Deep Neural Network Sleep Scoring Using Combined Motion and Heart Rate Variability Data
Abstract
1. Introduction
2. Methods
2.1. Dataset
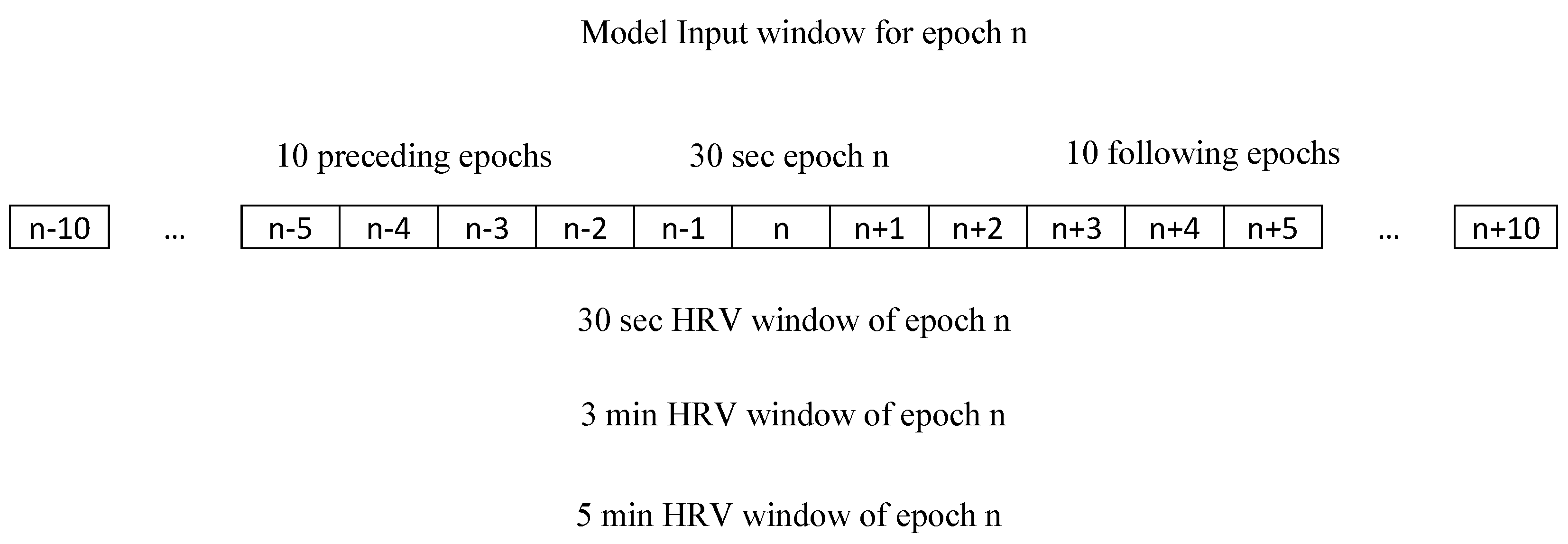
2.2. Heart Rate Variability Features
2.3. Participants
2.4. Deep Learning Algorithm Training and Selection
2.5. Statistical Analyses
3. Results
3.1. Comparison of Different HRV Period (Window) Lengths
3.2. Best Deep Learning Model’s Architecture
3.3. Overall Estimate of Sleep Parameters
3.4. Epoch-by-Epoch Comparisons
3.5. Performance in Estimating Sleep Parameters
3.6. REM and NREM Sleep Detection/Scoring
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Code Availability
Appendix A
| Actigraphy Scoring | |||
|---|---|---|---|
| 0 | 1 | ||
| PSG Scoring | 0 | TN | FP |
| 1 | FN | TP | |
References
- Wohlgemuth, W.K.; Edinger, P.J.D.; Fins, A.I.; Sullivan, R.J. How many nights are enough? The short-term stability of sleep parameters in elderly insomniacs and normal sleepers. Psychophysiology 1999, 36, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Moses, J.; Lubin, A.; Naitoh, P.; Johnson, L.C. Reliability of sleep measures. Psychophysiology 1972, 9, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Levendowski, D.; Zack, N.; Rao, S.; Wong, K.; Gendreau, M.; Kranzler, J.; Zavora, T.; Westbrook, P.R. Assessment of the test–retest reliability of laboratory polysomnography. Sleep Breath. 2009, 13, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Berney, L.R.; Blane, D.B. Collecting retrospective data: Accuracy of recall after 50 years judged against historical records. Soc. Sci. Med. 1997, 45, 1519–1525. [Google Scholar] [CrossRef]
- Wolfson, A.R.; Carskadon, M.A.; Acebo, C.; Seifer, R.; Fallone, G.; Labyak, S.E.; Martin, J.L. Evidence for the validity of a sleep habits survey for adolescents. Sleep 2003, 26, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Casaccia, S.; Braccili, E.; Scalise, L.; Revel, G.M. Experimental assessment of sleep-related parameters by passive infrared sensors: Measurement setup, feature extraction, and uncertainty analysis. Sensors 2019, 19, 3773. [Google Scholar] [CrossRef]
- Morgenthaler, T.; Alessi, C.; Friedman, L.; Owens, J.; Kapur, V.; Boehlecke, B.; Brown, T.; Chesson, A.; Coleman, J.; Lee-Chiong, T.; et al. Practice parameters for the use of actigraphy in the assessment of sleep and sleep disorders: An update for 2007. Sleep 2007, 30, 519–529. [Google Scholar] [CrossRef]
- van de Water, A.T.M.; Holmes, A.; Hurley, D.A. Objective measurements of sleep for non-laboratory settings as alternatives to polysomnography—A systematic review. J. Sleep Res. 2011, 20, 183–200. [Google Scholar] [CrossRef]
- Sadeh, A.; Sharkey, M.; Carskadon, M.A. Activity-based sleep-wake identification: An empirical test of methodological issues. Sleep 1994, 17, 201–207. [Google Scholar] [CrossRef]
- Cole, R.J.; Kripke, D.F.; Gruen, W.; Mullaney, D.J.; Gillin, J.C. Automatic Sleep/Wake Identification from Wrist Activity. Sleep 1992, 15, 461–469. [Google Scholar] [CrossRef]
- Webster, J.B.; Kripke, D.F.; Messin, S.; Mullaney, D.J.; Wyborney, G. An activity-based sleep monitor system for ambulatory use. Sleep 1982, 5, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Jean-Louis, G.; Kripke, D.F.; Mason, W.J.; Elliott, J.A.; Youngstedt, S.D. Sleep estimation from wrist movement quantified by different actigraphic modalities. J. Neurosci. Methods 2001, 105, 185–191. [Google Scholar] [CrossRef]
- Hersen, M. Clinician’s Handbook of Child Behavioral Assessment; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Stein, P.K.; Pu, Y. Heart rate variability, sleep and sleep disorders. Sleep Med. Rev. 2012, 16, 47–66. [Google Scholar] [CrossRef] [PubMed]
- ŽEmaitytė, D.; Varoneckas, G.; Sokolov, E. Heart Rhythm Control During Sleep in Ischemic Heart Disease. Psychophysiology 1984, 21, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Scholz, U.J.; Bianchi, A.M.; Cerutti, S.; Kubicki, S. Vegetative background of sleep: Spectral analysis of the heart rate variability. Physiol. Behav. 1997, 62, 1037–1043. [Google Scholar] [CrossRef]
- Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R.; Castriotta, R.J. Performance comparison of different interpretative algorithms utilized to derive sleep parameters from wrist actigraphy data. Chronobiol. Int. 2019, 36, 1752–1760. [Google Scholar] [CrossRef]
- Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R.; Castriotta, R.J. 1196 Machine learning derived-interpretative algorithm better differentiates sleep and wake epochs and estimates sleep parameters from wrist actigraphy data. Sleep 2020, 43, A457–A458. [Google Scholar] [CrossRef]
- Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R.; Castriotta, R.J. Performance assessment of new-generation Fitbit technology in deriving sleep parameters and stages. Chronobiol. Int. 2020, 37, 47–59. [Google Scholar] [CrossRef]
- Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R.; Castriotta, R.J. Accuracy of wristband fitbit models in assessing sleep: Systematic review and meta-analysis. J. Med. Internet Res. 2019, 21. [Google Scholar] [CrossRef]
- Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R. Application of deep learning to improve sleep scoring of wrist actigraphy. Sleep Med. 2020, 74, 235–241. [Google Scholar] [CrossRef]
- ANSI/CTA Standard: Definitions and Characteristics for Wearable Sleep Monitors; ANSI/CTA/NSF-2052.1; Consumer Technology Association: Arlington, Virginia, 2016.
- Chen, X.; Wang, R.; Zee, P.; Lutsey, P.L.; Javaheri, S.; Alcántara, C.; Jackson, C.L.; Williams, M.A.; Redline, S. Racial/Ethnic Differences in Sleep Disturbances: The Multi-Ethnic Study of Atherosclerosis (MESA). Sleep 2015, 38, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.; Redline, S. The national rleep research resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Tarvainen, M.P.; Niskanen, J.-P.; Lipponen, J.A.; Ranta-aho, P.O.; Karjalainen, P.A. Kubios HRV—Heart rate variability analysis software. Comput. Methods Programs Biomed. 2014, 113, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, F.; Ginsberg, J.P.; Shaffer, F. An overview of heart rate variability metrics and norms. Front. Public Heal. 2017, 5, 1–17. [Google Scholar] [CrossRef]
- Tarvainen, M.P.; Lipponen, J.; Niskanen, J.-P.; Ranta-aho, P.O. Kubios HRV User Guide, Kubios Oy. Available online: https://www.kubios.com/downloads/Kubios_HRV_Users_Guide.pdf (accessed on 2 August 2020).
- Baevsky, R.M.; Berseneva, A.P. Methodical Recommendations Use Kardivar System for Determination of the Stress Level and Estimation of the Body Adaptability Standards of Measurements and Physiological Interpretation. 2008. Available online: https://www.academia.edu/35296847/Methodical_recommendations_USE_KARDiVAR_SYSTEM_FOR_DETERMINATION_OF_THE_STRESS_LEVEL_AND_ESTIMATION_OF_THE_BODY_ADAPTABILITY_Standards_of_measurements_and_physiological_interpretation_Moscow_Prague_2008 (accessed on 2 August 2020).
- Fawaz, H.I.; Forestier, G.; Weber, J.; Idoumghar, L.; Muller, P.A. Deep learning for time series classification: A review. Data Min. Knowl. Discov. 2019, 33, 917–963. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, W.; Oates, T. Time series classification from scratch with deep neural networks: A strong baseline. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 1578–1585. [Google Scholar]
- Serrà, J.; Pascual, S.; Karatzoglou, A. Towards a universal neural network encoder for time series. In Proceedings of the CCIA 2018, 21st International Conference of the Catalan Association for Artificial Intelligence, Alt Empordà, Catalonia, Spain, 8–10 October 2018; pp. 120–129. [Google Scholar]
- Le Guennec, A.; Malinowski, S.; Tavenard, R. Data augmentation for time series classification using convolutional neural networks. In Proceedings of the ECML/PKDD Workshop on Advanced Analytics andLearning on Temporal Data, Riva Del Garda, Italy, 19–23 September 2016; Available online: https://halshs.archives-ouvertes.fr/halshs-01357973 (accessed on 2 August 2020).
- van Kuppevelt, D.; Meijer, C.; Huber, F.; van der Ploeg, A.; Georgievska, S.; van Hees, V.T. Mcfly: Automated deep learning on time series. SoftwareX 2020, 12, 100548. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 1998; pp. 255–258. [Google Scholar]
- Ordóñez, F.J.; Roggen, D. Deep convolutional and LSTM recurrent neural networks for multimodal wearable activity recognition. Sensors 2016, 16, 115. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI’ 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. 2015. Available online: www.tensorflow.org (accessed on 30 January 2020).
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 30 January 2020).
- Bland, J.M.; Altman, D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999, 8, 135–160. [Google Scholar] [CrossRef]
- Haghayegh, S.; Kang, H.-A.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R. A comprehensive guideline for bland-altman and intra class correlation calculations to properly compare two methods of measurement and interpret findings. Physiol. Meas. 2020. [Google Scholar] [CrossRef]
- Conley, S.; Knies, A.; Batten, J.; Ash, G.; Miner, B.; Hwang, Y.; Jeon, S.; Redeker, N.S. Agreement between actigraphic and polysomnographic measures of sleep in adults with and without chronic conditions: A systematic review and meta-analysis. Sleep Med. Rev. 2019, 46, 151–160. [Google Scholar] [CrossRef]
- Walch, O.; Huang, Y.; Forger, D.; Goldstein, C. Sleep stage prediction with raw acceleration and photoplethysmography heart rate data derived from a consumer wearable device. Sleep 2019, 42, 1–19. [Google Scholar] [CrossRef] [PubMed]

| Metric | Unit | Definition |
|---|---|---|
| Time Domain | ||
| Mean R-R | [ms] | Average of R-R intervals per window duration |
| SDRR | [ms] | Standard deviation of R-R intervals per window duration |
| Mean HR | [bpm] | Average heart rate per window duration |
| Min HR | [bpm] | Minimum heart rate calculated using 5 beat moving average per window duration |
| Max HR | [bpm] | Maximum heart rate calculated using 5 beat moving average per window duration |
| RMSSD | [ms] | Root mean square of differences between successive intervals per window duration |
| PNN50 | [%] | Number of successive R-R intervals differing by >50 ms divided by total number of R-R intervals per window duration |
| HRVti | - | Integral of the density of R-R interval histogram divided by height of the histogram per window duration |
| TINN | [ms] | Baseline width of R-R histogram per window duration |
| Frequency Domain | ||
| VLF Peak | [HZ] | Peak frequency of the very-low-frequency band (0.0–0.04 Hz) per window duration |
| LF Peak | [HZ] | Peak frequency of the low-frequency band (0.04–0.15 Hz) per window duration |
| HF Peak | [HZ] | Peak frequency of the high-frequency band (0.15–0.4 Hz) per window duration |
| VLF Power | [ms2] | Absolute power of the very-low-frequency band (0.0–0.04 Hz) per window duration |
| LF Power | [ms2] | Absolute power of the low-frequency band (0.04–0.15 Hz) per window duration |
| HF Power | [ms2] | Absolute power of the high-frequency band (0.15–0.4 Hz) per window duration |
| LF/HF | - | Ratio between low frequency and high frequency band power per window duration |
| Stress index | - | Square root of the Baevsky’s stress index per window duration [28] |
| Age | 67.5 ± 8.3 years |
| Gender | |
| Male | 108 (48.6%) |
| Female | 114 (51.4%) |
| Race | |
| White, Caucasian | 80 (36.0%) |
| Chinese American | 31 (13.9%) |
| Black, African-American | 64 (28.8%) |
| Hispanic | 47 (21.2%) |
| Told by doctor as having insomnia | |
| Yes | 9 (4.1%) |
| No | 213 (95.9%) |
| Trouble falling asleep past 4 weeks | |
| No, not in the past 4 weeks | 127 (57.2%) |
| Yes, less than once a week | 26 (11.7%) |
| Yes, 1 or 2 times a week | 36 (16.2%) |
| Yes, 3 or 4 times a week | 20 (9.0%) |
| Yes, 5 or more times a week | 13 (5.9%) |
| Chronotype | |
| Definitely a morning type | 78 (35.1%) |
| Rather more a morning than an evening type | 60 (27.0%) |
| Rather more an evening than a morning type | 29 (13.1%) |
| Definitely an evening type | 29 (13.1%) |
| Neither a morning nor an evening type | 26 (11.7%) |
| Sleep Metric | Metric Definition (Units) | Measurement Method | Measured Values Mean (SD) |
|---|---|---|---|
| Sleep Period Time (SPT) | Total recording time (min) | PSG | 509.7 (67.1) |
| Total Sleep Time (TST) | Total time scored as “sleep” (min) | PSG Actiwatch IA UCSD IA Deep Learning HA | 367.8 (65.3) 442.3 (62.9) 359.9 (70.2) 372.2 (81.5) |
| Sleep Efficiency (SE) | (TST/SPT) × 100 (%) | PSG Actiwatch IA UCSD IA Deep Learning HA | 72.6 (11.5) 86.9 (6.5) 70.8 (11.0) 73.3 (13.4) |
| Sleep Onset Latency (SOL) | Duration of time from lights off to first 10 min block when ≥50% of epochs scored as sleep (min) | PSG Actiwatch IA UCSD IA Deep Learning HA | 60.6 (39.9) 10.4 (19.5) 37.3 (33.5) 53.6 (42.0) |
| Wake After Sleep Onset (WASO) | Cumulative amount of time spent in wakefulness after sleep onset (min) | PSG Actiwatch IA UCSD IA Deep Learning HA | 82.5 (54.2) 58.5 (32.1) 115.5 (54.4) 86.7 (56.6) |
| Non-Rapid Eye Movement Sleep (NREM) | Time spent in Sleep Stages N1-N4 (min) | PSG Deep Learning HA | 293.7 (55.4) 328.1 (80.7) |
| Rapid Eye Movement Sleep (REM) | Time spent in Rapid Eye Movement Sleep (min) | PSG Deep Learning HA | 74.1 (24.3) 44.2 (25.3) |
| Features | Accuracy (%) | Sensitivity (%) | Specificity (%) | Cohen’s Kappa (%) |
|---|---|---|---|---|
| Activity count | 69.1 (59.7, 78.5) | 68.1 (52.1, 84.1) | 71.5 (63.9, 79.1) | 39.1 (30.0, 48.2) |
| Activity count + 30-s HRV | 73.3 (65.0, 81.6) | 81.6 (66.5, 96.7) | 52.5 (40.3, 64.7) | 37.4 (28.0, 46.8) |
| Activity count + 3-min HRV | 72.4 (66.0, 78.8) | 78.7 (66.1, 91.3) | 56.9 (44.8, 69.0) | 36.3 (28.8, 43.8) |
| Activity count + 5-min HRV | 74.9 (68.7, 81.1) | 79.5 (67.9, 91.1) | 63.7 (55.3, 72.1) | 43.8 (37.8, 49.8) |
| Interpretative Algorithm | Accuracy (%) | Sensitivity (%) | Specificity (%) | Cohen’s Kappa (%) |
|---|---|---|---|---|
| Sleep detection | ||||
| proprietary | 79.2 (77.2, 81.2) | 95.5 (94.6, 96.5) | 35.7 (32.2, 39.2) | 35.7 (31.6, 39.8) |
| UCSD | 77.8 (75.7, 79.8) | 83.3 (81.1, 85.5) | 61.8 (57.4, 66.1) | 42.7 (38.0, 47.4) |
| Deep learning HA | 84.5 (82.6, 86.3) | 89.5 (86.9, 92.2) | 70.0 (66.4, 73.5) | 58.7 (55.0, 62.4) |
| REM detection1 | ||||
| Deep learning HA | 88.1 (87.0, 89.1) | 39.6 (34.8, 44.4) | 96.6 (95.9, 97.3) | 40.9 (36.1, 45.7) |
| Non-REM detection2 | ||||
| Deep learning HA | 78.9 (77.4, 80.4) | 87.1 (84.1, 90.2) | 65.9 (62.7, 69.2) | 54.0 (50.8, 57.1) |
| Variable (Units) | Overall Bias (95% CI) [p Value] | Bland–Altman Limits of Agreement | MAE | |
|---|---|---|---|---|
| Lower Limit | Upper Limit | |||
| SOL (min) | ||||
| Actiwatch IA vs. PSG | 50.2 (41.0, 59.3) [<0.001] | −29.1 | 129.5 | 50.9 |
| UCSD IA vs. PSG | 23.3 (13.9, 32.6) [<0.001] | −58.0 | 104.5 | 31.8 |
| Deep Learning HA vs. PSG | 7.0 (−1.2, 15.2) [0.093] | −63.9 | 78.0 | 19.9 |
| Comparing bias of Deep Learning HA vs. bias of Actiwatch IA: d = 43.2, p < 0.001 Comparing bias of Deep Learning HA vs. bias of UCSD IA: d = 16.2, p < 0.001 | ||||
| WASO (min) | ||||
| Actiwatch IA vs. PSG | 24.0 (14.6, 33.4) [<0.001] | −57.2 | 105.2 | 35.4 |
| UCSD IA vs. PSG | −33.0 (−43.7, −22.4) [<0.001] | −125.2 | 59.1 | 44.8 |
| Deep Learning HA vs. PSG | −4.2 (−15.3, 6.9) [0.451] | −100.0 | 91.5 | 35.3 |
| Comparing bias of Deep Learning HA vs. bias of Actiwatch IA: d = 28.2, p < 0.001 Comparing bias of Deep Learning HA vs. bias of UCSD IA: d = 28.8, p < 0.001 | ||||
| TST (min) | ||||
| Actiwatch IA vs. PSG | −74.5 (−86.4, −62.7) [<0.001] | −177.1 | 28.1 | 76.0 |
| UCSD IA vs. PSG | 7.8 (−4.2, 19.8) [0.198] | −95.8 | 111.5 | 41.4 |
| Deep Learning HA vs. PSG | −4.4 (−17.0, 8.1) [0.483] | −113.0 | 104.1 | 40.1 |
| Comparing bias of Deep Learning HA vs. bias of Actiwatch IA: d = 70.1, p < 0.001 Comparing bias of Deep Learning HA vs. bias of UCSD IA: d = 12.3, p = 0.042 | ||||
| SE (%) | ||||
| Actiwatch IA vs. PSG | −14.3 (−16.5, −12.1) [<0.001] | −33.1 | 4.5 | 14.6 |
| UCSD IA vs. PSG | 1.8 (−0.6, 4.2) [0.134] | −18.9 | 22.5 | 8.2 |
| Deep Learning HA vs. PSG | −0.7 (−3.2, 1.8) [0.584] | −22.1 | 20.7 | 7.9 |
| Comparing bias of Deep Learning HA vs. bias of Actiwatch IA: d = 13.6, p < 0.001 Comparing bias of Deep Learning HA vs. bias of UCSD IA: d = 2.5, p = 0.032 | ||||
| REM sleep (min) | ||||
| Deep Learning HA vs. PSG | 29.9 (23.0, 36.8) [<0.001] | −29.5 | 89.3 | 33.9 |
| NREM sleep (min) | ||||
| Deep Learning HA vs. PSG | −34.3 (−47.5, −21.2) [<0.001] | −147.7 | 79.0 | 55.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R.; Castriotta, R.J. Deep Neural Network Sleep Scoring Using Combined Motion and Heart Rate Variability Data. Sensors 2021, 21, 25. https://doi.org/10.3390/s21010025
Haghayegh S, Khoshnevis S, Smolensky MH, Diller KR, Castriotta RJ. Deep Neural Network Sleep Scoring Using Combined Motion and Heart Rate Variability Data. Sensors. 2021; 21(1):25. https://doi.org/10.3390/s21010025
Chicago/Turabian StyleHaghayegh, Shahab, Sepideh Khoshnevis, Michael H. Smolensky, Kenneth R. Diller, and Richard J. Castriotta. 2021. "Deep Neural Network Sleep Scoring Using Combined Motion and Heart Rate Variability Data" Sensors 21, no. 1: 25. https://doi.org/10.3390/s21010025
APA StyleHaghayegh, S., Khoshnevis, S., Smolensky, M. H., Diller, K. R., & Castriotta, R. J. (2021). Deep Neural Network Sleep Scoring Using Combined Motion and Heart Rate Variability Data. Sensors, 21(1), 25. https://doi.org/10.3390/s21010025





