated in a hand grasp neuroprosthesis. Med. Eng. Phys. 2004, 26, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Haugland, M.K.; Sinkjær, T. Cutaneous whole nerve recordings used for correction of footdrop in hemiplegic man. IEEE Trans. Rehabil. Eng. 1995, 3, 307–317. [Google Scholar] [CrossRef]
- Plachta, D.T.T.; Gierthmuehlen, M.; Cota, O.; Espinosa, N.; Boeser, F.; Herrera, T.C.; Stieglitz, T.; Zentner, J. Blood pressure control with selective vagal nerve stimulation and minimal side effects. J. Neural Eng. 2014, 11, 036011. [Google Scholar] [CrossRef] [PubMed]
- Harreby, K.R.; Sevcencu, C.; Struijk, J.J. Early seizure detection in rats based on vagus nerve activity. Med. Biol. Eng. Comput. 2011, 49, 143–151. [Google Scholar] [CrossRef]
- Nielsen, T.N.; Struijk, J.J.; Harreby, K.R.; Sevcencu, C. Early detection of epileptic seizures in pigs based on vagus nerve activity. In Converging Clinical and Engineering Research on Neurorehabilitation; Springer: Berlin, Germany, 2013. [Google Scholar]
- Cloutier, A.; Yang, J. Design, control, and sensory feedback of externally powered hand prostheses: A literature review. Crit. Rev. Biomed. Eng. 2013, 41. [Google Scholar] [CrossRef]
- Del Valle, J.; Navarro, X. Interfaces with the peripheral nerve for the control of neuroprostheses. Int. Rev. Neurobiol. 2013, 109, 63–83. [Google Scholar]
- Brindley, G.S. The first 500 patients with sacral anterior root stimulator implants: General description. Paraplegia 1994, 32, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Lyons, G.M.; Sinkjær, T.; Burridge, J.H.; Wilcox, D.J. A review of portable FES-based neural orthoses for the correction of drop foot. IEEE Trans. Neural Syst. Rehabil. Eng. 2002, 10, 260–279. [Google Scholar] [CrossRef] [PubMed]
- Waters, R.L.; McNeal, D.R.; Faloon, W.; Clifford, B. Functional electrical stimulation of the peroneal nerve for hemiplegia. Long-term clinical follow-up. J. Bone Jt. Surg. Ser. A 1985, 67, 792–793. [Google Scholar] [CrossRef]
- Dweiri, Y.M.; Eggers, T.E.; Gonzalez-Reyes, L.E.; Drain, J.; McCallum, G.A.; Durand, D.M. Stable detection of movement intent from peripheral nerves: Chronic study in dogs. Proc. IEEE 2017, 105, 50–65. [Google Scholar] [CrossRef]
- Stein, R.B.; Nichols, T.R.; Jhamandas, J.; Davis, L.; Charles, D. Stable long-term recordings from cat peripheral nerves. Brain Res. 1977, 128, 21–38. [Google Scholar] [CrossRef]
- Popović, D.B.; Stein, R.B.; Jovanović, K.L.; Dai, R.; Kostov, A.; Armstrong, W.W. Sensory nerve recording for closed-loop control to restore motor functions. IEEE Trans. Biomed. Eng. 1993, 40, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Struijk, J.J.; Thomsen, M.; Larsen, J.O.; Sinkjaer, T. Cuff electrodes for long-term recording of natural sensory information. IEEE Eng. Med. Biol. Mag. 1999, 18, 91–98. [Google Scholar] [CrossRef]
- Rodriguez, F.J.; Ceballos, D.; Schu, M.; Valero, A.; Valderrama, E.; Stieglitz, T.; Navarro, X. Polyimide cuff electrodes for peripheral nerve stimulation. J. Neurosci. Methods 2000, 98, 105–118. [Google Scholar] [CrossRef]
- Grill, W.M.; Mortimer, J.T. Neural and connective tissue response to long-term implantation of multiple contact nerve cuff electrodes. J. Biomed. Mater. Res. 2000, 50, 215–226. [Google Scholar] [CrossRef]
- Vasudevan, S.; Patel, K.; Welle, C. Rodent model for assessing the long term safety and performance of peripheral nerve recording electrodes. J. Neural Eng. 2017, 14, 16008. [Google Scholar] [CrossRef]
- Christie, B.P.; Freeberg, M.; Memberg, W.D.; Pinault, G.J.; Hoyen, H.A.; Tyler, D.J.; Triolo, R.J. Long-term stability of stimulating spiral nerve cuff electrodes on human peripheral nerves. J. Neuroeng. Rehabil. 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Navarro, X.; Valderrama, E.; Stieglitz, T.; Schüttler, M. Selective fascicular stimulation of the rat sciatic nerve with multipolar polyimide cuff electrodes. Restor. Neurol. Neurosci. 2001, 18, 9–21. [Google Scholar] [PubMed]
- Tarler, M.D.; Mortimer, J.T. Selective and independent activation of four motor fascicles using a four contact nerve-cuff electrode. IEEE Trans. Neural Syst. Rehabil. Eng. 2004, 12, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Veraart, C.; Grill, W.M.; Mortimer, J.T. Selective control of muscle activation with a multipolar nerve cuff electrode. IEEE Trans. Biomed. Eng. 1993, 40, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.S.; Griffith, P.; Sweeney, J.; Scarpine, V.; Bidnar, M.; McLane, J.; Robinson, C. Multielectrode nerve cuff stimulation of the median nerve produces selective movements in a raccoon animal model. J. Spinal Cord Med. 1997, 20, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Grill, W.M.; Mortimer, J.T. The effect of stimulus pulse duration on selectivity of neural stimulation. IEEE Trans. Biomed. Eng. 1996, 43, 161–166. [Google Scholar] [CrossRef]
- Raspopovic, S.; Carpaneto, J.; Udina, E.; Navarro, X.; Micera, S. On the identification of sensory information from mixed nerves by using single-channel cuff electrodes. J. Neuroeng. Rehabil. 2010, 7, 17. [Google Scholar] [CrossRef]
- Tesfayesus, W.; Durand, D.M. Blind source separation of peripheral nerve recordings. J. Neural Eng. 2007, 4, S157. [Google Scholar] [CrossRef]
- Metcalfe, B.; Chew, D.; Clarke, C.; Donaldson, N.; Taylor, J. Fibre-selective discrimination of physiological ENG using velocity selective recording: Report on pilot rat experiments. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society EMBC 2014, Chicago, IL, USA, 26–30 August 2014; pp. 2645–2648. [Google Scholar]
- Metcalfe, B.W.; Chew, D.J.; Clarke, C.T.; Donaldson, N.D.N.; Taylor, J.T. A new method for spike extraction using velocity selective recording demonstrated with physiological ENG in Rat. J. Neurosci. Methods 2015, 251, 47–55. [Google Scholar] [CrossRef]
- Metcalfe, B.; Nielsen, T.; Taylor, J. Velocity selective recording: A demonstration of effectiveness on the vagus nerve in pig. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society EMBS, Honolulu, HI, USA, 18–21 July 2018; Volume 2018, pp. 2945–2948. [Google Scholar]
- Schuettler, M.; Donaldson, N.; Seetohul, V.; Taylor, J. Fibre-selective recording from the peripheral nerves of frogs using a multi-electrode cuff. J. Neural Eng. 2013, 10, 036016. [Google Scholar] [CrossRef]
- Yoshida, K.; Kurstjens, G.A.M.; Hennings, K. Experimental validation of the nerve conduction velocity selective recording technique using a multi-contact cuff electrode. Med. Eng. Phys. 2009, 31, 1261–1270. [Google Scholar] [PubMed]
- Wodlinger, B.; Durand, D.M. Recovery of neural activity from nerve cuff electrodes. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 4653–4656. [Google Scholar]
- Eggers, T.E.; Dweiri, Y.M.; McCallum, G.A.; Durand, D.M. Model-based Bayesian signal extraction algorithm for peripheral nerves. J. Neural Eng. 2017, 14, 056009. [Google Scholar] [PubMed]
- Wodlinger, B.; Durand, D.M. Localization and recovery of peripheral neural sources with beamforming algorithms. IEEE Trans. Neural Syst. Rehabil. Eng. 2009, 17, 461–468. [Google Scholar] [PubMed]
- Zariffa, J.; Popovic, M.R. Localization of active pathways in peripheral nerves: A simulation study. IEEE Trans. Neural Syst. Rehabil. Eng. 2009, 17, 53–62. [Google Scholar] [PubMed]
- Zariffa, J.; Nagai, M.K.; Schuettler, M.; Stieglitz, T.; Daskalakis, Z.J.; Popovic, M.R. Use of an experimentally derived leadfield in the peripheral nerve pathway discrimination problem. IEEE Trans. Neural Syst. Rehabil. Eng. 2011, 19, 147–156. [Google Scholar] [PubMed]
- Koh, R.G.L.; Nachman, A.I.; Zariffa, J. Use of spatiotemporal templates for pathway discrimination in peripheral nerve recordings: A simulation study. J. Neural Eng. 2017, 14, 016013. [Google Scholar] [PubMed]
- Koh, R.G.L.; Nachman, A.I.; Zariffa, J. Classification of naturally evoked compound action potentials in peripheral nerve spatiotemporal recordings. Sci. Rep. 2019, 9, 11145. [Google Scholar]
- Koh, R.G.L.; Balas, M.; Nachman, A.I.; Zariffa, J. Selective peripheral nerve recordings from nerve cuff electrodes using convolutional neural networks. J. Neural Eng. 2020, 17, 016042. [Google Scholar]
- Grill, W.M.; Mortimer, J.T. Electrical properties of implant encapsulation tissue. Ann. Biomed. Eng. 1994, 22, 23–33. [Google Scholar]
- Polikov, V.S.; Tresco, P.A.; Reichert, W.M. Response of brain tissue to chronically implanted neural electrodes. J. Neurosci. Methods 2005, 148, 1–18. [Google Scholar]
- Merrill, D.R.; Tresco, P.A. Impedance characterization of microarray recording electrodes in vitro. IEEE Trans. Biomed. Eng. 2005, 52, 1960–1965. [Google Scholar] [PubMed]
- Garai, P.; Koh, R.G.L.; Schuettler, M.; Stieglitz, T.; Zariffa, J. Influence of anatomical detail and tissue conductivity variations in simulations of multi-contact nerve cuff recordings. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Silveira, C.; Brunton, E.; Spendiff, S.; Nazarpour, K. Influence of nerve cuff channel count and implantation site on the separability of afferent ENG. J. Neural Eng. 2018, 15, 046004. [Google Scholar] [CrossRef]
- CIBC. Seg3D: Volumetric Image Segmentation and Visualization; Scientific Computing and Imaging Institute (SCI): Salt Lake City, UT, USA, 2016. [Google Scholar]
- Fang, Q.; Boas, D.A. Tetrahedral mesh generation from volumetric binary and grayscale images. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI, Boston, MA, USA, 28 June–1 July 2009. [Google Scholar]
- Scientific Computing and Imaging Institute (SCI). SCIRun: A Scientific Computing Problem Solving Environment; Scientific Computing and Imaging Institute (SCI): Salt Lake City, UT, USA, 2015. [Google Scholar]
- Weinstein, D.; Zhukov, L.; Johnson, C. Lead-field bases for electroencephalography source imaging. Ann. Biomed. Eng. 2000, 28, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Yoo, P.B.; Durand, D.M. Selective recording of the canine hypoglossal nerve using a multicontact flat interface nerve electrode. IEEE Trans. Biomed. Eng. 2005, 52, 1461–1469. [Google Scholar] [CrossRef] [PubMed]
- Parrini, S.; Delbeke, J.; Romero, E.; Legat, V.; Veraart, C. Hybrid finite elements and spectral method for computation of the electric potential generated by a nerve cuff electrode. Med. Biol. Eng. Comput. 1999, 37, 733–736. [Google Scholar] [CrossRef]
- Schwarz, J.R.; Reid, G.; Bostock, H. Action potentials and membrane currents in the human node of Ranvier. Pflügers Arch. Eur. J. Physiol. 1995, 430, 283–292. [Google Scholar] [CrossRef]
- Struijk, J.J.; Thomsen, M. Tripolar nerve cuff recording: Stimulus artifact, EMG and the recorded nerve signal. In Proceedings of the 17th International Conference of the Engineering in Medicine and Biology Society, Montreal, QC, Canada, 20–23 September 1995; Volume 2, pp. 1105–1106. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; Volume 16, pp. 265–283. [Google Scholar]
- Schwemmer, M.A.; Skomrock, N.D.; Sederberg, P.B.; Ting, J.E.; Sharma, G.; Bockbrader, M.A.; Friedenberg, D.A. Meeting brain–computer interface user performance expectations using a deep neural network decoding framework. Nat. Med. 2018, 24, 1669–1676. [Google Scholar] [CrossRef]
- Zariffa, J.; Nagai, M.K.; Daskalakis, Z.J.; Popovic, M.R. Influence of the number and location of recording contacts on the selectivity of a nerve cuff electrode. IEEE Trans. Neural Syst. Rehabil. Eng. 2009, 17, 420–427. [Google Scholar]
- Moffitt, M.A.; McIntyre, C.C. Model-based analysis of cortical recording with silicon microelectrodes. Clin. Neurophysiol. 2005, 116, 2240–2250. [Google Scholar] [CrossRef]
- Vetter, R.J.; Williams, J.C.; Hetke, J.F.; Nunamaker, E.A.; Kipke, D.R. Chronic neural recording using silicon-substrate microelectrode arrays implanted in cerebral cortex. IEEE Trans. Biomed. Eng. 2004, 51, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Grill, W.M.; Mortimer, J.T. Temporal stability of nerve cuff electrode recruitment properties. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Montreal, QC, Canada, 20–23 September 1995; Volume 17, pp. 1089–1090. [Google Scholar]
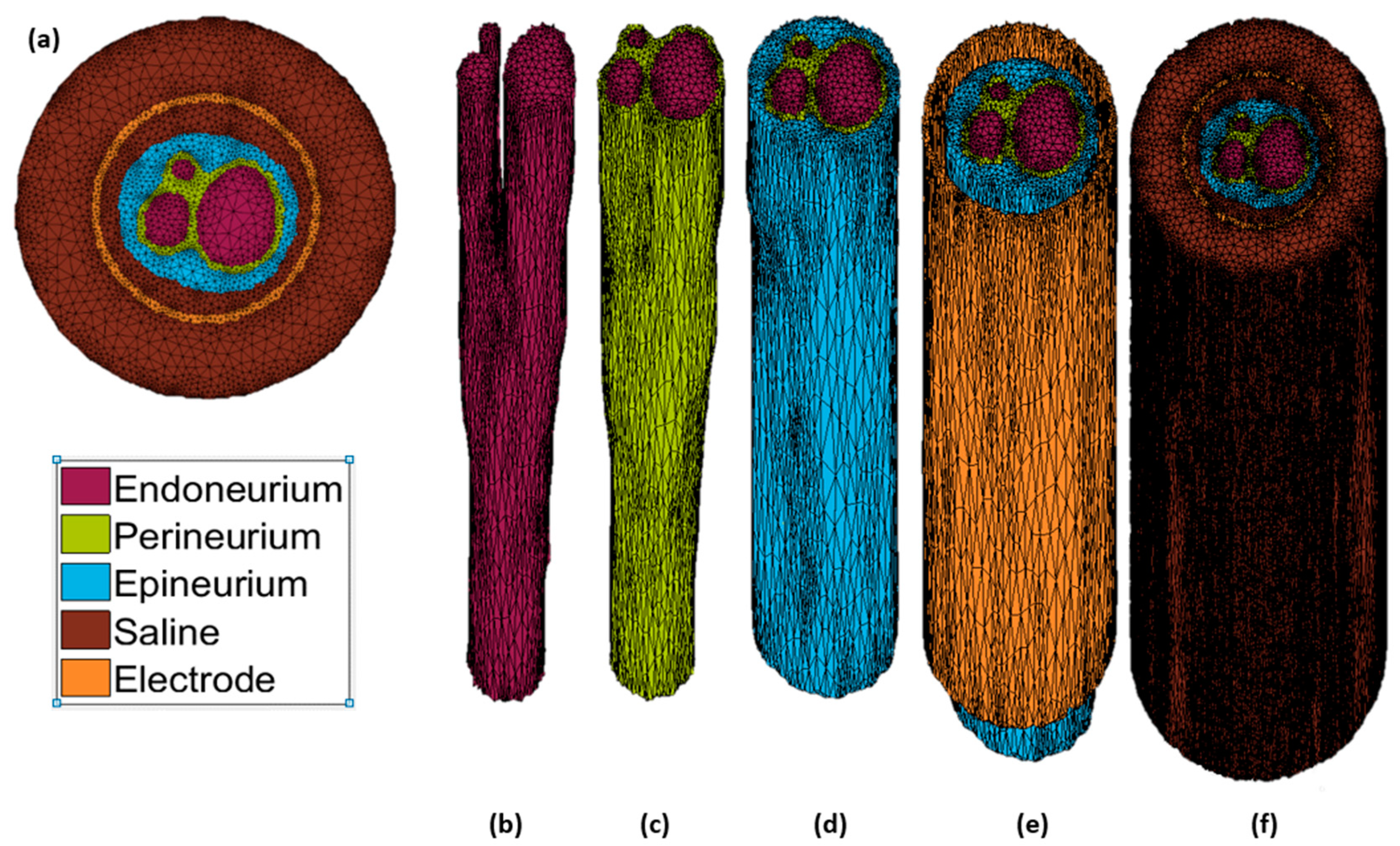
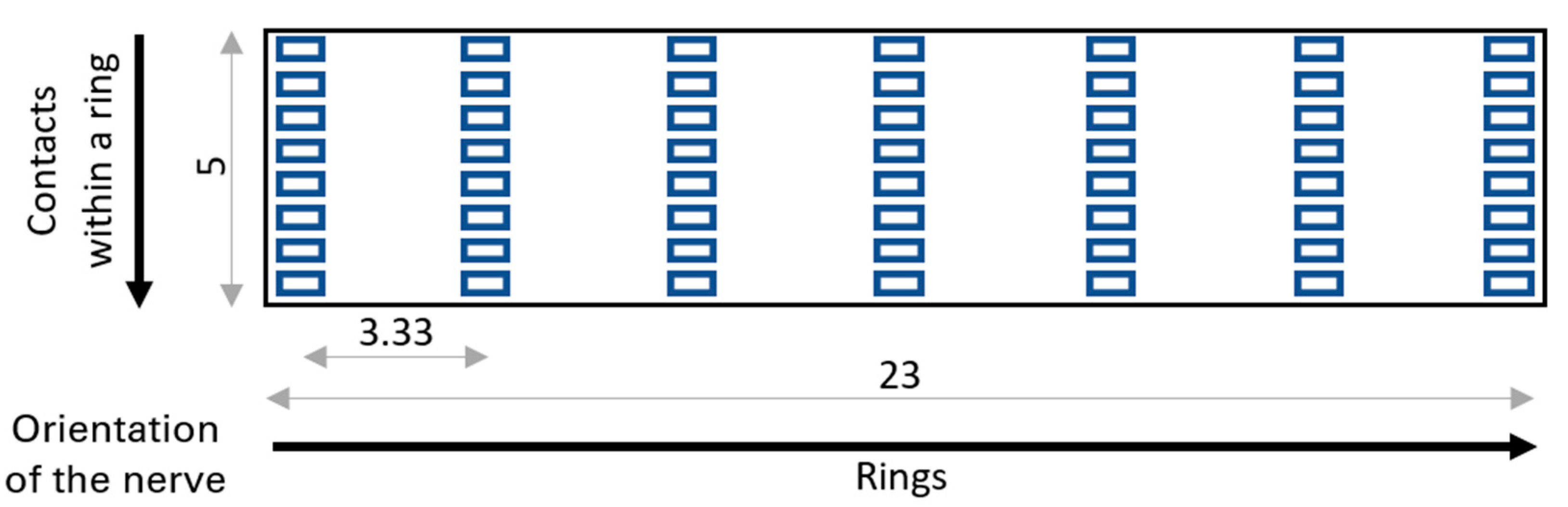
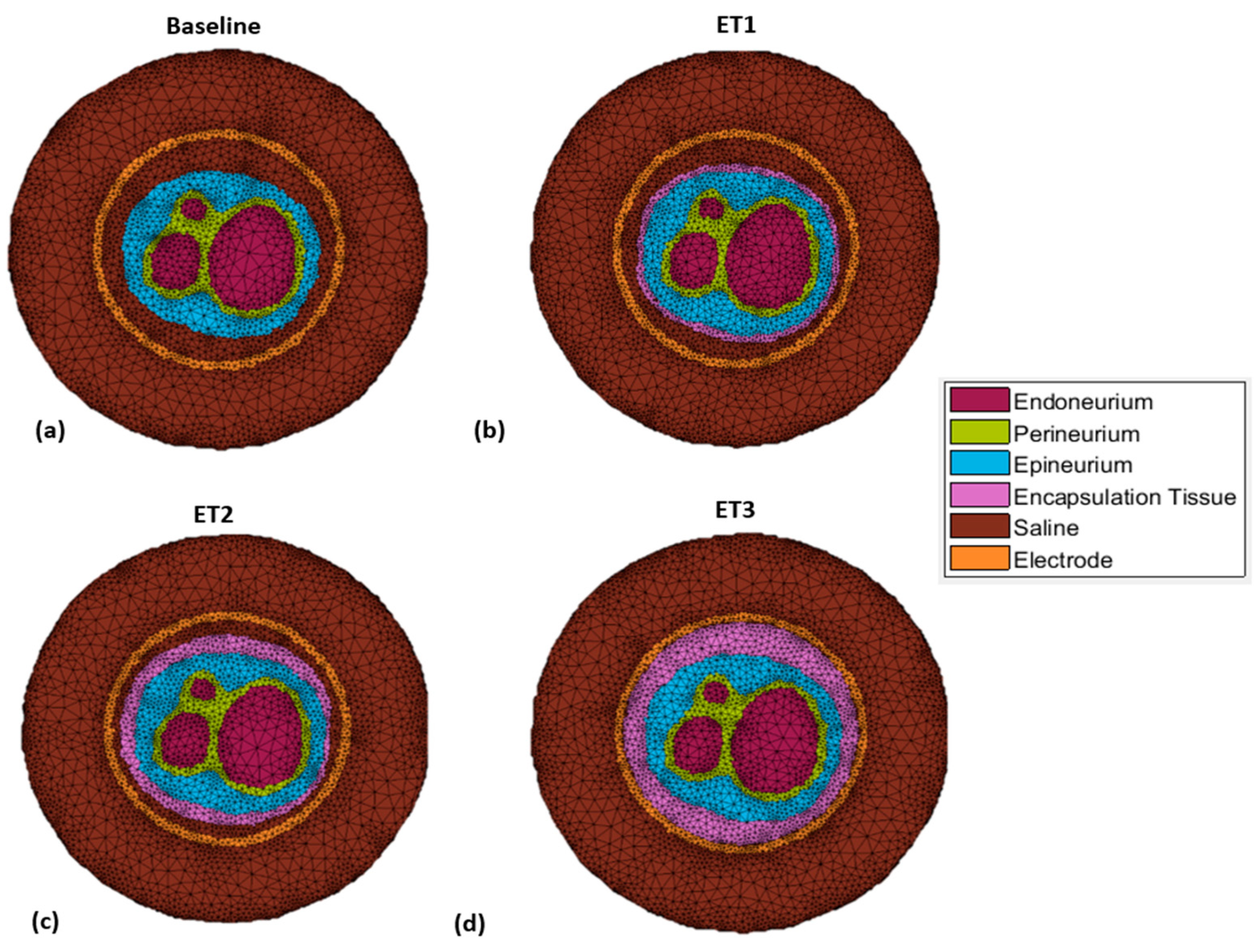
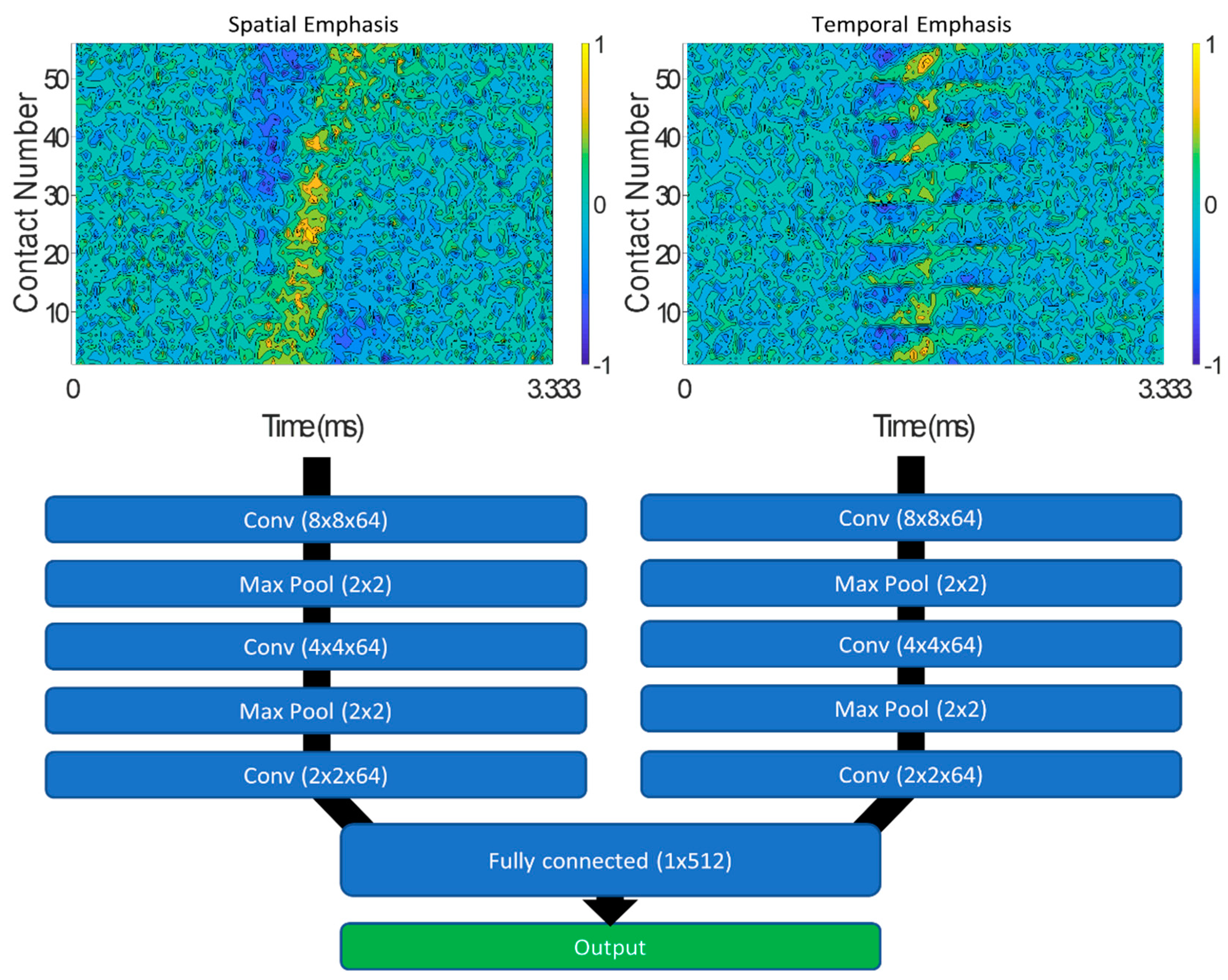

| Parameter | Value | References |
|---|---|---|
| Nerve Length | 26 mm | [46] |
| Cuff Length | 23 mm | [46] |
| Cuff Width | 60 µm | [46] |
| Cuff Radius | 800 µm | [46] |
| Endoneurium conductivity (radial) | 8.26 × 10−2 S m−1 | [38,46,52] |
| Endoneurium conductivity (longitudinal) | 5.71 × 10−1 S m−1 | [38,46,52] |
| Perineurium conductivity (all directions) | 2.10 × 10−3 S m−1 | [38,46,52] |
| Epineurium conductivity (all directions) | 8.26 × 10−2 S m−1 | [38,46,52] |
| Encapsulation tissue conductivity (all directions) | 6.59 × 10−2 S m−1 | [38,53] |
| Saline Conductivity (all directions) | 2.00 × 10−1 S m−1 | [38,46,52] |
| Cuff Conductivity (all directions) | 1.00 × 10−7 S m−1 | [38,46,52] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sammut, S.; Koh, R.G.L.; Zariffa, J. Compensation Strategies for Bioelectric Signal Changes in Chronic Selective Nerve Cuff Recordings: A Simulation Study. Sensors 2021, 21, 506. https://doi.org/10.3390/s21020506
Sammut S, Koh RGL, Zariffa J. Compensation Strategies for Bioelectric Signal Changes in Chronic Selective Nerve Cuff Recordings: A Simulation Study. Sensors. 2021; 21(2):506. https://doi.org/10.3390/s21020506
Chicago/Turabian StyleSammut, Stephen, Ryan G. L. Koh, and José Zariffa. 2021. "Compensation Strategies for Bioelectric Signal Changes in Chronic Selective Nerve Cuff Recordings: A Simulation Study" Sensors 21, no. 2: 506. https://doi.org/10.3390/s21020506
APA StyleSammut, S., Koh, R. G. L., & Zariffa, J. (2021). Compensation Strategies for Bioelectric Signal Changes in Chronic Selective Nerve Cuff Recordings: A Simulation Study. Sensors, 21(2), 506. https://doi.org/10.3390/s21020506





