Lysenin Channels as Sensors for Ions and Molecules
Abstract
1. Introduction
2. Lysenin Channels as Multivalent Ion Sensors
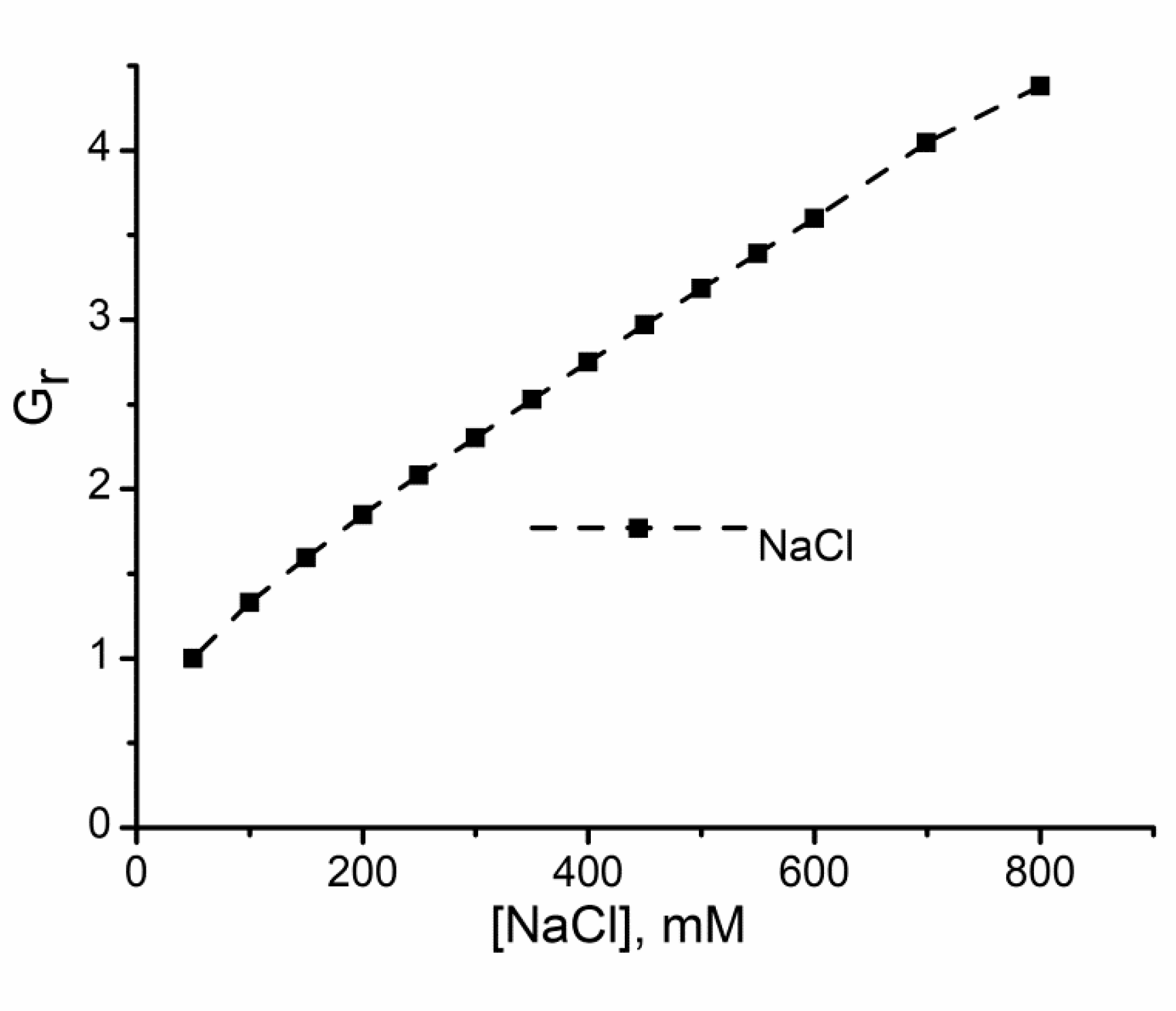
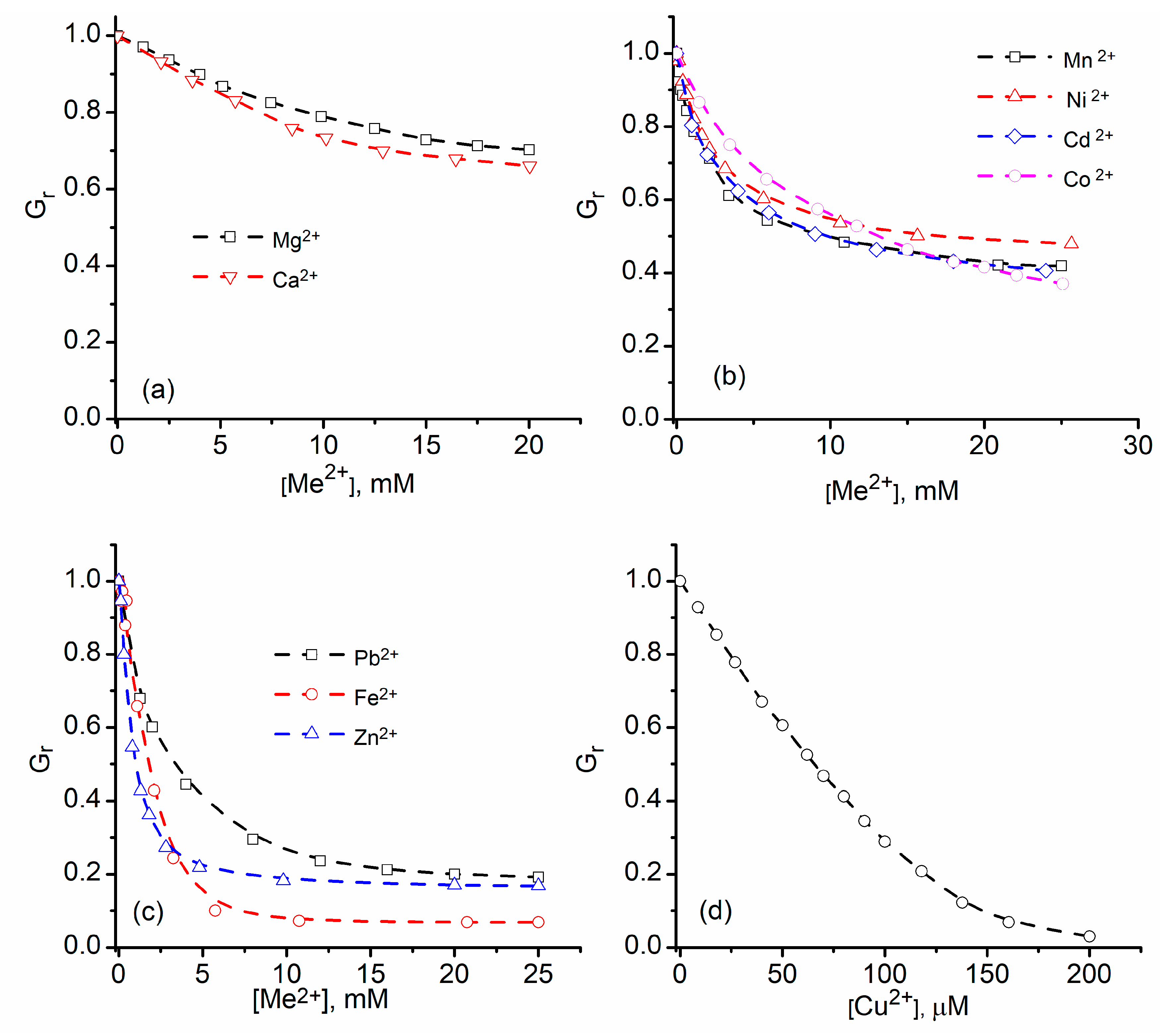
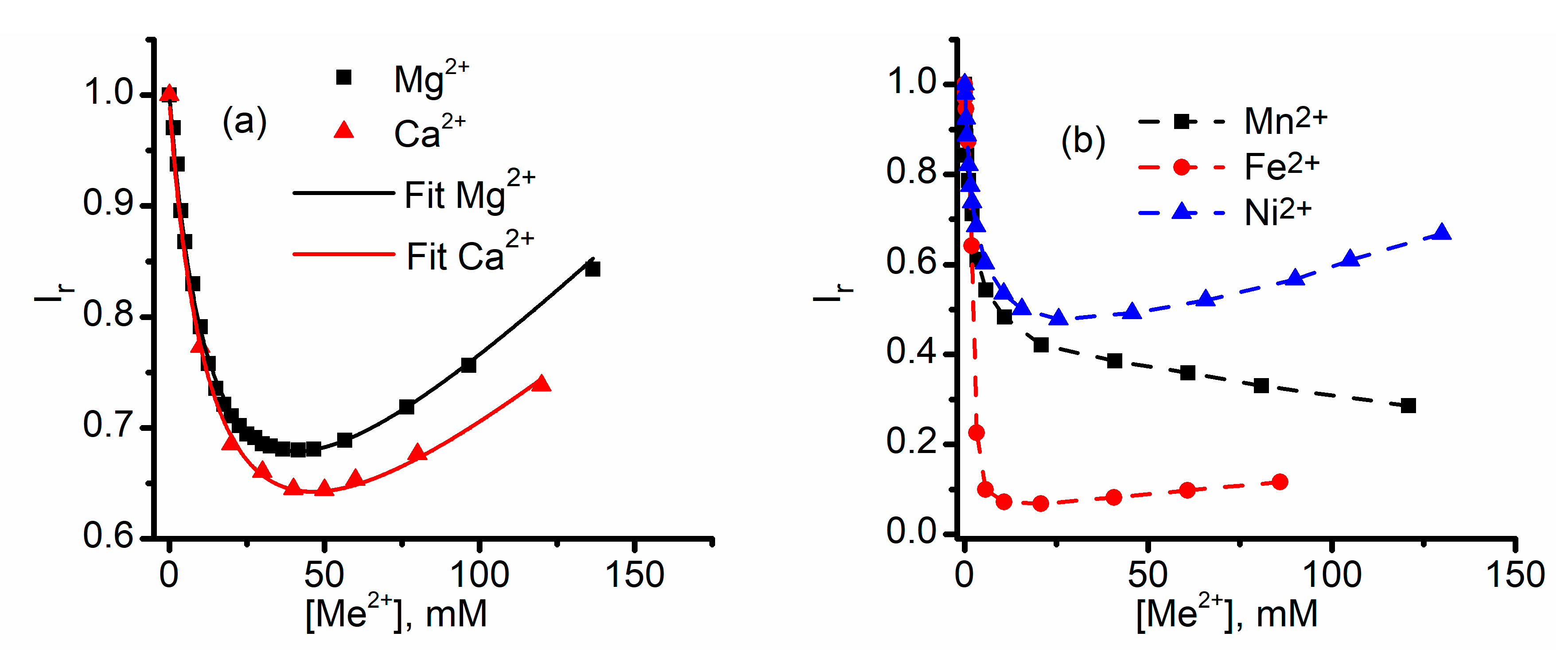
2.1. Divalent Metal Cations Modulate the Macroscopic Conductance of Lysenin Channels in a Concentration-Dependent Manner
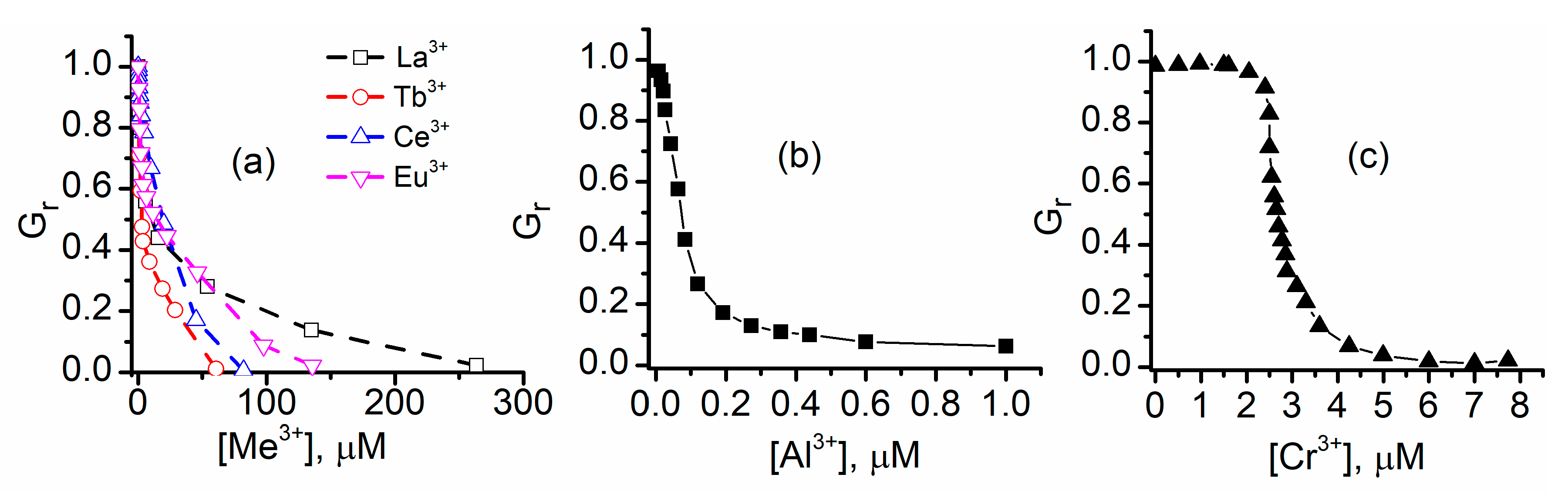
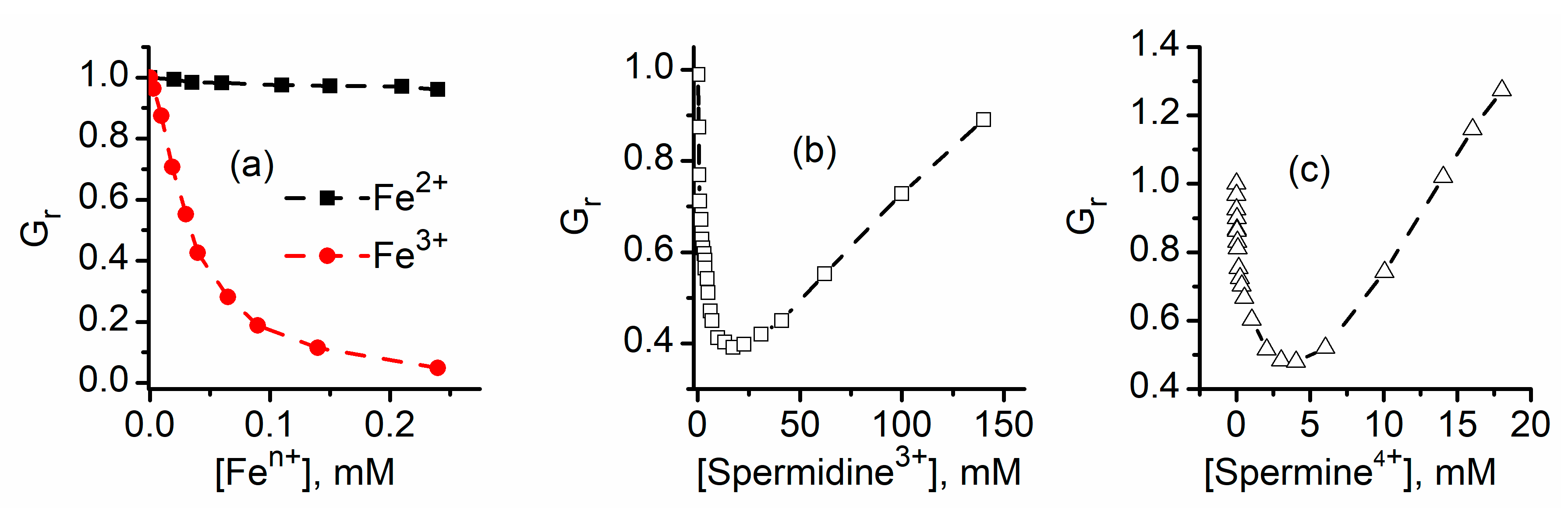
2.2. Trivalent Metal Cations Strongly Inhibit the Macroscopic Conductance of Lysenin Channels
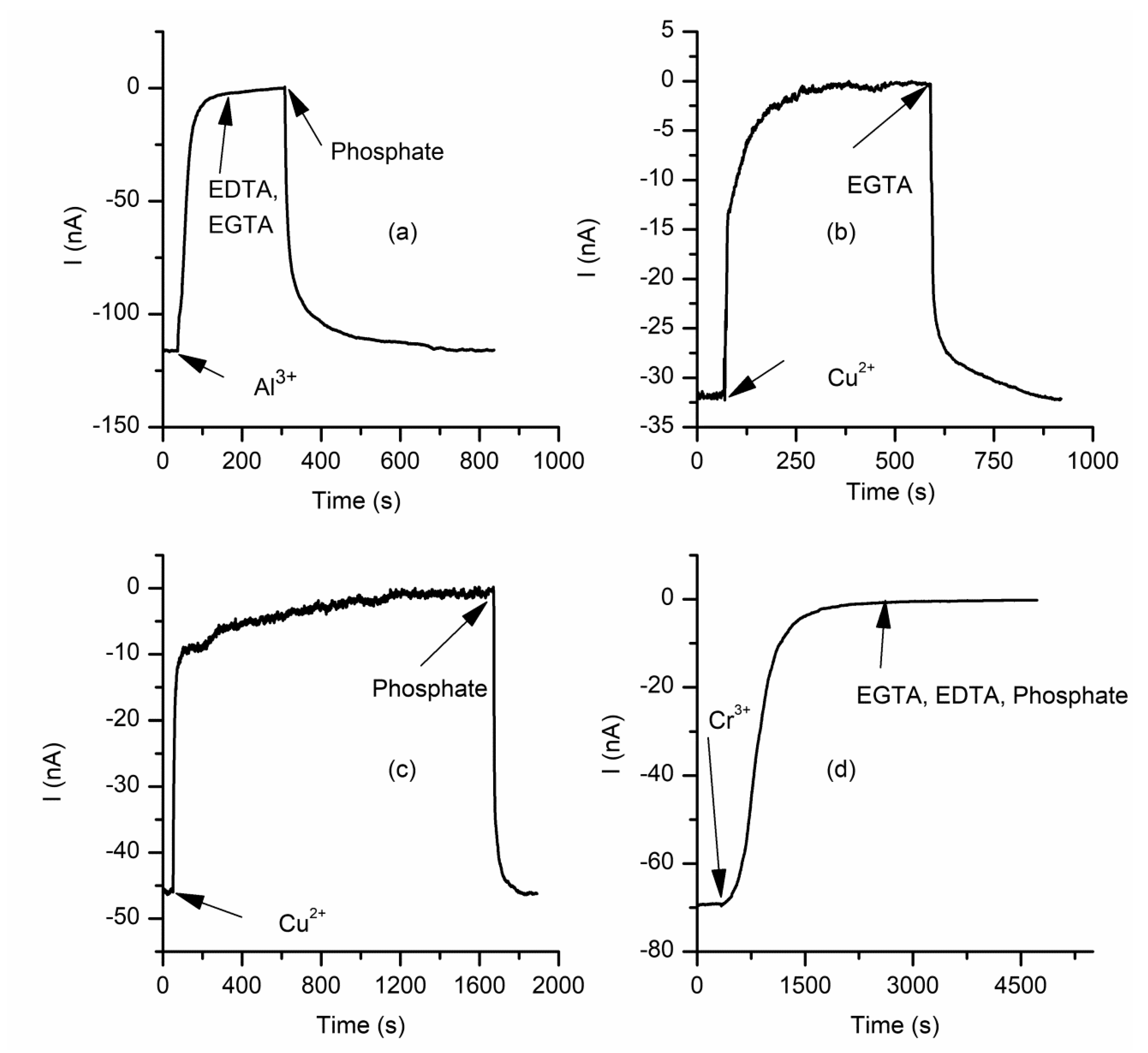
2.3. The Changes in Macroscopic Conductance Elicited by Multivalent Cations Are Reversible
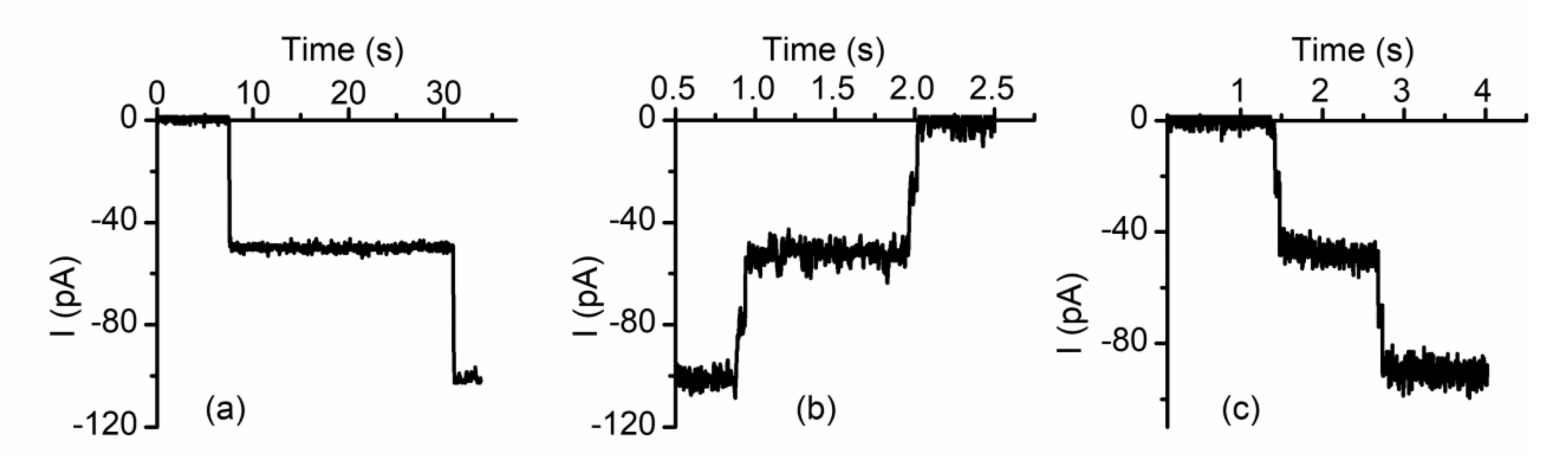
2.4. Lysenin Channels Undergo Ligand-Induced Gating Upon Exposure to Multivalent Cations
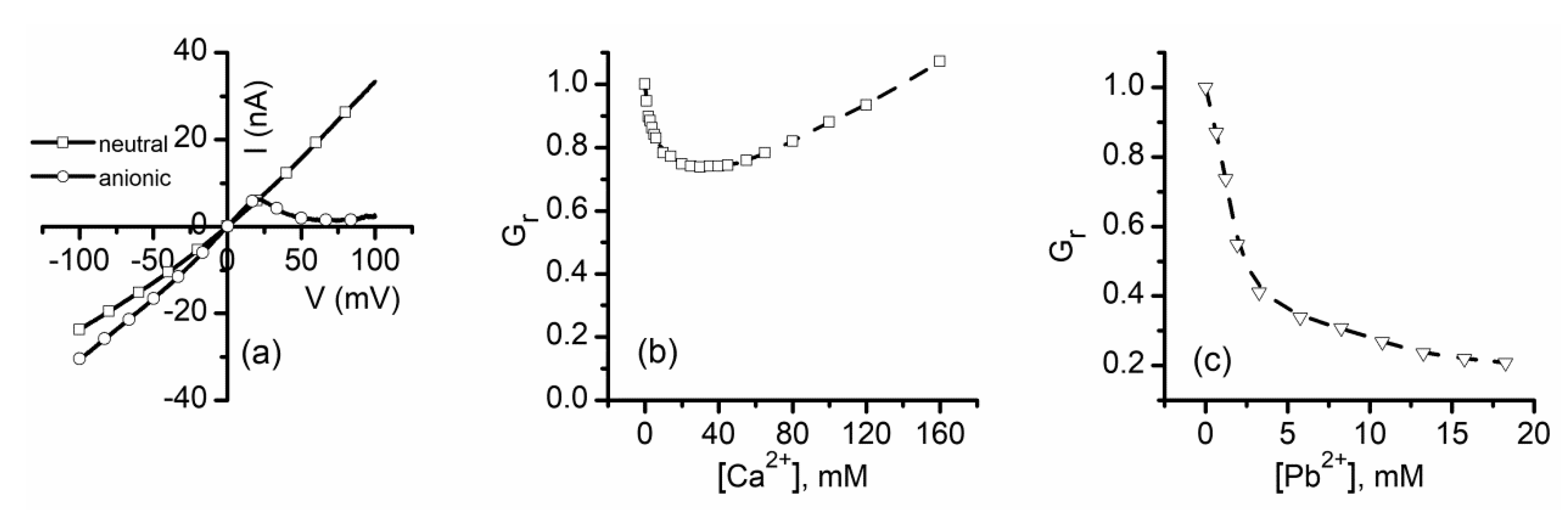
2.5. Which One Matters, Charge, or Size?
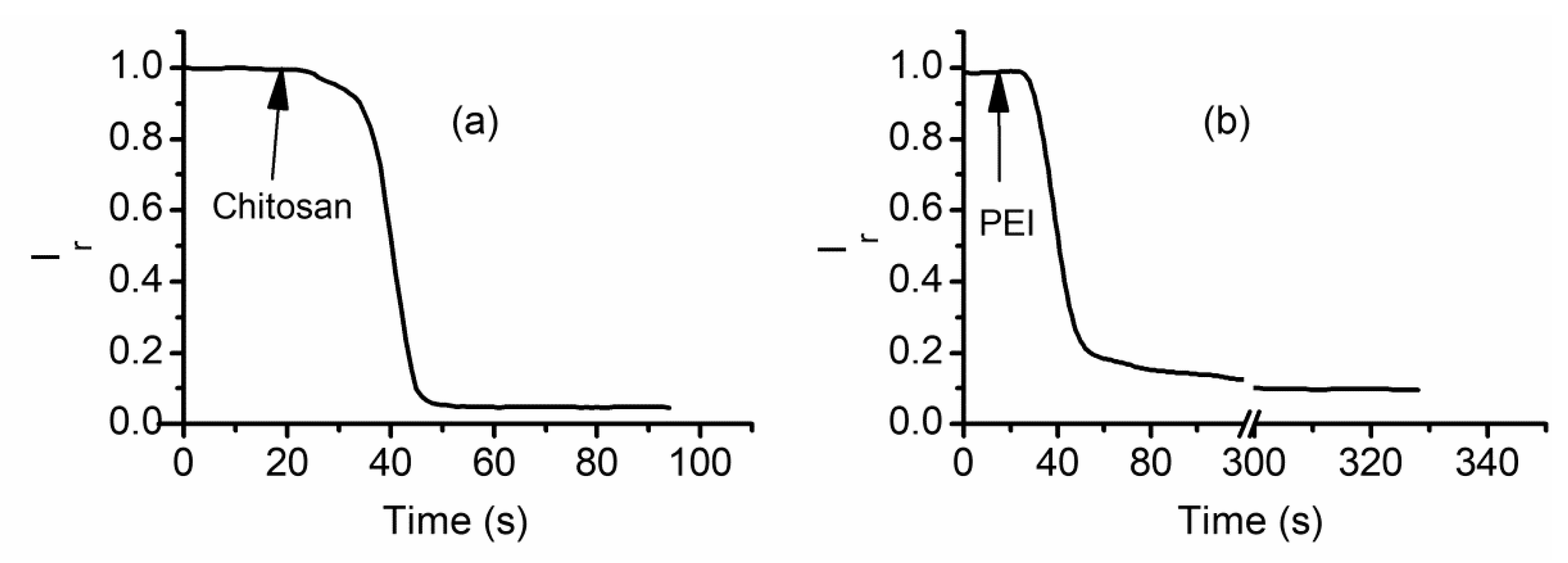
2.6. Cationic Polymers Irreversibly Block Lysenin Channels
2.7. Ligand and Voltage Gating of Lysenin Channels Are Not Coupled
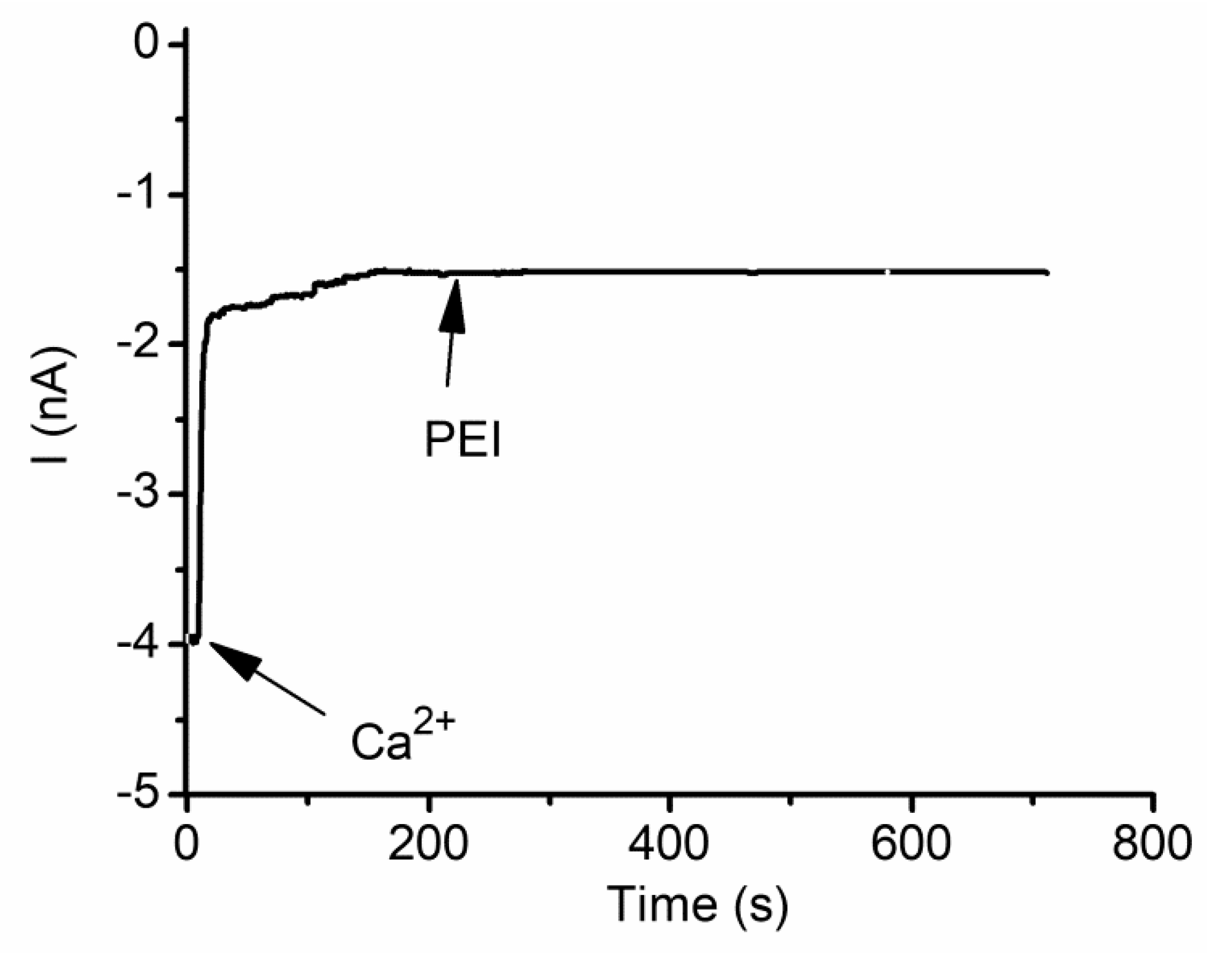
2.8. Cationic Ions and Polymers May Compete for the Binding Sites
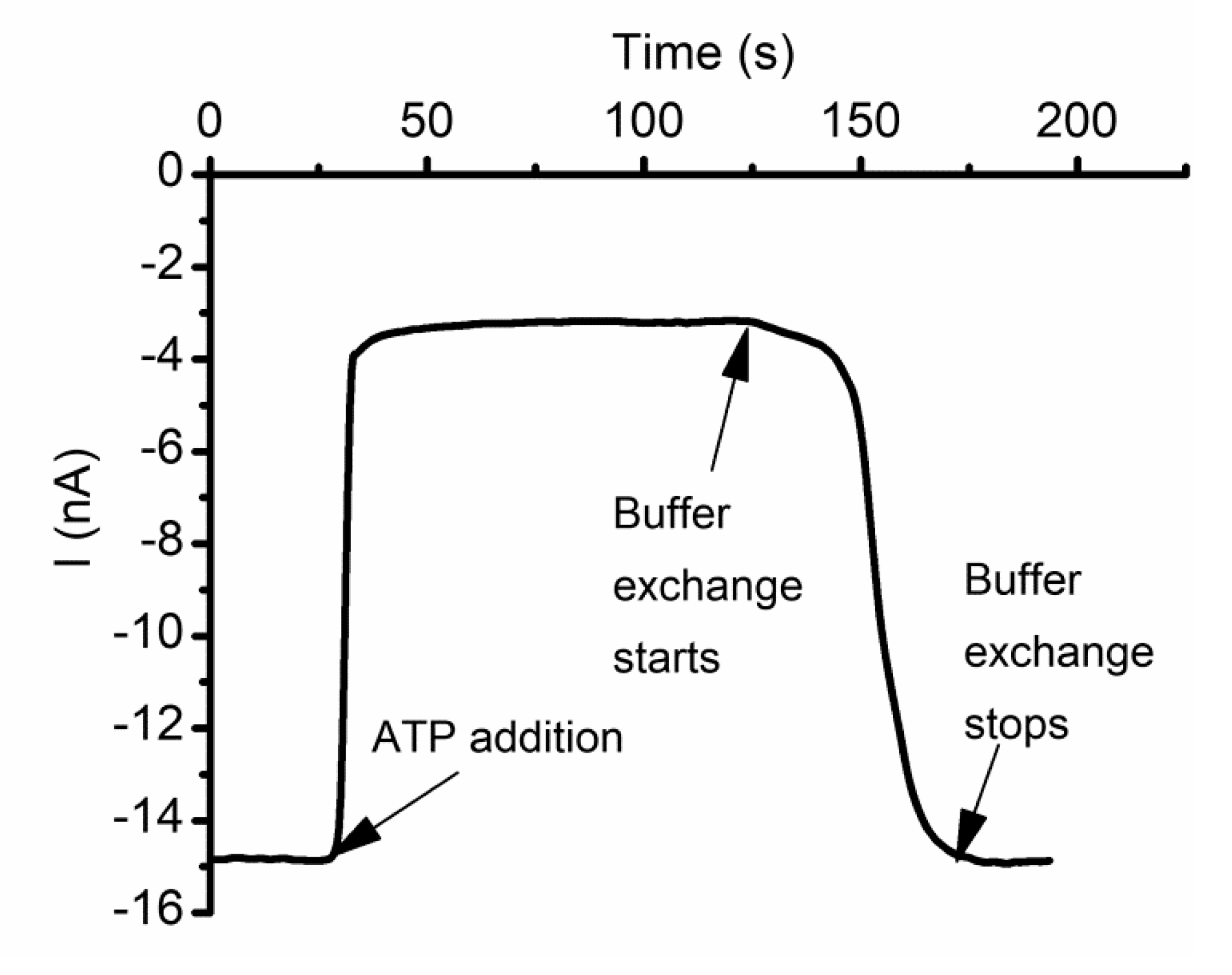
2.9. Lysenin Interactions with Purines
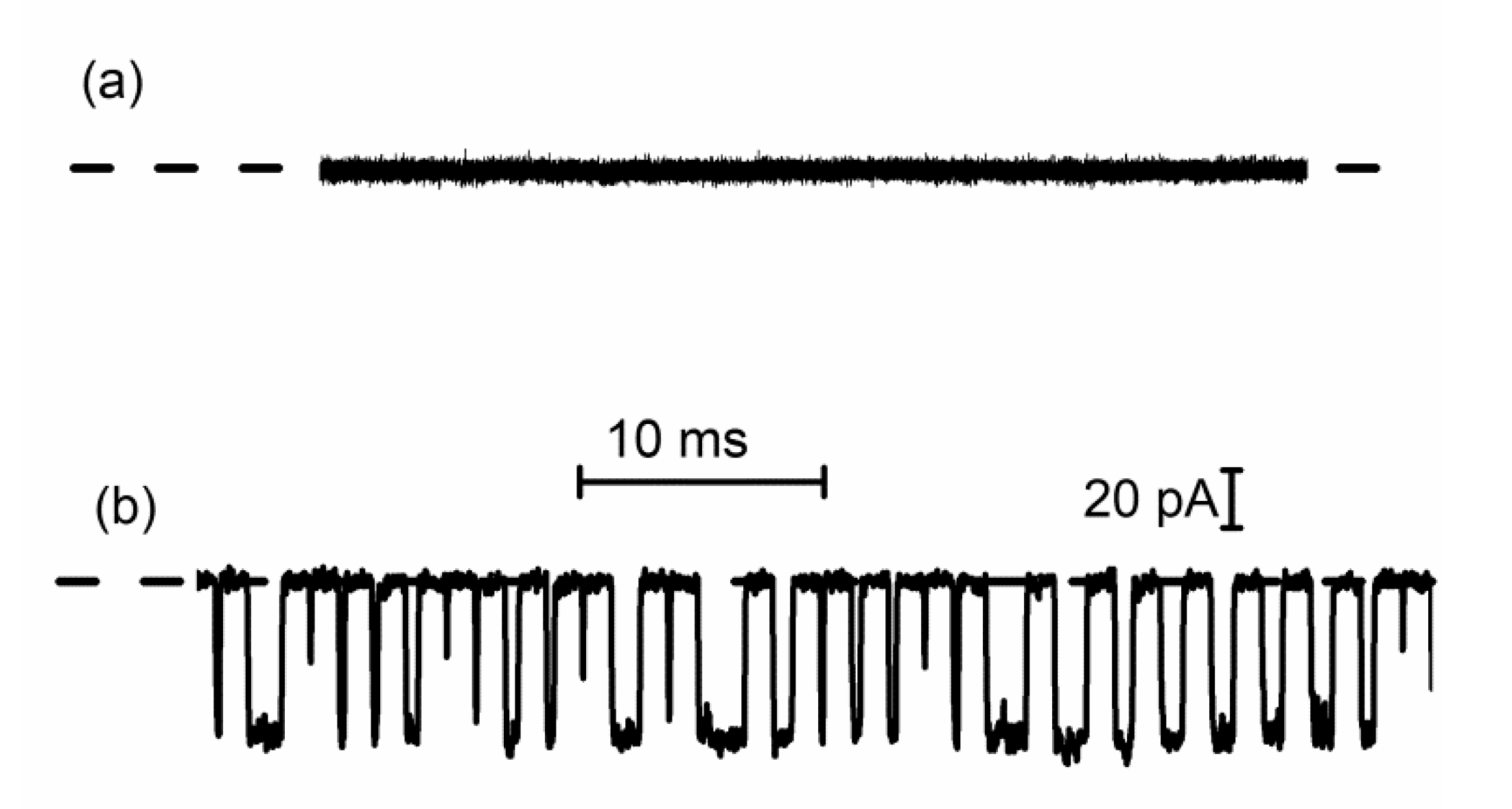
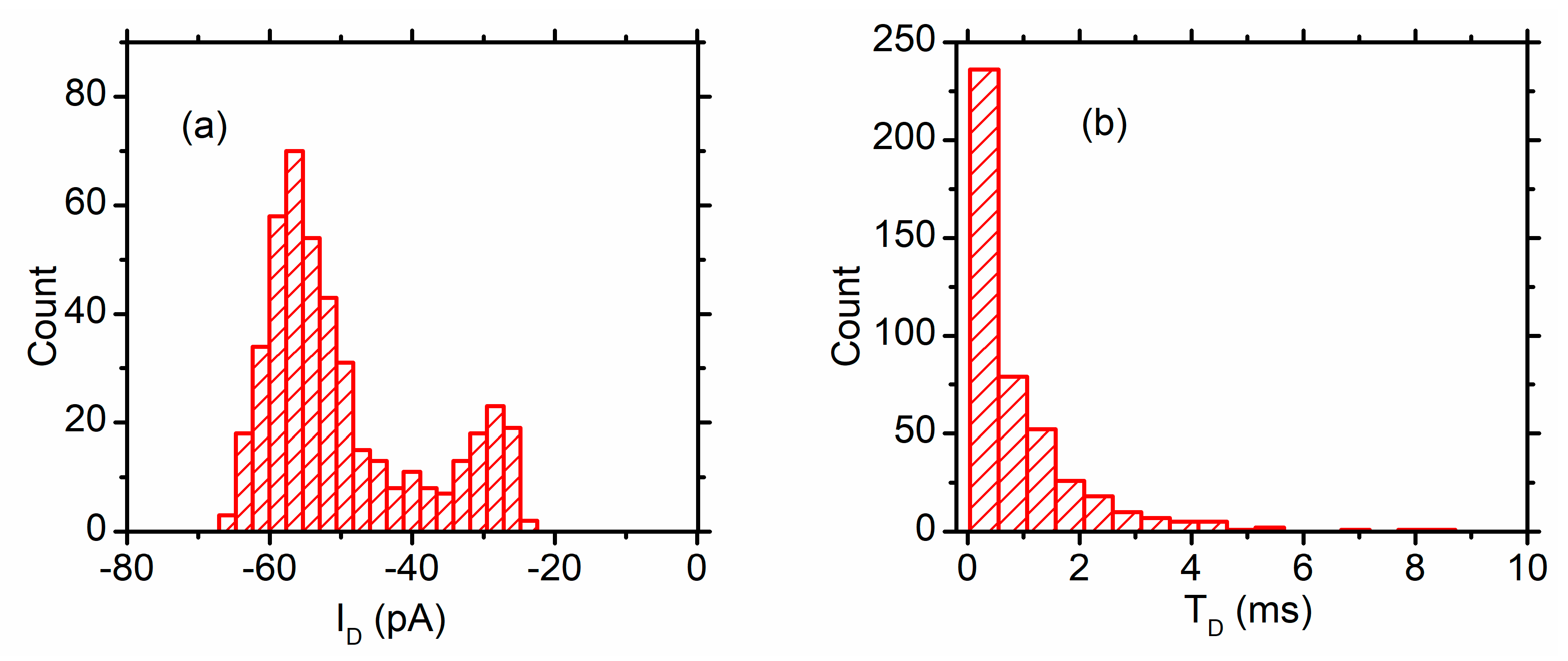
3. Lysenin Channels as Stochastic Sensors: Translocation of Macromolecules
3.1. DNA Translocation Experiments
3.2. Peptide Translocation
4. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
Appendix A
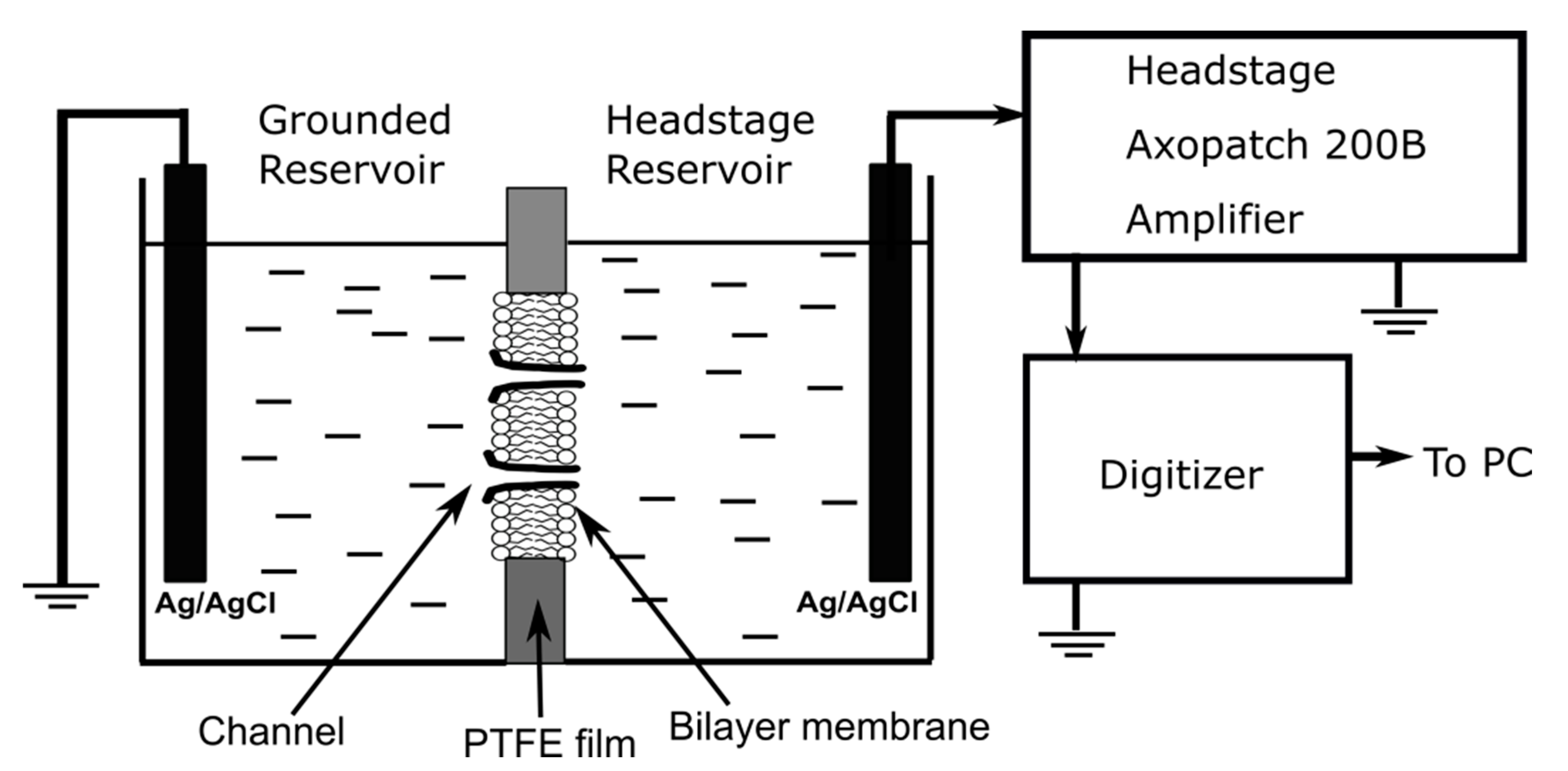
Experimental Details
References
- Panchal, R.G.; Smart, M.L.; Bowser, D.N.; Williams, D.A.; Petrou, S. Pore-Forming Proteins and their Application in Biotechnology. Curr. Pharm. Biotechnol. 2002, 3, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, M.; Hirano, A.; Buhlmann, P.; Umezawa, Y. Design and Application of Ion-Channel Sensors Based on Biological and Artificial Receptors. Bull. Chem. Soc. Jpn. 2002, 75, 187–201. [Google Scholar] [CrossRef]
- Krishnamurthy, V.; Monfared, S.M.; Cornell, B. Ion-Channel Biosensors—Part I: Construction, Operation, and Clinical Studies. IEEE Trans. Nanotechnol. 2010, 9, 303–312. [Google Scholar] [CrossRef]
- Steller, L.; Kreir, M.; Salzer, R. Natural and artificial ion channels for biosensing platforms. Anal. Bioanal. Chem. 2012, 402, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Ayub, M.; Bayley, H. Engineered transmembrane pores. Curr. Opin. Chem. Biol. 2016, 34, 117–126. [Google Scholar] [CrossRef]
- Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 2017, 12, 619–630. [Google Scholar] [CrossRef]
- Kisovec, M.; Rezelj, S.; Knap, P.; Cajnko, M.M.; Caserman, S.; Flasker, A.; Znidarsic, N.; Repic, M.; Mavri, J.; Ruan, Y.; et al. Engineering a pH responsive pore forming protein. Sci. Rep. 2017, 7, 42231. [Google Scholar] [CrossRef] [PubMed]
- Cressiot, B.; Ouldali, H.; Pastoriza-Gallego, M.; Bacri, L.; Van der Goot, F.G.; Pelta, J. Aerolysin, a Powerful Protein Sensor for Fundamental Studies and Development of Upcoming Applications. ACS Sens. 2019, 4, 530–548. [Google Scholar] [CrossRef]
- Majd, S.; Yusko, E.C.; Billeh, Y.N.; Macrae, M.X.; Yang, J.; Mayer, M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010, 21, 439–476. [Google Scholar] [CrossRef]
- Bayley, H.; Braha, O.; Gu, L.-Q. Stochastic Sensing with Protein Pores. Adv. Mater. 2000, 12, 139–142. [Google Scholar] [CrossRef]
- Bayley, H.; Cremer, P.S. Stochastic sensors inspired by biology. Nature 2001, 413, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Block, B.M.; Stacey, W.C.; Jones, S.W. Surface Charge and Lanthanum Block of Calcium Current in Bullfrog Sympathetic Neurons. Biophys. J. 1998, 74, 2278–2284. [Google Scholar] [CrossRef]
- Braha, O.; Gu, L.-Q.; Zhou, L.; Xiaofeng, L.; Cheley, S.; Bayley, H. Simultaneous stochastic sensing of divalent metal ions. Nat. Biotechnol. 2000, 18, 1005–1007. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M.; Cota, G. Calcium block of Na+ channels and its effect on closing rate. Proc. Natl. Acad. Sci. USA 1999, 96, 4154–4157. [Google Scholar] [CrossRef] [PubMed]
- Kasianowicz, J.J.; Burden, D.L.; Han, L.C.; Cheley, S.; Bayley, H. Genetically Engineered Metal Ion Binding Sites on the Outside of a Channel’s Transmembrane β-Barrel. Biophys. J. 1999, 76, 837–845. [Google Scholar] [CrossRef]
- Zhang, Y.; Niu, X.; Brelidze, T.I.; Magleby, K.L. Ring of Negative Charge in BK Channels Facilitates Block by Intracellular Mg2+ and Polyamines through Electrostatics. J. Gen. Physiol. 2006, 128, 185–202. [Google Scholar] [CrossRef]
- Elinder, F.; Arhem, P. Metal effects on ion channel gating. Q. Rev. Biophys. 2004, 36, 373–427. [Google Scholar] [CrossRef]
- Rotem, D.; Jayasinghe, L.; Salichou, M.; Bayley, H. Protein detection by nanopores equipped with aptamers. J. Am. Chem. Soc. 2012, 134, 2781–2787. [Google Scholar] [CrossRef]
- Stefureac, R.I.; Madampage, C.A.; Andrievskaia, O.; Lee, J.S. Nanopore analysis of the interaction of metal ions with prion proteins and peptides. Biochem. Cell Biol. 2010, 88, 347–358. [Google Scholar] [CrossRef]
- Andersen, O.S.; Ingolfsson, H.I.; Lundbaek, J.A. Ion Channels. In Wiley Encyclopedia of Chemical Biology; Wiley Publishing: Hoboken, NJ, USA, 2008; pp. 1–14. [Google Scholar]
- Bezanilla, F. Ion Channels: From Conductance to Structure. Neuron 2008, 60, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Phung, T.; Zhang, Y.; Dunlop, J.; Dalziel, J. Bilayer lipid membranes supported on Teflon Filters: A functional environment for ion channels. Biosens. Bioelectron. 2011, 26, 3127–3135. [Google Scholar] [CrossRef]
- Studer, A.; Demarche, S.; Langenegger, D.; Tiefenauer, L. Integration and recording of a reconstituted voltage-gated sodium channel in planar lipid bilayers. Biosens. Bioelectron. 2011, 26, 1924–1928. [Google Scholar] [CrossRef]
- Subrahmanyam, S.; Piletsky, S.A.; Turner, A.P.F. Application of Natural Receptors in Sensors and Assays. Anal. Chem. 2002, 74, 3942–3951. [Google Scholar] [CrossRef]
- Bashford, C.L. Pore-Forming Toxins: Attack and Defence at the Cell Surface. Cell. Mol. Biol. Lett. 2001, 6, 328–333. [Google Scholar]
- Delcour, A.H. Structure and Function of Pore-Forming—Barrels from Bacteria. J. Mol. Microbiol. Biotechnol. 2002, 4, 1–10. [Google Scholar] [PubMed]
- Gilbert, R.J. Pore-forming toxins. Cell. Mol. Life Sci. 2002, 59, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Bryant, S.L.; Thomas, C.; Richtsmeier, D.; Pu, X.; Tinker, J.; Fologea, D. Stochastic sensing of Angiotensin II with lysenin channels. Sci. Rep. 2017, 7, 2448. [Google Scholar] [CrossRef]
- Butler, T.Z.; Gundlach, J.H.; Troll, M. Ionic Current Blockades from DNA and RNA Molecules in the α-Hemolysin Nanopore. Biophys. J. 2007, 93, 3229–3240. [Google Scholar] [CrossRef]
- Derrington, I.M.; Craig, J.M.; Stava, E.; Laszlo, A.H.; Ross, B.C.; Brinkerhoff, H.; Nova, I.C.; Doering, K.; Tickman, B.I.; Ronaghi, M.; et al. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 2015, 33, 1073–1075. [Google Scholar] [CrossRef]
- Haque, F.; Li, J.; Wu, H.C.; Liang, X.J.; Guo, P. Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA. Nano Today 2013, 8, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Ying, Y.-L.; Hu, Z.-L.; Liao, D.-F.; Tian, H.; Long, Y.-T. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 2016, 11, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Yu, J.; Wang, Y.-Q.; Ying, Y.-L.; Long, Y.-T. Driven Translocation of Polynucleotides through an Aerolysin Nanopore. Anal. Chem. 2016, 88, 5046–5049. [Google Scholar] [CrossRef] [PubMed]
- Bayley, H. Nanopore sequencing: From imagination to reality. Clin. Chem. 2015, 61, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Gurnev, P.A.; Nestorovich, E.M. Channel-Forming Bacterial Toxins in Biosensing and Macromolecule Delivery. Toxins 2014, 6, 2483–2540. [Google Scholar] [CrossRef] [PubMed]
- Shakor, A.-B.A.; Czurylo, E.A.; Sobota, A. Lysenin, a unique sphingomyelin-binding protein. FEBS Lett. 2003, 542, 1–6. [Google Scholar] [CrossRef]
- Yamaji-Hasegawa, A.; Makino, A.; Baba, T.; Senoh, Y.; Kimura-Suda, H.; Sato, S.B.; Terada, N.; Ohno, S.; Kiyokawa, E.; Umeda, M.; et al. Oligomerization and pore formation of a sphingomyelin-specific toxin, lysenin. J. Biol. Chem. 2003, 278, 22762–22770. [Google Scholar] [CrossRef]
- Bruhn, H.; Winkelmann, J.; Andersen, C.; Andra, J.; Leippe, M. Dissection of the mechanisms of cytolytic and antibacterial activity of lysenin, a defense protein of the annelid Eisenia fetida. Dev. Comp. Immunol. 2006, 30, 597–606. [Google Scholar] [CrossRef]
- Ide, T.; Aoki, T.; Takeuchi, Y.; Yanagida, T. Lysenin forms a voltage-dependent channel in artificial lipid bilayer membranes. Biochem. Biophys. Res. Commun. 2006, 346, 288–292. [Google Scholar] [CrossRef]
- Kwiatkowska, K.; Hordejuk, R.; Szymczyk, P.; Kulma, M.; Abdel-Shakor, A.-B.; Plucienniczak, A.; Dolowy, K.; Szewczyk, A.; Sobota, A. Lysenin-His, a sphingomyelin-recognizing toxin, requires tryptophan 20 for cation-selective channel assembly but not for membrane binding. Mol. Membr. Biol. 2007, 24, 121–134. [Google Scholar] [CrossRef]
- Shogomori, H.; Kobayashi, T. Lysenin: A sphingomyelin specific pore-forming toxin. Biochim. Biophys. Acta 2008, 1780, 612–618. [Google Scholar] [CrossRef]
- Yamaji, A.; Sekizawa, Y.; Emoto, K.; Sakuraba, H.; Inoue, K. Lysenin, A novel sphingomyelin-specific binding protein. J. Biol. Chem. 1998, 273, 5300–5306. [Google Scholar] [CrossRef] [PubMed]
- De Colibus, L.; Sonnen, A.F.; Morris, K.J.; Siebert, C.A.; Abrusci, P.; Plitzko, J.; Hodnik, V.; Leippe, M.; Volpi, E.; Anderluh, G.; et al. Structures of lysenin reveal a shared evolutionary origin for pore-forming proteins and its mode of sphingomyelin recognition. Structure 2012, 20, 1498–1507. [Google Scholar] [CrossRef] [PubMed]
- Kulma, M.; Dadlez, M.; Kwiatkowska, K. Insight into the Structural Dynamics of the Lysenin During Prepore-to-Pore Transition Using Hydrogen—Deuterium Exchange Mass Spectrometry. Toxins 2019, 11, 462. [Google Scholar] [CrossRef]
- Munguira, I.L.B.; Takahashi, H.; Casuso, I.; Scheuring, S. Lysenin Toxin Membrane Insertion is pH-Dependent but Independent of Neighboring Lysenins. Biophys. J. 2017, 113, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Bokori-Brown, M.; Martin, T.G.; Naylor, C.E.; Basak, A.K.; Titball, R.W.; Savva, C.G. Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nat. Commun. 2016, 7, 11293. [Google Scholar] [CrossRef]
- Yilmaz, N.; Kobayashi, T. Visualization of Lipid Membrane Reorganization Induced by a Pore-Forming Toxin Using High-Speed Atomic Force Microscopy. ACS Nano 2015, 9, 7960–7967. [Google Scholar] [CrossRef]
- Yilmaz, N.; Yamada, T.; Greimel, P.; Uchihashi, T.; Ando, T.; Kobayashi, T. Real-Time Visualization of Assembling of a Sphingomyelin-Specific Toxin on Planar Lipid Membranes. Biophys. J. 2013, 105, 1397–1405. [Google Scholar] [CrossRef]
- Yilmaz, N.; Yamaji-Hasegawa, A.; Hullin-Matsuda, F.; Kobayashi, T. Molecular mechanisms of action of sphingomyelin-specific pore-forming toxin, lysenin. Semin. Cell Dev. Biol. 2018, 73, 188–198. [Google Scholar] [CrossRef]
- Podobnik, M.; Savory, P.; Rojko, N.; Kisovec, M.; Wood, N.; Hambley, R.; Pugh, J.; Wallace, E.J.; McNeill, L.; Bruce, M.; et al. Crystal structure of an invertebrate cytolysin pore reveals unique properties and mechanism of assembly. Nat. Commun. 2016, 7, 11598. [Google Scholar] [CrossRef]
- Kulma, M.; Herec, M.; Grudzinski, W.; Anderluh, G.; Gruszecki, W.I.; Kwiatkowska, K.; Sobota, A. Sphingomyelin-rich domains are sites of lysenin oligomerization: Implications for raft studies. Biochim. Biophys. Acta Biomembr. 2010, 1798, 471–481. [Google Scholar] [CrossRef]
- Fologea, D.; Krueger, E.; Lee, R.; Naglak, M.; Mazur, Y.; Henry, R.; Salamo, G. Controlled gating of lysenin pores. Biophys. Chem. 2010, 146, 25–29. [Google Scholar] [CrossRef]
- Fologea, D.; Krueger, E.; Mazur, Y.I.; Stith, C.; Okuyama, Y.; Henry, R.; Salamo, G.J. Bi-stability, hysteresis, and memory of voltage-gated lysenin channels. Biochim. Biophys. Acta Biomembr. 2011, 1808, 2933–2939. [Google Scholar] [CrossRef]
- Ishitsuka, R.; Sato, S.B.; Kobayashi, T. Imaging Lipid Rafts. J. Biochem. 2005, 137, 249–254. [Google Scholar] [CrossRef]
- Krueger, E.; Bryant, S.; Shrestha, N.; Clark, T.; Hanna, C.; Pink, D.; Fologea, D. Intramembrane congestion effects on lysenin channel voltage-induced gating. Eur. Biophys. J. 2016, 45, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, R.; Kobayashi, T. Lysenin: A new tool for investigating membrane lipid organization. Anat. Sci. Int. 2004, 79, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Hirano, M.; Takeuchi, Y.; Kobayashi, T.; Yanagida, T.; Ide, T. Single channel properties of lysenin measured in artificial lipid bilayers and their application to biomolecule detection. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 920–925. [Google Scholar] [CrossRef]
- Bryant, S.; Shrestha, N.; Carnig, P.; Kosydar, S.; Belzeski, P.; Hanna, C.; Fologea, D. Purinergic control of lysenin’s transport and voltage-gating properties. Purinergic Signal. 2016, 12, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Bryant, S.L.; Eixenberger, J.E.; Rossland, S.; Apsley, H.; Hoffmann, C.; Shrestha, N.; McHugh, M.; Punnoose, A.; Fologea, D. ZnO nanoparticles modulate the ionic transport and voltage regulation of lysenin nanochannels. J. Nanobiotechnol. 2017, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Fologea, D.; Al Faori, R.; Krueger, E.; Mazur, Y.I.; Kern, M.; Williams, M.; Mortazavi, A.; Henry, R.; Salamo, G.J. Potential analytical applications of lysenin channels for detection of multivalent ions. Anal. Bioanal. Chem. 2011, 401, 1871–1879. [Google Scholar] [CrossRef]
- Fologea, D.; Krueger, E.; Al Faori, R.; Lee, R.; Mazur, Y.I.; Henry, R.; Arnold, M.; Salamo, G.J. Multivalent ions control the transport through lysenin channels. Biophys. Chem. 2010, 152, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Fologea, D.; Krueger, E.; Rossland, S.; Bryant, S.; Foss, W.; Clark, T. Cationic Polymers Inhibit the Conductance of Lysenin Channels. Sci. World J. 2013, 316758. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [PubMed]
- Bezrukov, S.M. Ion Channels as Molecular Coulter Counters to Probe Metabolite Transport. J. Membr. Biol. 2000, 174, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.W.; Akeson, M. Nanopores and nucleic acids: Prospects for ultrarapid sequencing. TIBTECH 2000, 18, 147–151. [Google Scholar] [CrossRef]
- Li, J.; Gershow, M.; Stein, D.; Brandin, E.; Golovchenko, J.A. DNA Molecules and Configurations in a Solid-state Nanopore Microscope. Nat. Mater. 2003, 2, 611–615. [Google Scholar] [CrossRef]
- Storm, A.J.; Chen, J.H.; Ling, X.S.; Zandbergen, H.W.; Dekker, C. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2003, 2, 537–540. [Google Scholar] [CrossRef]
- Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef]
- Stefureac, R.; Waldner, L.; Howard, P.; Lee, J.S. Nanopore Analysis of a Small 86-Residue Protein. Small 2008, 4, 59–63. [Google Scholar] [CrossRef]
- Stoloff, D.H.; Wanunu, M. Recent trends in nanopores for biotechnology. Curr. Opin. Biotechnol. 2013, 24, 699–704. [Google Scholar] [CrossRef]
- Traversi, F.; Raillon, C.; Benameur, S.M.; Liu, K.; Khlybov, S.; Tosun, M.; Krasnozhon, D.; Kis, A.; Radenovic, A. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat. Biotechnol. 2013, 8, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Dallman, T.; Rubino, S.; Rabsch, W.; Mwaigwisya, S.; Wain, J.; O’Grady, J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2014, 33, 296–300. [Google Scholar] [CrossRef]
- Laszlo, A.H.; Derrington, I.M.; Ross, B.C.; Brinkerhoff, H.; Adey, A.; Nova, I.C.; Craig, J.M.; Langford, K.W.; Samson, J.M.; Daza, R. Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 2014, 32, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Pastoriza-Gallego, M.; Breton, M.-F.; Discala, F.; Auvray, L.; Betton, J.-M.; Pelta, J. Evidence of Unfolded Protein Translocation through a Protein Nanopore. ACS Nano 2014, 8, 11350–11360. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-based Fourth-generation DNA Sequencing Technology. Genom. Proteom. Bioinform. 2015, 13, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Huang, Q.; Zhao, Y.; Zhou, D.; Ying, C.; Wang, D. Precise fabrication of a 5 nm graphene nanopore with a helium ion microscope for biomolecule detection. Nanotechnology 2017, 28, 045302. [Google Scholar] [CrossRef] [PubMed]
- Butler, T.Z.; Pavlenok, M.; Derrington, I.M.; Niederweis, M.; Gundlach, J.H. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl. Acad. Sci. USA 2008, 105, 20647–20652. [Google Scholar] [CrossRef]
- Wang, S.; Haque, F.; Rychahou, P.G.; Evers, B.M.; Guo, P. Engineered Nanopore of Phi29 DNA-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano 2013, 7, 9814–9822. [Google Scholar] [CrossRef]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef]
- Belzeski, P.; Shrestha, N.; Bryant, S.; Tinker, J.; Thomas, C.A.; Richtsmeier, D.; Fologea, D. Sensing ssDNA Molecules with Single Lysenin Channels. Biophys. J. 2017, 112, 153a–154a. [Google Scholar] [CrossRef][Green Version]
- Stefureac, R.; Long, Y.; Kraatz, H.-B.; Howard, P.; Lee, J.S. Transport of α—Helical Peptides through α—Hemolysin and Aerolysin Pores. Biochemistry 2006, 45, 9172–9179. [Google Scholar] [CrossRef] [PubMed]
- Diederichs, T.; Pugh, G.; Dorey, A.; Xing, Y.; Burns, J.R.; Hung Nguyen, Q.; Tornow, M.; Tampe, R.; Howorka, S. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 2019, 10, 5018. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Wang, S.; Zhao, Z.; Zhou, Z.; Haque, F.; Guo, P. Fingerprinting of Peptides with a Large Channel of Bacteriophage Phi29 DNA Packaging Motor. Small 2016, 12, 4572–4578. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.R.; Barcena-Uribarri, I.; Modi, N.; Kleinekathofer, U.; Benz, R.; Winterhalter, M.; Mahendran, K.R. Pulling Peptides across Nanochannels: Resolving Peptide Binding and Translocation through the Hetero-oligomeric Channel from Nocardia farcinica. ACS Nano 2012, 6, 10699–10707. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Jayawardhana, D.A.; Wang, D.; Guan, X. Study of Peptide Transport through Engineered Protein Channels. J. Phys. Chem. B 2009, 113, 3572–3578. [Google Scholar] [CrossRef] [PubMed]
- Elliott, I.; Batty, E.M.; Ming, D.; Robinson, M.T.; Nawtaisong, P.; de Cesare, M.; Newton, P.N.; Bowden, R. Oxford Nanopore MinION Sequencing Enables Rapid Whole Genome Assembly of Rickettsia typhi in a Resource-Limited Setting. Am. J. Trop. Med. Hyg. 2020, 102, 408–414. [Google Scholar] [CrossRef]
- Patel, A.; Belykh, E.; Miller, E.J.; George, L.L.; Martirosyan, N.L.; Byvaltsev, V.A.; Preul, M.C. MinION rapid sequencing: Review of potential applications in neurosurgery. Surg. Neurol. Int. 2018, 9, 157. [Google Scholar]
- Lamichhane, U.; Islam, T.; Prasad, S.; Weingart, H.; Mahendran, K.R.; Winterhalter, M. Peptide translocation through the mesoscopic channel: Binding kinetics at the single molecule level. Eur. Biophys. J. 2013, 42, 363–369. [Google Scholar] [CrossRef]
- Mereuta, L.; Roy, M.; Asandei, A.; Lee, J.K.; Park, Y.; Andricioaei, I.; Luchian, T. Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Sci. Rep. 2014, 4, 3885. [Google Scholar] [CrossRef]
- Plesa, C.; Dekker, C. Data analysis methods for solid-state nanopores. Nanotechnology 2015, 26, 084003. [Google Scholar] [CrossRef]
- Fologea, D.; Ledden, B.; McNabb, D.S.; Li, J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007, 91, 0539011. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Thomas, C.A.; Richtsmeier, D.; Bogard, A.; Hermann, R.; Walker, M.; Abatchev, G.; Brown, R.J.; Fologea, D. Temporary Membrane Permeabilization via the Pore-Forming Toxin Lysenin. Toxins 2020, 12, 343. [Google Scholar] [CrossRef]


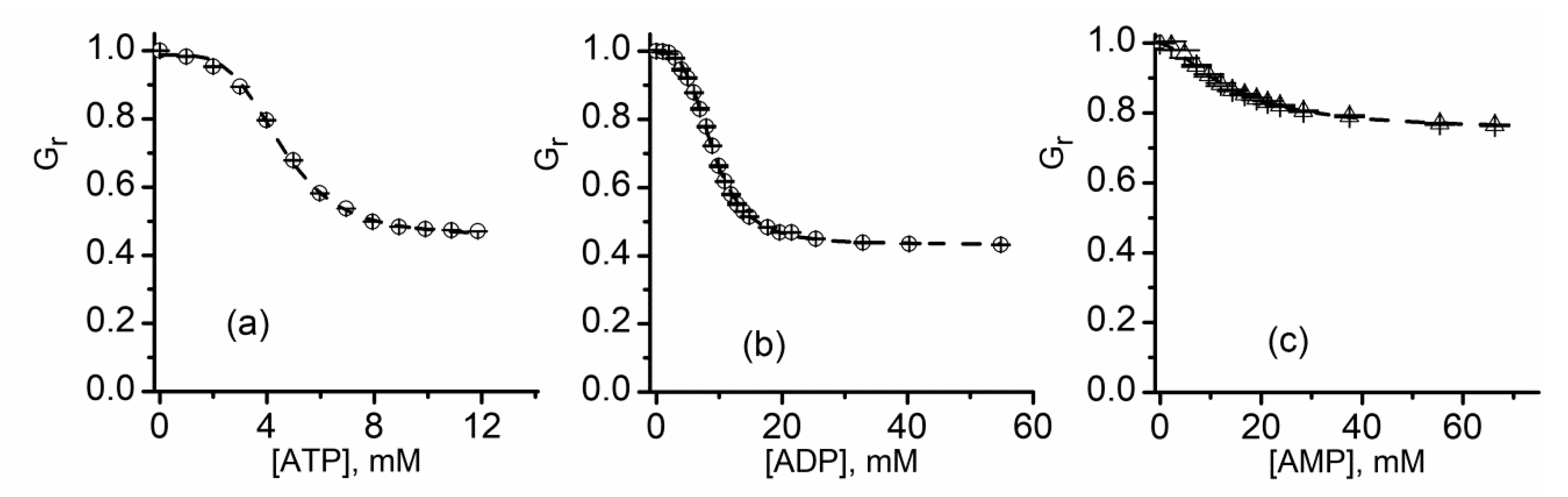
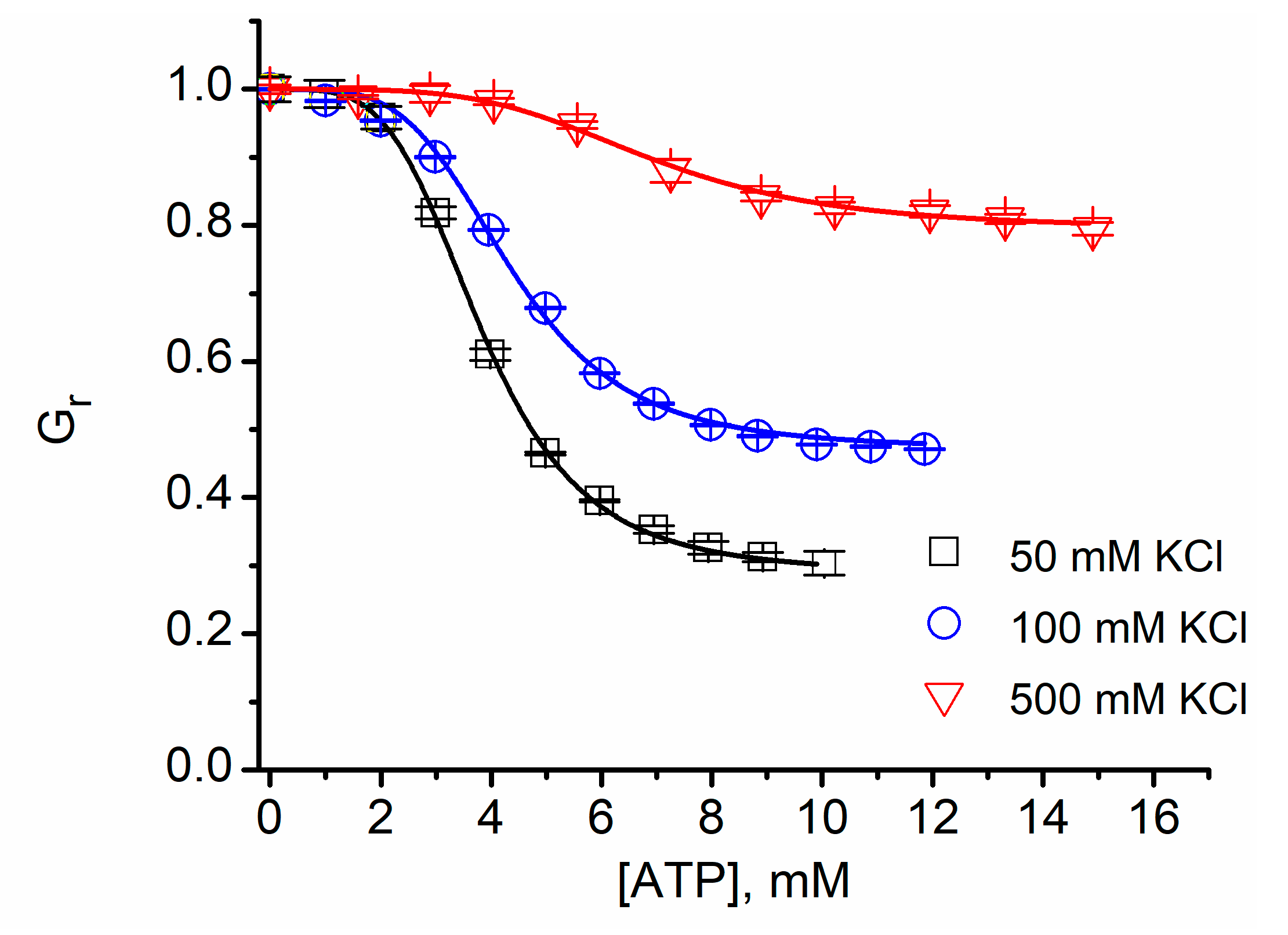
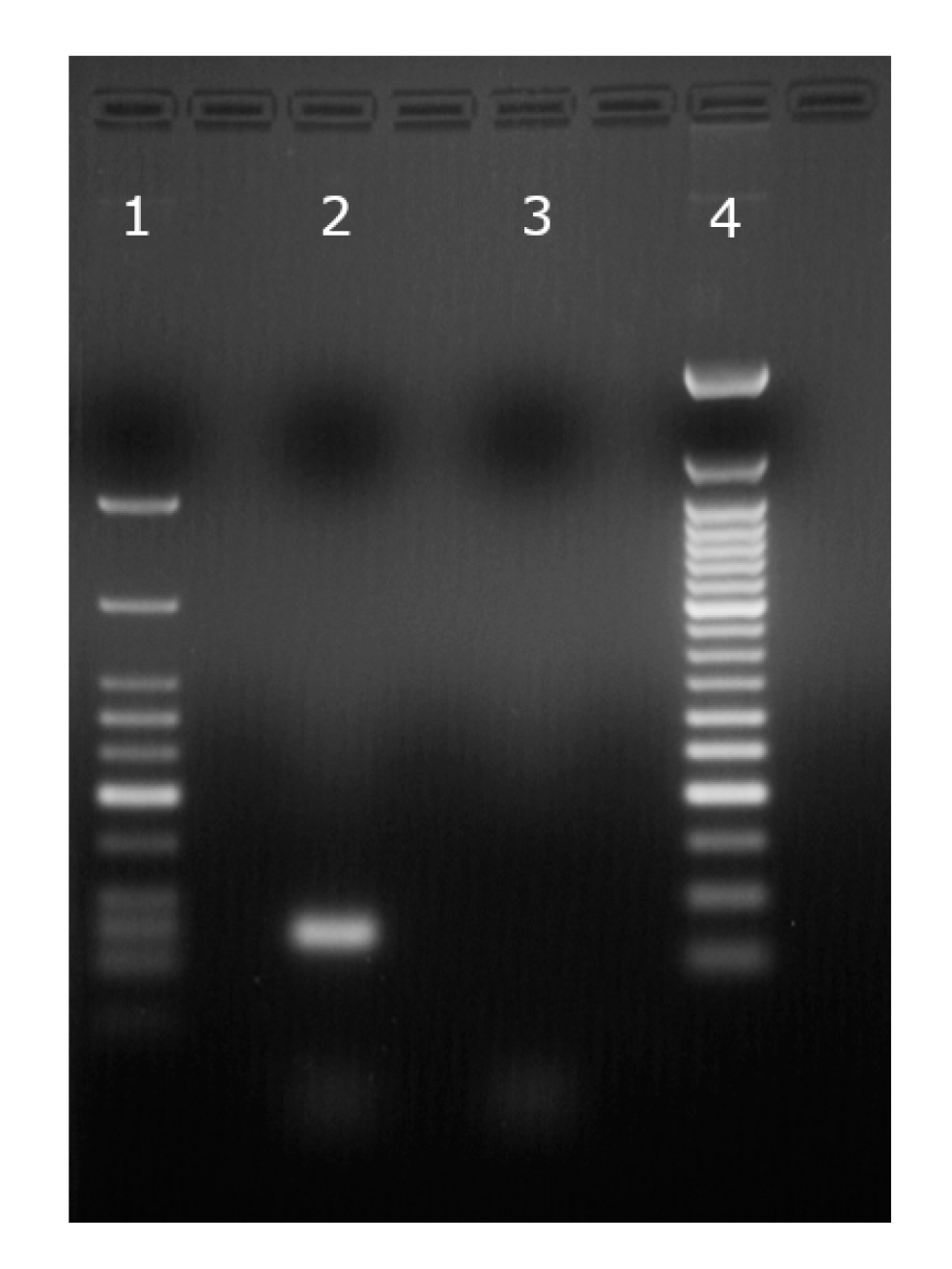

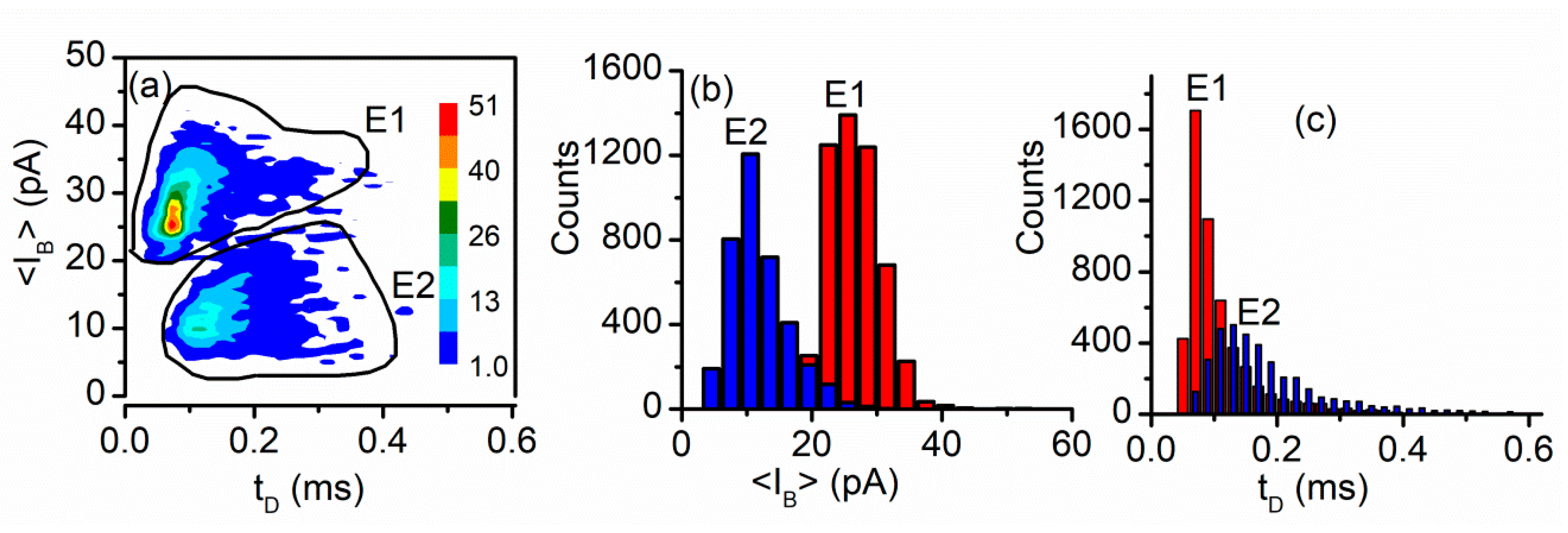
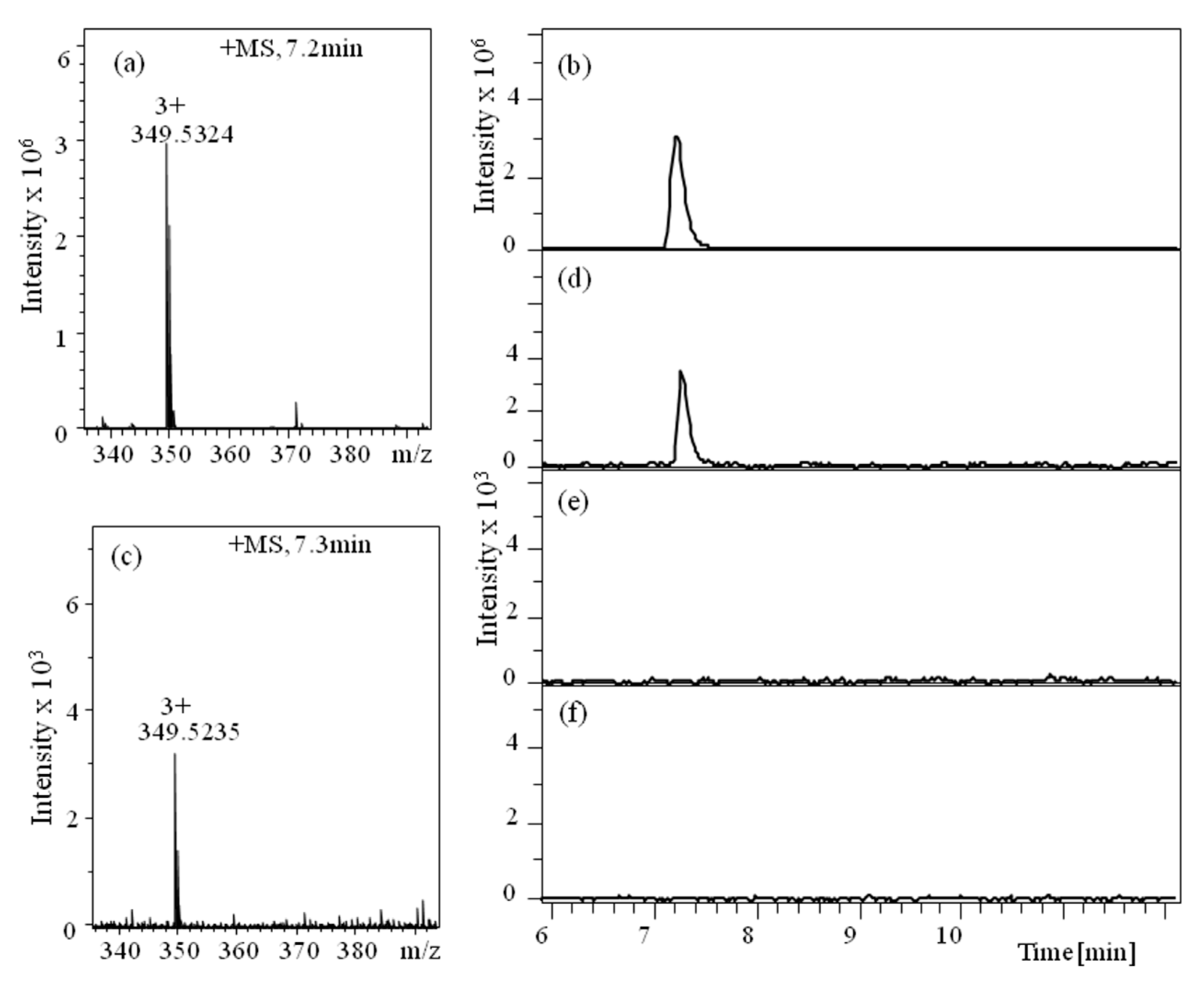
| IC50 (mM, ±SD) | n | |
|---|---|---|
| ATP | 4.53 ± 0.07 | 4.15 ± 0.2 |
| ADP | 8.92 ± 0.07 | 3.43 ± 0.16 |
| AMP | 13.43 ± 0.08 | 1.62 ± 0.17 |
| KCl (mM) | IC50 (mM, ±SD) | n |
|---|---|---|
| 50 | 3.83 ± 0.05 | 4.11 ± 0.16 |
| 135 | 4.36 ± 0.07 | 4.14 ± 0.2 |
| 500 | 6.94 ± 0.07 | 4.1 ± 0.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogard, A.; Abatchev, G.; Hutchinson, Z.; Ward, J.; Finn, P.W.; McKinney, F.; Fologea, D. Lysenin Channels as Sensors for Ions and Molecules. Sensors 2020, 20, 6099. https://doi.org/10.3390/s20216099
Bogard A, Abatchev G, Hutchinson Z, Ward J, Finn PW, McKinney F, Fologea D. Lysenin Channels as Sensors for Ions and Molecules. Sensors. 2020; 20(21):6099. https://doi.org/10.3390/s20216099
Chicago/Turabian StyleBogard, Andrew, Gamid Abatchev, Zoe Hutchinson, Jason Ward, Pangaea W. Finn, Fulton McKinney, and Daniel Fologea. 2020. "Lysenin Channels as Sensors for Ions and Molecules" Sensors 20, no. 21: 6099. https://doi.org/10.3390/s20216099
APA StyleBogard, A., Abatchev, G., Hutchinson, Z., Ward, J., Finn, P. W., McKinney, F., & Fologea, D. (2020). Lysenin Channels as Sensors for Ions and Molecules. Sensors, 20(21), 6099. https://doi.org/10.3390/s20216099







