GhoMR: Multi-Receptive Lightweight Residual Modules for Hyperspectral Classification
Abstract
1. Introduction
- A novel lightweight multi-receptive feature extraction module called GhoMR is proposed for HSI classification,
- A GhoMR utilizes complex feature extraction strategy using several internal RFs, connected in a residual fashion,
- To reduce the number of trainable parameters, Ghost modules are used, which uses low-cost transformations to address feature redundancy in CNNs,
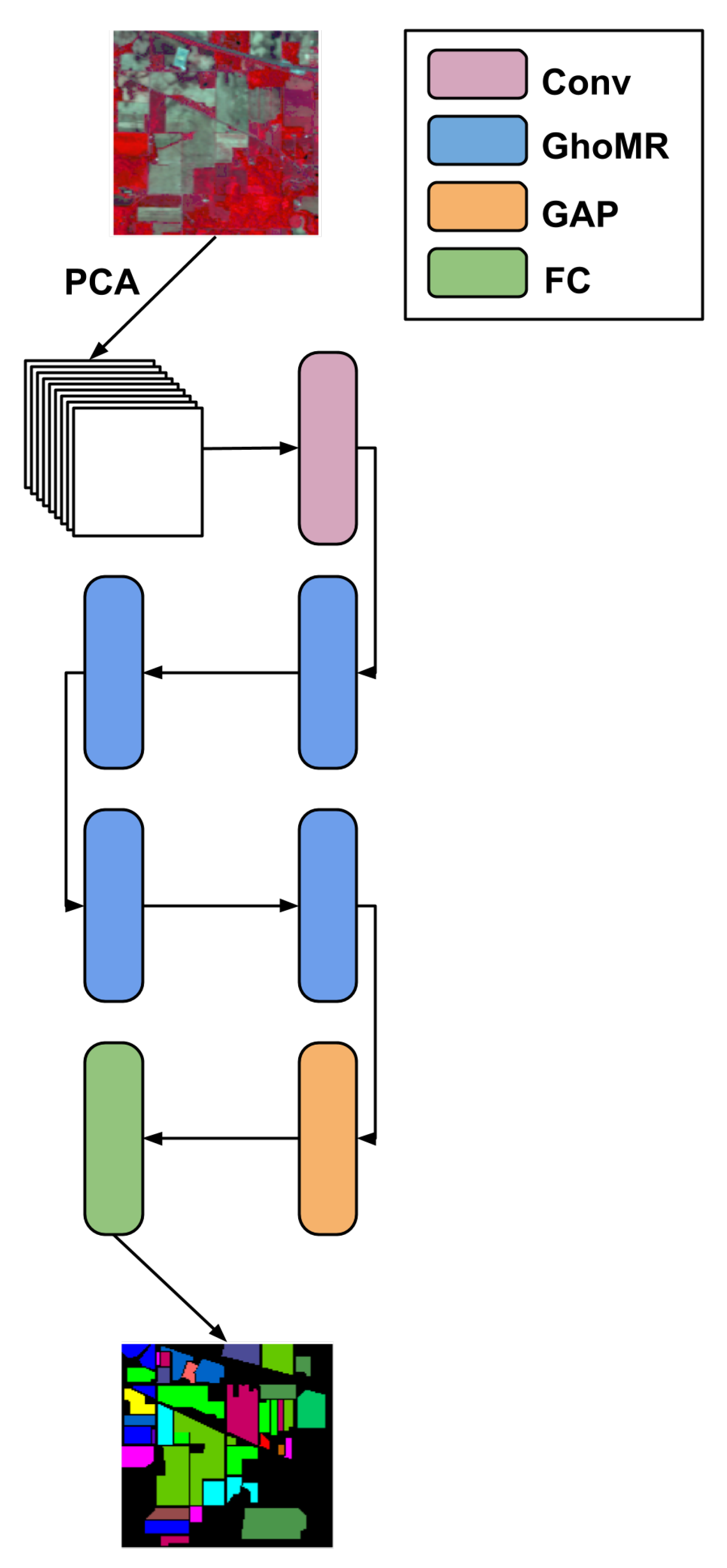
- An architecture called GhoMR-Net is designed using multiple GhoMR blocks to perform experiments on three public HSI datasets,
- Comparisons are shown, which verifies that the proposed GhoMR gives better or comparable results than state-of-the-art techniques.
2. Methodology
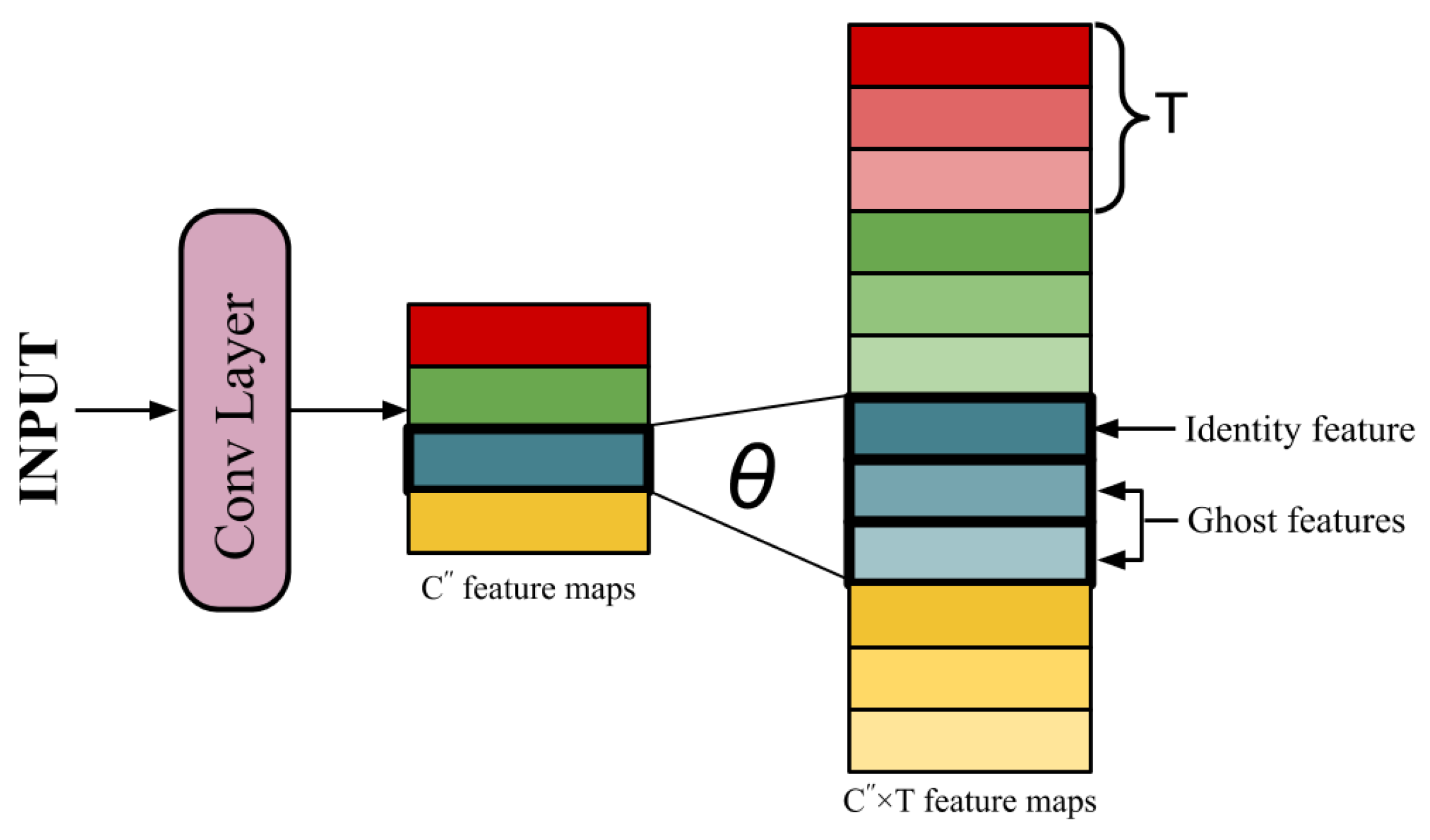
2.1. Brief Description of Ghost Modules
- The first step involves simple convolutional operations as described above. Keeping all hyper-parameters constant, kernels are used to generate a set of intrinsic feature maps , where . As a result, the total number of parameters in the network reduces to .
- The reduction of parameters leads to the loss of significant information. To make up for the remaining features, new feature maps are derived from each of the existing features by performing T low-cost operations (Ghost transformations) on them. These derived features are called Ghost features. This equation can be represented aswhere is the ith feature map in and is the jth linear operation deriving a Ghost feature from . Thus, and . Among the T Ghost transformations applied on , one operation is kept as identity operation to retain the original feature map. The remaining operations generates the ghost features. Thus, now a total of features are generated, such that .
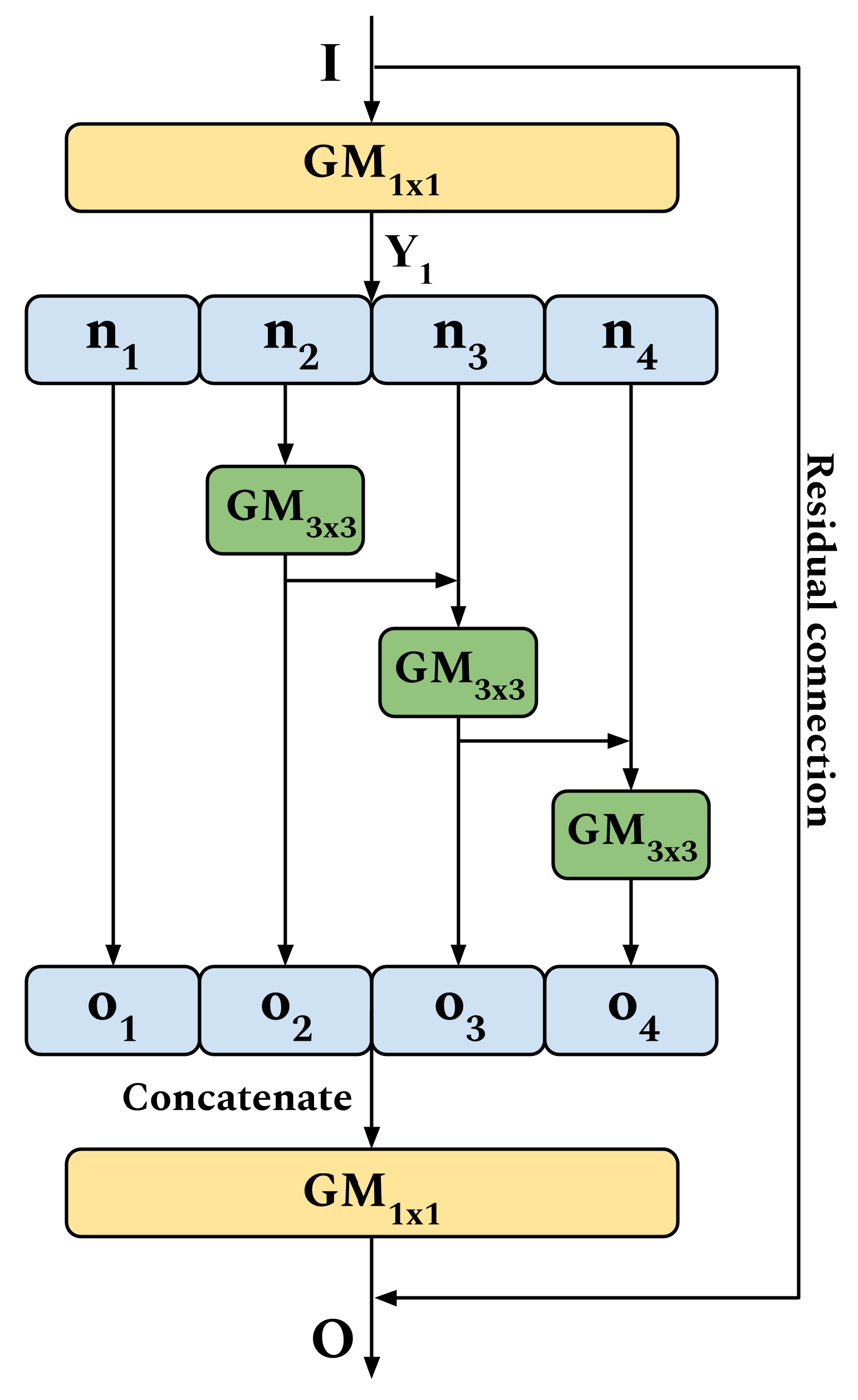
2.2. GhoMR—Proposed Multi-Receptive Module for HSI Classification
- At first, a GM using kernels is used to extract the feature block .Note, these kernels are not the Ghost filters, but are used to generate the original feature maps. For the Ghost filters, experiments with different sizes () are performed, which is discussed in Section 3.
- In the next step, the N feature maps of are split into four subsets, denoted by , where . Except , each subset is passed through a GM. The output of the previous GM, is fused hierarchically using element-wise summation with the current subset , to produce the set of features . The equations supporting this operation arewhere + refers to element-wise summation. Note, the GM for the first split is omitted in order to reuse features and reduce parameters in the module.
- Finally, the output maps , , and , are concatenated on their depth to form a singular feature block containing all the information. This is further passed through a GM and fused with input I through a residual connection to produce the final output O. This operation is expressed aswhere ⊕ refers to concatenation and + denotes element-wise summation.
3. Experiments and Discussion
3.1. Datasets
- Indian Pines (IP)—The images in this dataset were collected in 1992, over the Indian Pines test site in north-western Indiana using the AVIRIS [67] sensor. The HSI cube has a spatial dimension of pixels with 224 spectral bands in the wavelength range of 400 to 2500 nm, among which 24 bands corresponding to regions of water absorption were eliminated. Among the pixels, are annotated with ground truth from a set of 16 different vegetation classes.
- University of Pavia (UP)—This dataset was acquired in 2001, over the university campus at Pavia, Northern Italy, using the ROSIS sensor. It has a spatial dimension of pixels and 103 spectral bands in wavelength between 430 to 860 nm. The ground truth is a set of 9 urban land-cover classes, and approx. of the total pixels are annotated with this information.
- Salinas Scene (SA)—This dataset was collected over Salinas Valley, California, in 1998 using the AVRIS sensor. The spatial dimension is pixels and the spectral information is encoded in 224 bands with a wavelength in the range of 360 to 2500 nm. Similar to IP, 20 spectral bands due to water absorption are discarded. The ground truth contains 16 different classes from vegetables, bare soils, and vineyard fields.
3.2. Experimental Protocols
- First experiment calculates the class-wise accuracies, OA, AA, and Kappa for IP, UP, and SA datasets using and training data. The 3D spectral-spatial inputs have spatial dimensions for all three datasets. The value of T and are kept 2 and 3 respectively.
- In the second experiment, OA, AA, and Kappa are measured on the three datasets for different values of T and , such that and . A comparative study between all the six combinations of T and is performed. This experiment is conducted on 10% training data with 3D input cubes of spatial dimension .
- In the third experiment, the proposed architecture is compared with the following state-of-the-art techniques—SVM [24], 2D-CNN [51], 3D-CNN [52], M3D-CNN [56], Two-CNN [55], SSRN [58], HybridSN [59], SENet [63] (with global average pooling and max pooling) and FuSENet [63]. Comparisons are shown for both and training data, keeping input spatial dimension of .
- The fourth experiment measures the OA, AA, and Kappa on lesser training data ( and ) and smaller spatial dimensions ( and ) of input patches. The parameters T and are kept 2 and 3 respectively.
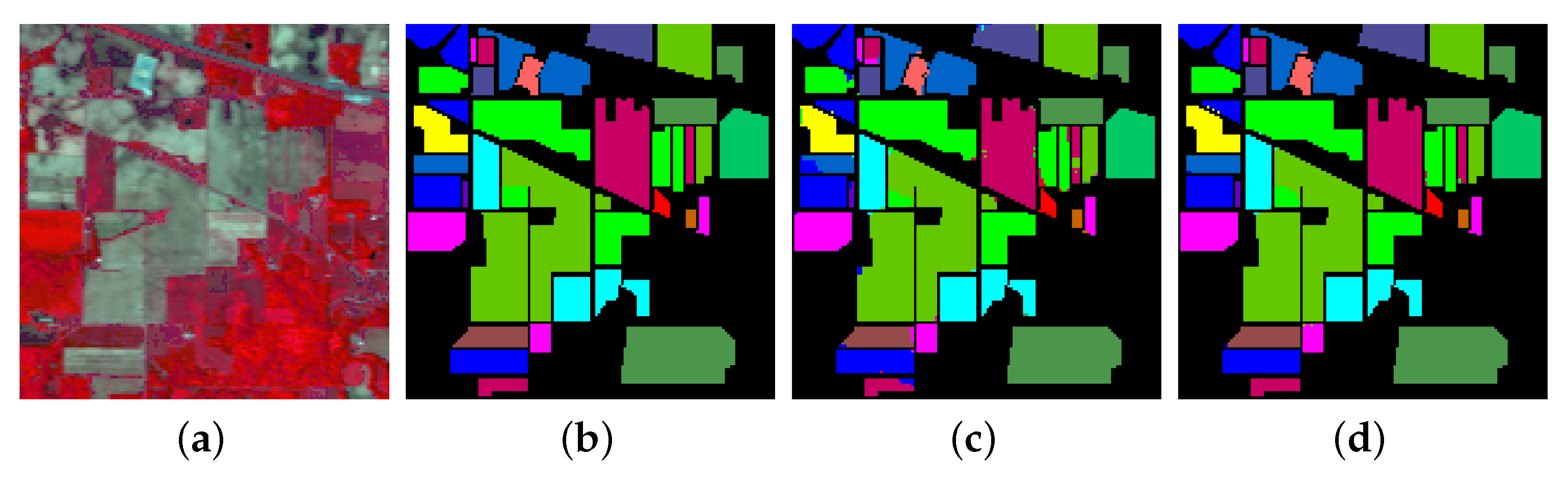
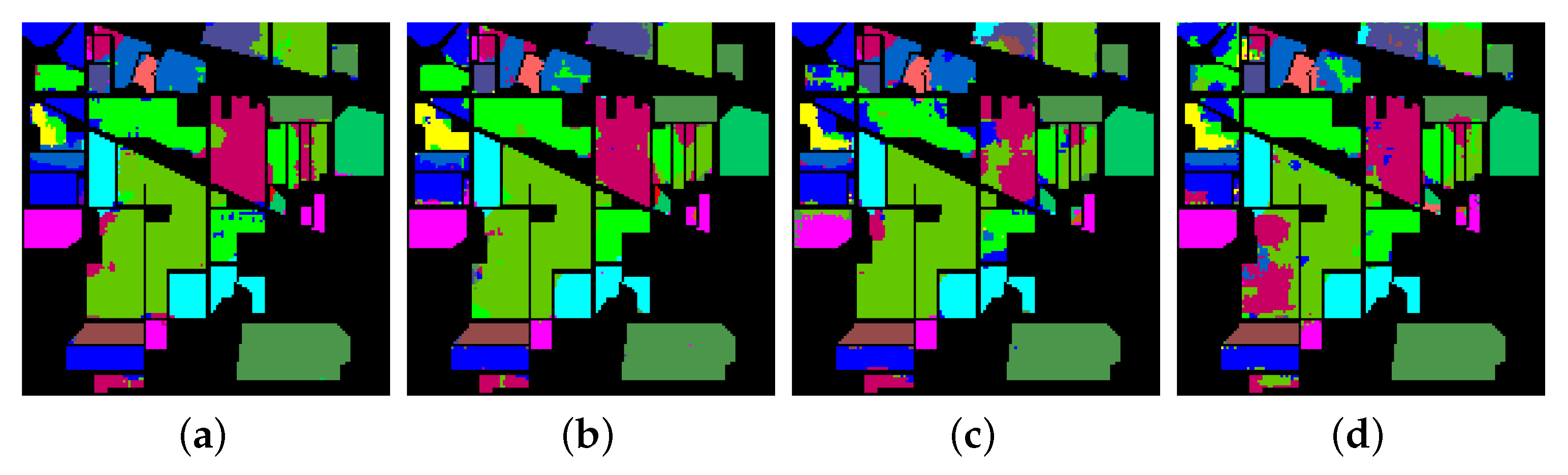
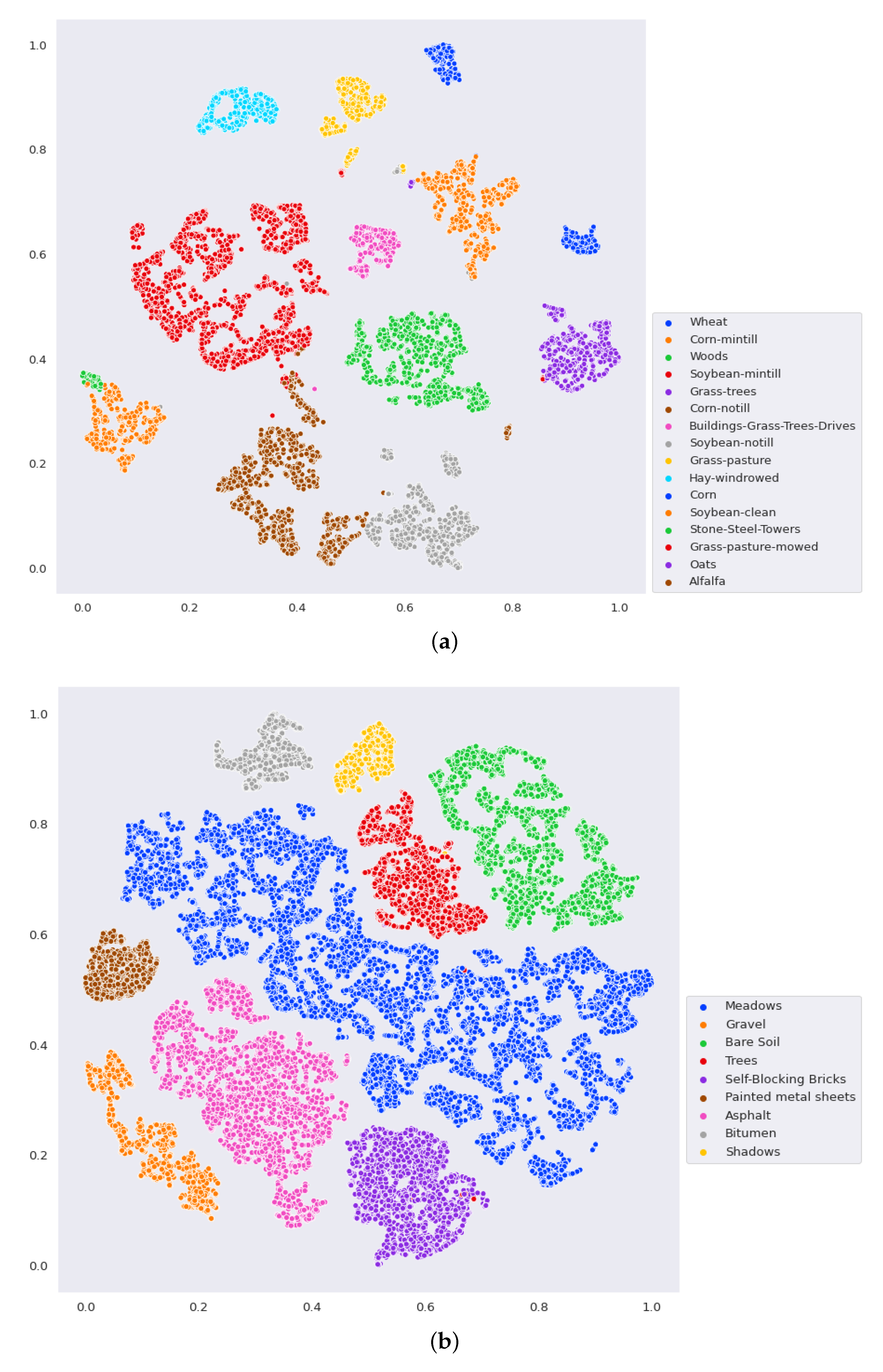
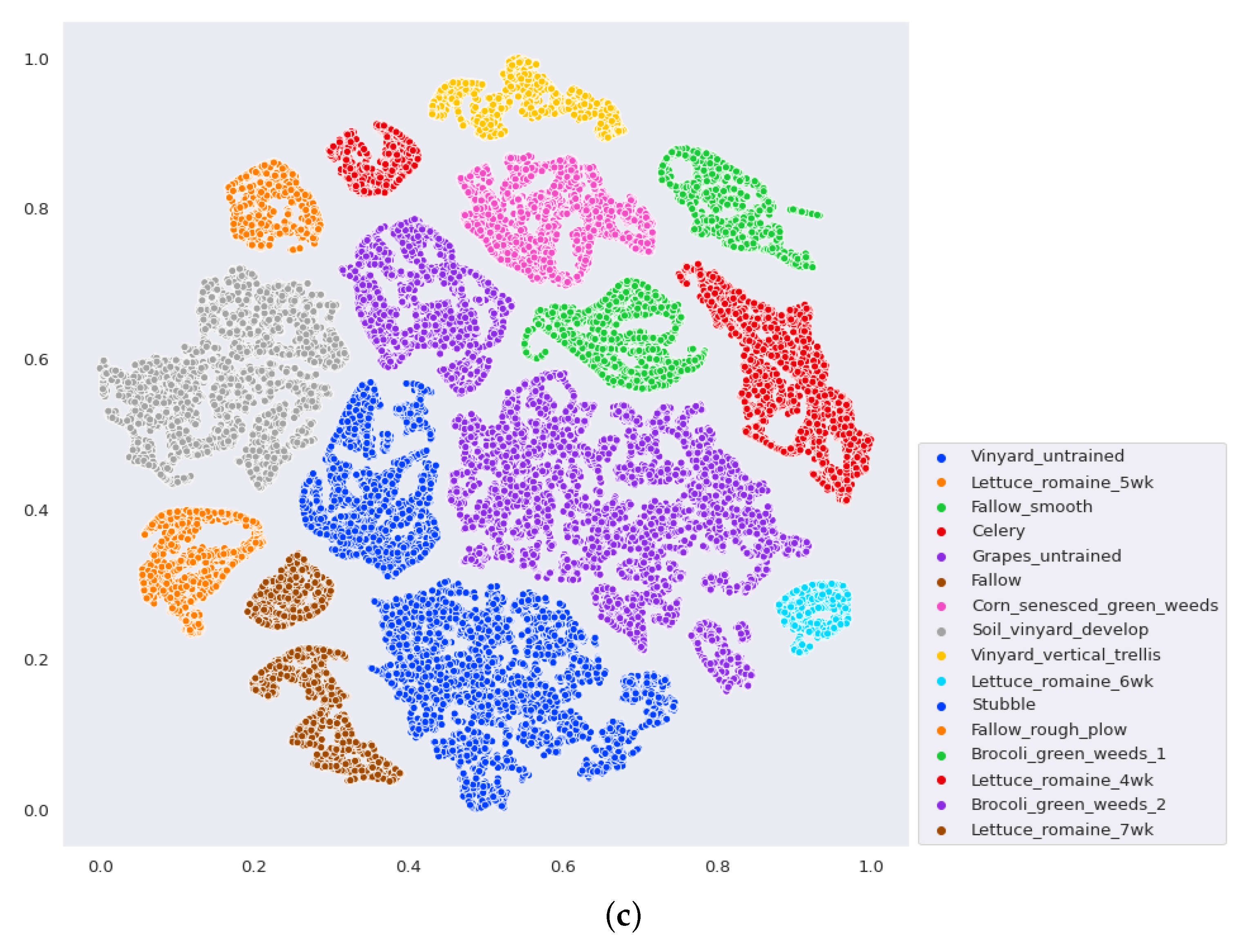
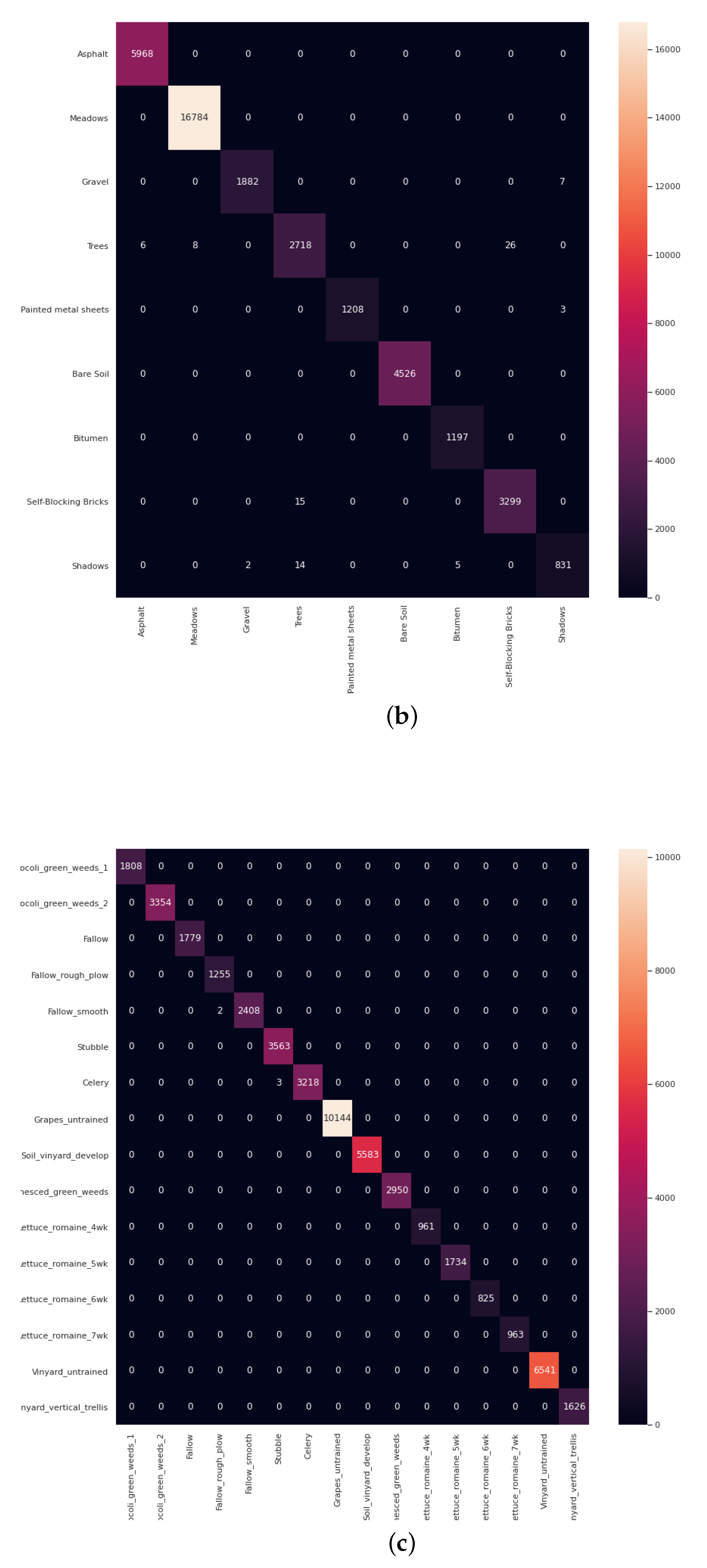
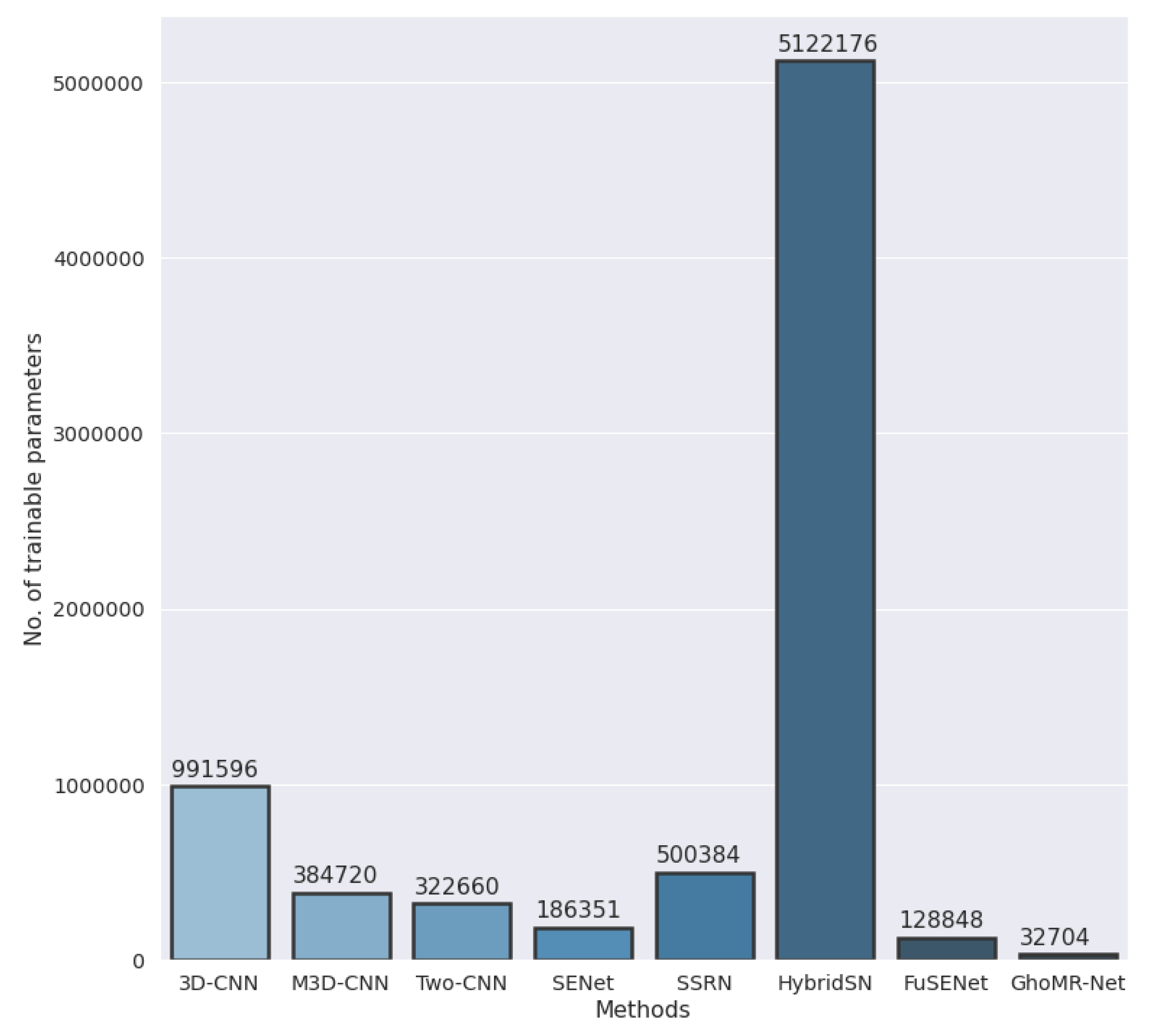
- The final experiment demonstrates the effectiveness of GhoMR-Net using t-SNE visualization [71] and confusion matrices. Moreover, the number of trainable parameters in the network is compared with other state-of-the-art architectures.
3.3. Classification Results and Visualizations
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Park, B.; Lu, R. Hyperspectral Imaging Technology in Food and Agriculture; Springer: Berlin, Germany, 2015. [Google Scholar]
- Goodenough, D.G.; Chen, H.; Gordon, P.; Niemann, K.O.; Quinn, G. Forest applications with hyperspectral imaging. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Munich, Germany, 22–27 July 2012; pp. 7309–7312. [Google Scholar]
- Tusa, E.; Laybros, A.; Monnet, J.M.; Dalla Mura, M.; Barré, J.B.; Vincent, G.; Dalponte, M.; Feret, J.B.; Chanussot, J. Fusion of hyperspectral imaging and LiDAR for forest monitoring. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 32, pp. 281–303. [Google Scholar]
- Liang, H. Advances in multispectral and hyperspectral imaging for archaeology and art conservation. Appl. Phys. A 2012, 106, 309–323. [Google Scholar] [CrossRef]
- Calin, M.A.; Parasca, S.V.; Savastru, D.; Manea, D. Hyperspectral imaging in the medical field: Present and future. Appl. Spectrosc. Rev. 2014, 49, 435–447. [Google Scholar] [CrossRef]
- Huang, H.; Liu, L.; Ngadi, M.O. Recent developments in hyperspectral imaging for assessment of food quality and safety. Sensors 2014, 14, 7248–7276. [Google Scholar] [CrossRef] [PubMed]
- Ardouin, J.P.; Lévesque, J.; Rea, T.A. A demonstration of hyperspectral image exploitation for military applications. In Proceedings of the 10th International Conference on Information Fusion, Quebec, QC, Canada, 9–12 July 2007; pp. 1–8. [Google Scholar]
- Edelman, G.; Gaston, E.; Van Leeuwen, T.; Cullen, P.; Aalders, M. Hyperspectral imaging for non-contact analysis of forensic traces. Forensic Sci. Int. 2012, 223, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Villa, A.; Benediktsson, J.A.; Chanussot, J.; Jutten, C. Hyperspectral image classification with independent component discriminant analysis. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4865–4876. [Google Scholar] [CrossRef]
- Licciardi, G.; Marpu, P.R.; Chanussot, J.; Benediktsson, J.A. Linear versus nonlinear PCA for the classification of hyperspectral data based on the extended morphological profiles. IEEE Geosci. Remote Sens. Lett. 2011, 9, 447–451. [Google Scholar] [CrossRef]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Trans. Geosci. Remote Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Xu, J.; Zhu, X.X. Joint and Progressive Subspace Analysis (JPSA) with Spatial-Spectral Manifold Alignment for Semi-Supervised Hyperspectral Dimensionality Reduction. arXiv 2020, arXiv:2009.10003. [Google Scholar]
- Liu, H.; Xia, K.; Li, T.; Ma, J.; Owoola, E. Dimensionality Reduction of Hyperspectral Images Based on Improved Spatial–Spectral Weight Manifold Embedding. Sensors 2020, 20, 4413. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Xu, J.; Zhu, X.X. Learning to propagate labels on graphs: An iterative multitask regression framework for semi-supervised hyperspectral dimensionality reduction. ISPRS J. Photogramm. Remote Sens. 2019, 158, 35–49. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Q.; Li, X. A Fast Neighborhood Grouping Method for Hyperspectral Band Selection. IEEE Trans. Geosci. Remote Sens. 2020. [Google Scholar] [CrossRef]
- Lorenzo, P.R.; Tulczyjew, L.; Marcinkiewicz, M.; Nalepa, J. Hyperspectral band selection using attention-based convolutional neural networks. IEEE Access 2020, 8, 42384–42403. [Google Scholar] [CrossRef]
- Sun, W.; Peng, J.; Yang, G.; Du, Q. Fast and latent low-rank subspace clustering for hyperspectral band selection. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3906–3915. [Google Scholar] [CrossRef]
- Han, Z.; Hong, D.; Gao, L.; Zhang, B.; Chanussot, J. Deep Half-Siamese Networks for Hyperspectral Unmixing. IEEE Geosci. Remote Sens. Lett. 2020. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. An augmented linear mixing model to address spectral variability for hyperspectral unmixing. IEEE Trans. Image Process. 2018, 28, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- Khajehrayeni, F.; Ghassemian, H. Hyperspectral unmixing using deep convolutional autoencoders in a supervised scenario. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 567–576. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Su, H.; Du, Q. Local binary patterns and extreme learning machine for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3681–3693. [Google Scholar] [CrossRef]
- Kang, X.; Li, C.; Li, S.; Lin, H. Classification of hyperspectral images by Gabor filtering based deep network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 11, 1166–1178. [Google Scholar] [CrossRef]
- Fang, L.; He, N.; Li, S.; Plaza, A.J.; Plaza, J. A new spatial–spectral feature extraction method for hyperspectral images using local covariance matrix representation. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3534–3546. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Benediktsson, J.A.; Palmason, J.A.; Sveinsson, J.R. Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 2005, 43, 480–491. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Muñoz-Marí, J.; Vila-Francés, J.; Calpe-Maravilla, J. Composite kernels for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2006, 3, 93–97. [Google Scholar] [CrossRef]
- Li, J.; Marpu, P.R.; Plaza, A.; Bioucas-Dias, J.M.; Benediktsson, J.A. Generalized composite kernel framework for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2013, 51, 4816–4829. [Google Scholar] [CrossRef]
- Tang, Y.Y.; Lu, Y.; Yuan, H. Hyperspectral image classification based on three-dimensional scattering wavelet transform. IEEE Trans. Geosci. Remote Sens. 2014, 53, 2467–2480. [Google Scholar] [CrossRef]
- Jia, S.; Shen, L.; Li, Q. Gabor feature-based collaborative representation for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2014, 53, 1118–1129. [Google Scholar]
- Chen, Y.; Nasrabadi, N.M.; Tran, T.D. Hyperspectral image classification using dictionary-based sparse representation. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3973–3985. [Google Scholar] [CrossRef]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spectral–spatial hyperspectral image classification via multiscale adaptive sparse representation. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7738–7749. [Google Scholar] [CrossRef]
- Fang, L.; Wang, C.; Li, S.; Benediktsson, J.A. Hyperspectral image classification via multiple-feature-based adaptive sparse representation. IEEE Trans. Instrum. Meas. 2017, 66, 1646–1657. [Google Scholar] [CrossRef]
- Rasti, B.; Hong, D.; Hang, R.; Ghamisi, P.; Kang, X.; Chanussot, J.; Benediktsson, J.A. Feature extraction for hyperspectral imagery: The evolution from shallow to deep. arXiv 2020, arXiv:2003.02822. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Stateline, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, San Juan, Puerto Rico, 17–19 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 21–37. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Basha, S.S.; Ghosh, S.; Babu, K.K.; Dubey, S.R.; Pulabaigari, V.; Mukherjee, S. Rccnet: An efficient convolutional neural network for histological routine colon cancer nuclei classification. In Proceedings of the 15th International Conference on Control, Automation, Robotics and Vision, Singapore, 18–21 November 2018; pp. 1222–1227. [Google Scholar]
- Makantasis, K.; Karantzalos, K.; Doulamis, A.; Doulamis, N. Deep supervised learning for hyperspectral data classification through convolutional neural networks. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Milan, Italy, 26–31 July 2015; pp. 4959–4962. [Google Scholar]
- Hamida, A.B.; Benoit, A.; Lambert, P.; Amar, C.B. 3-D deep learning approach for remote sensing image classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4420–4434. [Google Scholar] [CrossRef]
- Zhu, J.; Fang, L.; Ghamisi, P. Deformable convolutional neural networks for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2018, 15, 1254–1258. [Google Scholar] [CrossRef]
- Hao, S.; Wang, W.; Ye, Y.; Li, E.; Bruzzone, L. A deep network architecture for super-resolution-aided hyperspectral image classification with classwise loss. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4650–4663. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.Q.; Chan, J.C.W. Learning and transferring deep joint spectral–spatial features for hyperspectral classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4729–4742. [Google Scholar] [CrossRef]
- He, M.; Li, B.; Chen, H. Multi-scale 3D deep convolutional neural network for hyperspectral image classification. In Proceedings of the IEEE International Conference on Image Processing, Beijing, China, 17–20 September 2017; pp. 3904–3908. [Google Scholar]
- Zhang, H.; Li, Y.; Jiang, Y.; Wang, P.; Shen, Q.; Shen, C. Hyperspectral classification based on lightweight 3-D-CNN with transfer learning. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5813–5828. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, J.; Luo, Z.; Chapman, M. Spectral–spatial residual network for hyperspectral image classification: A 3-D deep learning framework. IEEE Trans. Geosci. Remote Sens. 2017, 56, 847–858. [Google Scholar] [CrossRef]
- Roy, S.K.; Krishna, G.; Dubey, S.R.; Chaudhuri, B.B. HybridSN: Exploring 3-D–2-D CNN feature hierarchy for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2019, 17, 277–281. [Google Scholar] [CrossRef]
- Kang, X.; Zhuo, B.; Duan, P. Dual-path network-based hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2018, 16, 447–451. [Google Scholar] [CrossRef]
- Yu, Y.; Gong, Z.; Wang, C.; Zhong, P. An unsupervised convolutional feature fusion network for deep representation of remote sensing images. IEEE Geosci. Remote Sens. Lett. 2017, 15, 23–27. [Google Scholar] [CrossRef]
- Song, W.; Li, S.; Fang, L.; Lu, T. Hyperspectral image classification with deep feature fusion network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3173–3184. [Google Scholar] [CrossRef]
- Roy, S.K.; Dubey, S.R.; Chatterjee, S.; Chaudhuri, B.B. FuSENet: Fused squeeze-and-excitation network for spectral-spatial hyperspectral image classification. IET Image Process. 2020, 14, 1653–1661. [Google Scholar] [CrossRef]
- Gao, S.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P.H. Res2net: A new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2019. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Tian, Q.; Guo, J.; Xu, C.; Xu, C. GhostNet: More features from cheap operations. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 1580–1589. [Google Scholar]
- Wei, B.; Shen, X.; Yuan, Y. Remote Sensing Scene Classification Based on Improved GhostNet. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2020; Volume 1621, p. 012091. [Google Scholar]
- Green, R.O.; Eastwood, M.L.; Sarture, C.M.; Chrien, T.G.; Aronsson, M.; Chippendale, B.J.; Faust, J.A.; Pavri, B.E.; Chovit, C.J.; Solis, M.; et al. Imaging spectroscopy and the airborne visible/infrared imaging spectrometer (AVIRIS). Remote Sens. Environ. 1998, 65, 227–248. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the Machine Learning Research, Lille, France, 7–9 July 2015; Volume 37, pp. 448–456. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]



| IP | UP | SA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Training | Test | Accuracy | Name | Training | Test | Accuracy | Name | Training | Test | Accuracy |
| Alfalfa | 9 | 37 | Asphalt | 1326 | 5305 | Brocoli_green_weeds_1 | 402 | 1607 | |||
| Corn-notill | 285 | 1143 | Meadows | 3730 | 14,919 | Brocoli_green_weeds_2 | 745 | 2981 | |||
| Corn-mintill | 166 | 664 | Gravel | 420 | 1679 | Fallow | 395 | 1581 | |||
| Corn | 47 | 190 | Trees | 613 | 2451 | Fallow_rough_plow | 279 | 1115 | |||
| Grass-pasture | 97 | 386 | Painted metal sheets | 269 | 1076 | Fallow_smooth | 536 | 2142 | |||
| Grass-trees | 146 | 584 | Bare Soil | 1006 | 4023 | Stubble | 792 | 3167 | |||
| Grass-pasture-mowed | 6 | 22 | Bitumen | 266 | 1064 | Celery | 716 | 2863 | |||
| Hay-windrowed | 96 | 382 | Self-Blocking Bricks | 736 | 2946 | Grapes_untrained | 2254 | 9017 | |||
| Oats | 4 | 16 | Shadows | 189 | 758 | Soil_vinyard_develop | 1240 | 4963 | |||
| Soybean-notill | 194 | 778 | Corn_senesced_green_weeds | 656 | 2622 | ||||||
| Soybean-mintill | 491 | 1964 | Lettuce_romaine_4wk | 214 | 854 | ||||||
| Soybean-clean | 118 | 475 | Lettuce_romaine_5wk | 385 | 1542 | ||||||
| Wheat | 41 | 164 | Lettuce_romaine_6wk | 183 | 733 | ||||||
| Woods | 253 | 1012 | Lettuce_romaine_7wk | 214 | 856 | ||||||
| Buildings-Grass-Trees-Drives | 77 | 309 | Vinyard_untrained | 1453 | 5815 | ||||||
| Stone-Steel-Towers | 19 | 74 | Vinyard_vertical_trellis | 361 | 1446 | ||||||
| OA | 2049 | 8200 | OA | 8555 | 34,221 | OA | 10,825 | 43,304 | |||
| Kappa | Kappa | Kappa | |||||||||
| AA | AA | AA | |||||||||
| Training time | 3 min 34 s | Training time | 13 min 50 s | Training time | 17 min 52 s | ||||||
| IP | UP | SA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Training | Test | Accuracy | Name | Training | Test | Accuracy | Name | Training | Test | Accuracy |
| Alfalfa | 5 | 41 | Asphalt | 663 | 5968 | Brocoli_green_weeds_1 | 201 | 1808 | |||
| Corn-notill | 143 | 1285 | Meadows | 1865 | 16,784 | Brocoli_green_weeds_2 | 372 | 3354 | |||
| Corn-mintill | 83 | 747 | Gravel | 210 | 1889 | Fallow | 197 | 1779 | |||
| Corn | 24 | 213 | Trees | 306 | 2758 | Fallow_rough_plow | 139 | 1255 | |||
| Grass-pasture | 48 | 435 | Painted metal sheets | 134 | 1211 | Fallow_smooth | 268 | 2410 | |||
| Grass-trees | 73 | 657 | Bare Soil | 503 | 4526 | Stubble | 396 | 3563 | |||
| Grass-pasture-mowed | 3 | 25 | Bitumen | 133 | 1197 | Celery | 358 | 3221 | |||
| Hay-windrowed | 48 | 430 | Self-Blocking Bricks | 368 | 3314 | Grapes_untrained | 1127 | 10,144 | |||
| Oats | 2 | 18 | Shadows | 95 | 852 | Soil_vinyard_develop | 620 | 5583 | |||
| Soybean-notill | 97 | 875 | Corn_senesced_green_weeds | 328 | 2950 | ||||||
| Soybean-mintill | 245 | 2210 | Lettuce_romaine_4wk | 107 | 961 | ||||||
| Soybean-clean | 59 | 534 | Lettuce_romaine_5wk | 193 | 1734 | ||||||
| Wheat | 20 | 185 | Lettuce_romaine_6wk | 91 | 825 | ||||||
| Woods | 126 | 1139 | Lettuce_romaine_7wk | 107 | 963 | ||||||
| Buildings-Grass-Trees-Drives | 39 | 347 | Vinyard_untrained | 727 | 6541 | ||||||
| Stone-Steel-Towers | 9 | 84 | Vinyard_vertical_trellis | 181 | 1626 | ||||||
| OA | 1024 | 9225 | OA | 4277 | 38,499 | OA | 5412 | 48,717 | |||
| Kappa | Kappa | Kappa | |||||||||
| AA | AA | AA | |||||||||
| Training time | 2 min 58 s | Training time | 11 min 20 s | Training time | 14 min 20 s | ||||||
| T | IP | UP | SA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OA | Kappa | AA | OA | Kappa | AA | OA | Kappa | AA | ||
| 3 | ||||||||||
| 2 | 5 | |||||||||
| 7 | ||||||||||
| 3 | ||||||||||
| 4 | 5 | |||||||||
| 7 | ||||||||||
| Training | Methods | IP | UP | SA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OA | Kappa | AA | OA | Kappa | AA | OA | Kappa | AA | ||
| 10% | SVM | |||||||||
| 2D-CNN | ||||||||||
| 3D-CNN | ||||||||||
| M3D-CNN | ||||||||||
| Two-CNN | ||||||||||
| SENet (GMP) | ||||||||||
| SENet (GAP) | ||||||||||
| FuSENet | ||||||||||
| SSRN | ||||||||||
| HybridSN | ||||||||||
| GhoMR-Net | ||||||||||
| 20% | SVM | |||||||||
| 2D-CNN | ||||||||||
| 3D-CNN | ||||||||||
| M3D-CNN | ||||||||||
| Two-CNN | ||||||||||
| SENet (GMP) | ||||||||||
| SENet (GAP) | ||||||||||
| FuSENet | ||||||||||
| SSRN | ||||||||||
| HybridSN | ||||||||||
| GhoMR-Net | ||||||||||
| Training Samples | Spatial Size | IP | UP | SA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OA | Kappa | AA | OA | Kappa | AA | OA | Kappa | AA | ||
| 5% | 13 × 13 | |||||||||
| 11 × 11 | ||||||||||
| 3% | 13 × 13 | |||||||||
| 11 × 11 | ||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, A.; Saha, I.; Scherer, R. GhoMR: Multi-Receptive Lightweight Residual Modules for Hyperspectral Classification. Sensors 2020, 20, 6823. https://doi.org/10.3390/s20236823
Das A, Saha I, Scherer R. GhoMR: Multi-Receptive Lightweight Residual Modules for Hyperspectral Classification. Sensors. 2020; 20(23):6823. https://doi.org/10.3390/s20236823
Chicago/Turabian StyleDas, Arijit, Indrajit Saha, and Rafał Scherer. 2020. "GhoMR: Multi-Receptive Lightweight Residual Modules for Hyperspectral Classification" Sensors 20, no. 23: 6823. https://doi.org/10.3390/s20236823
APA StyleDas, A., Saha, I., & Scherer, R. (2020). GhoMR: Multi-Receptive Lightweight Residual Modules for Hyperspectral Classification. Sensors, 20(23), 6823. https://doi.org/10.3390/s20236823






