Data-Analytics Modeling of Electrical Impedance Measurements for Cell Culture Monitoring
Abstract
1. Introduction
2. Materials and Methods
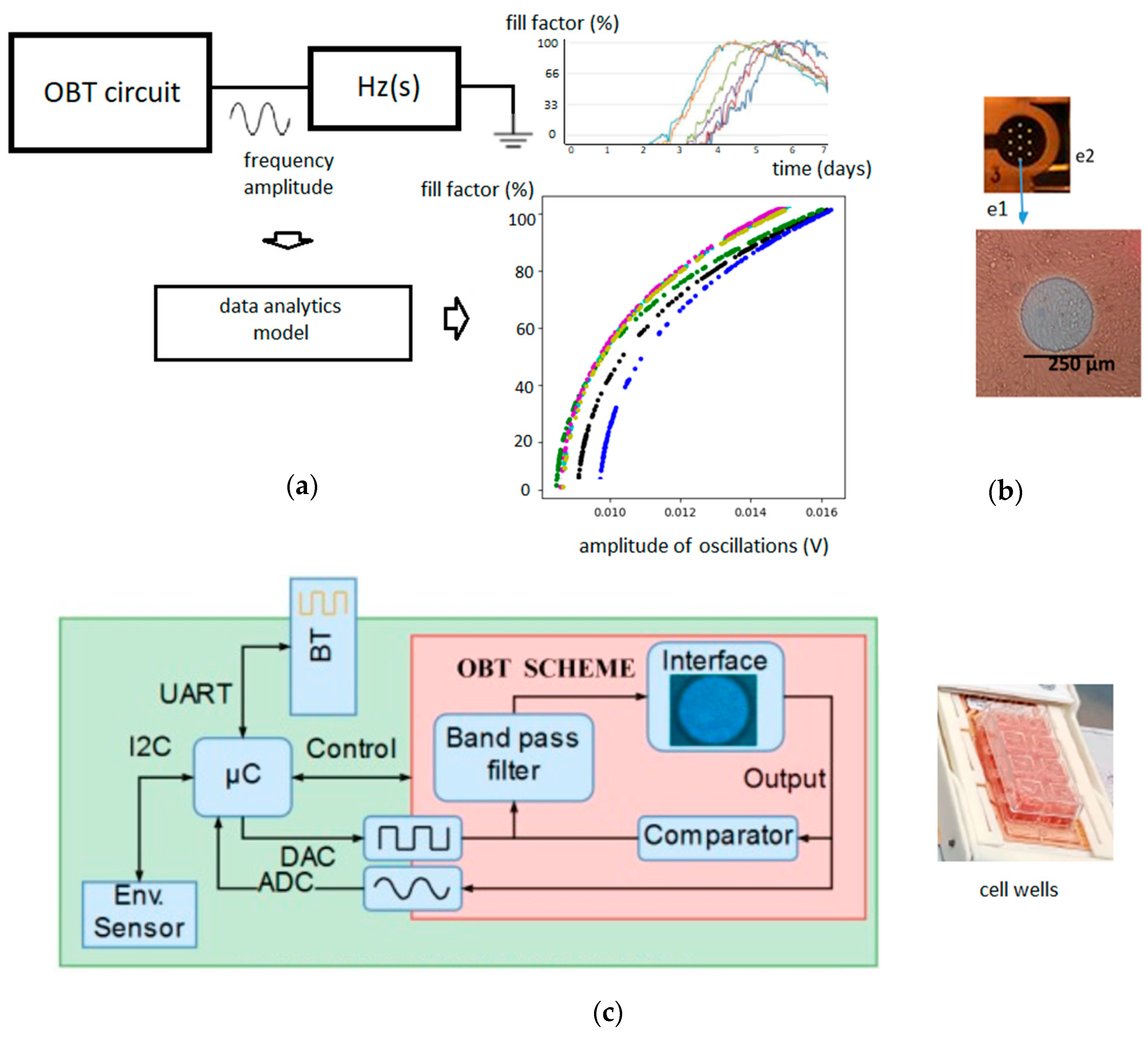
2.1. Oscillation-Based Technique
2.2. Datasets Used
2.3. Data Processing and Modeling
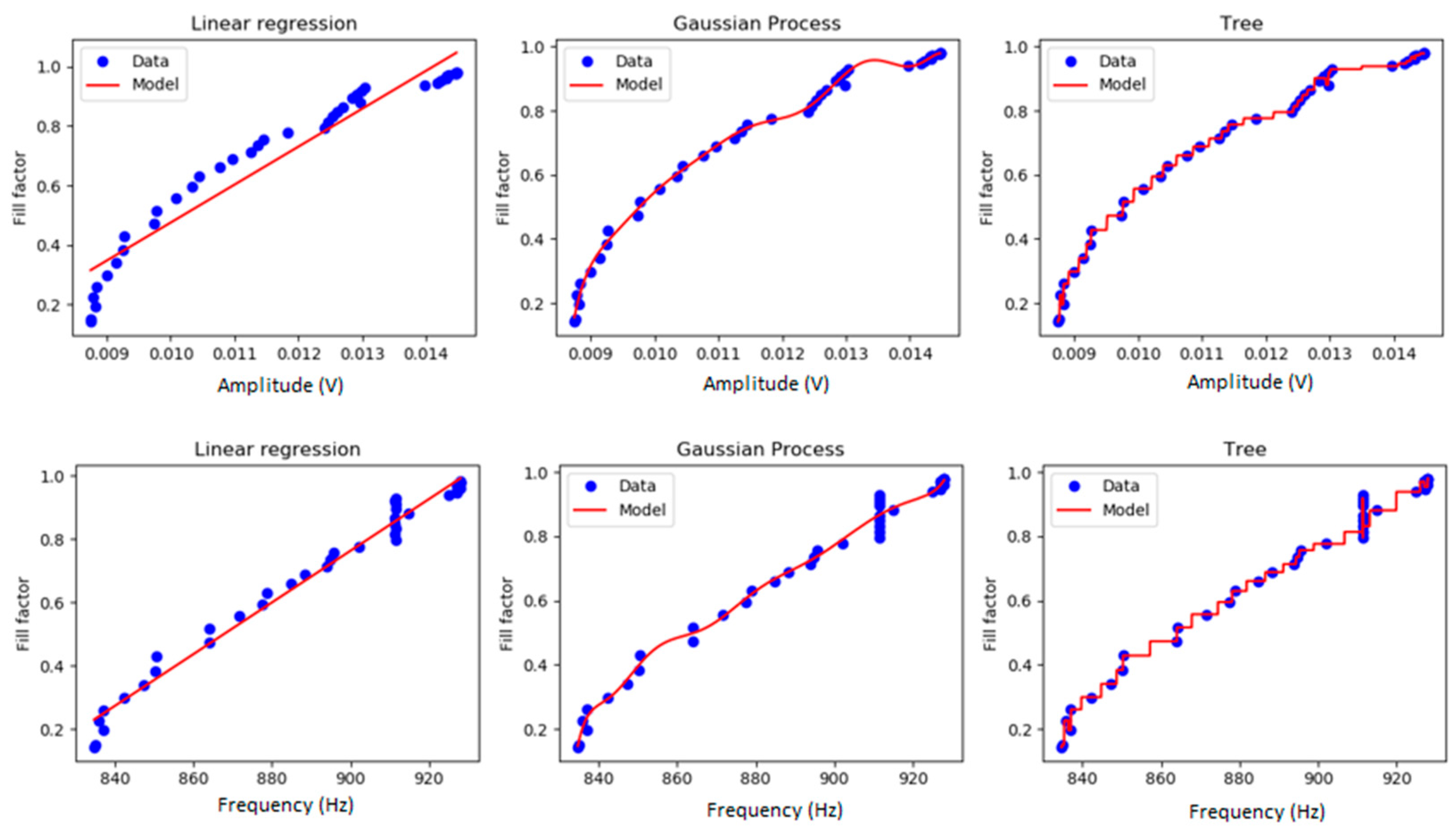
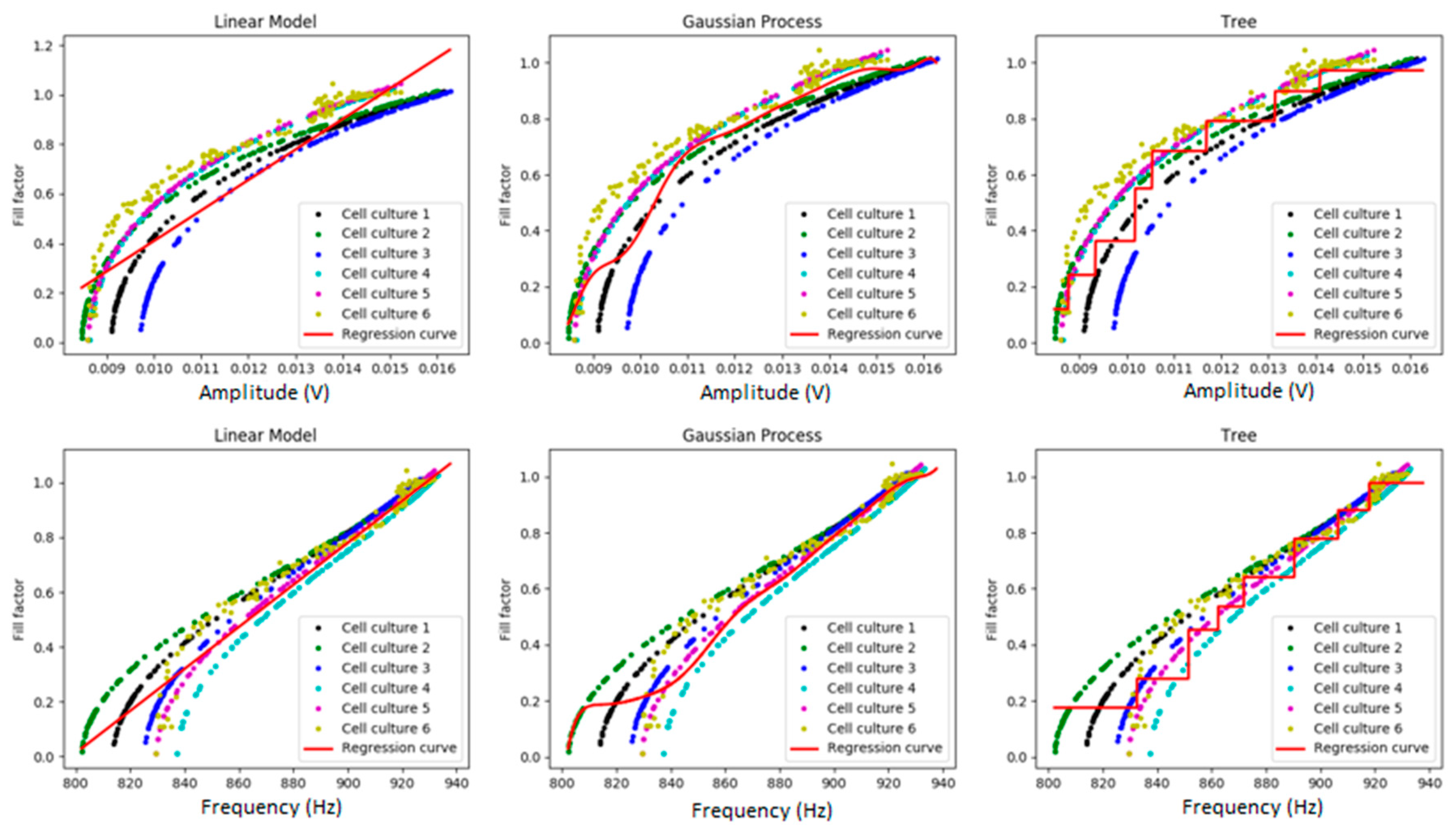
2.3.1. Linear Regression Model
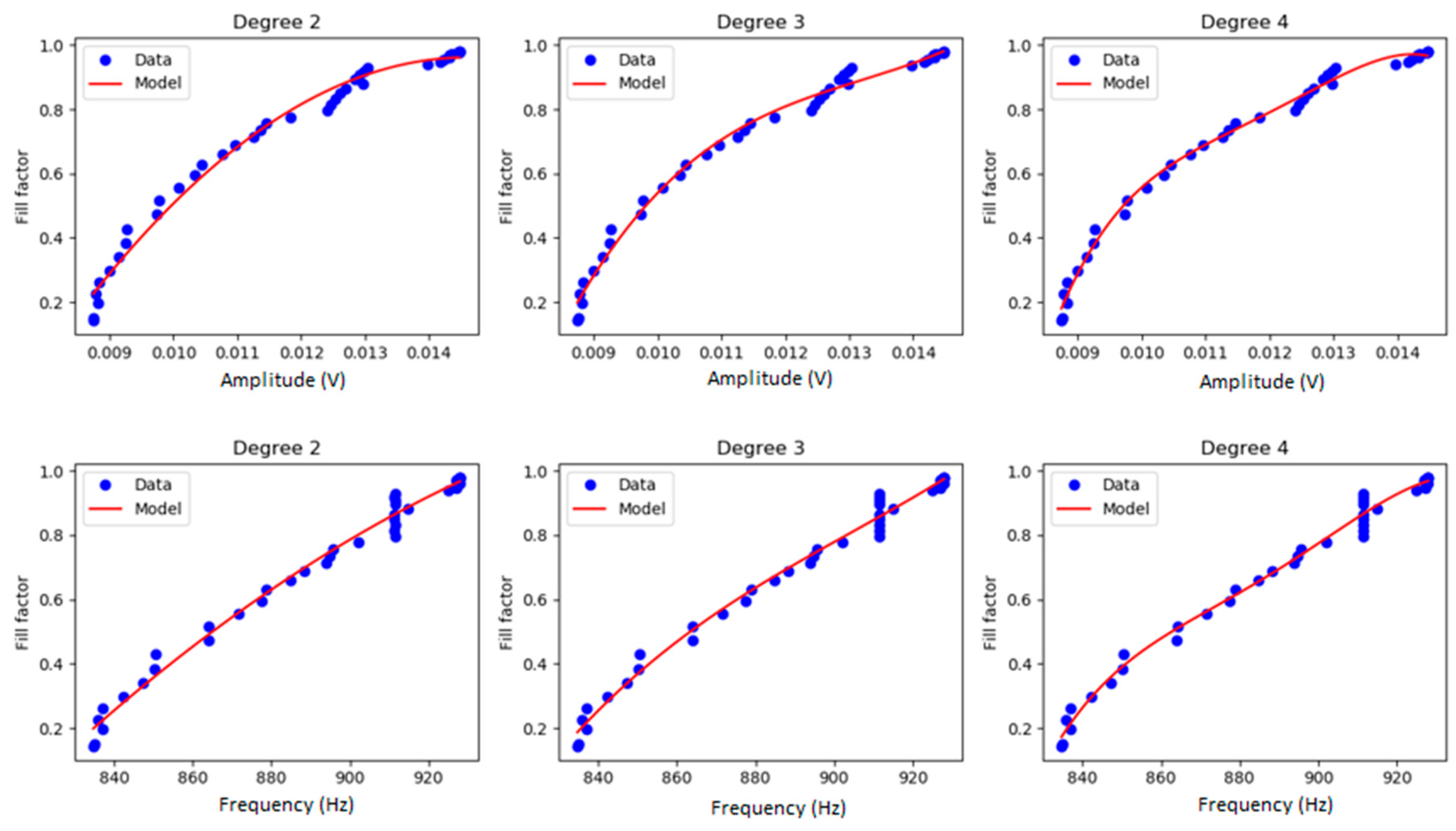
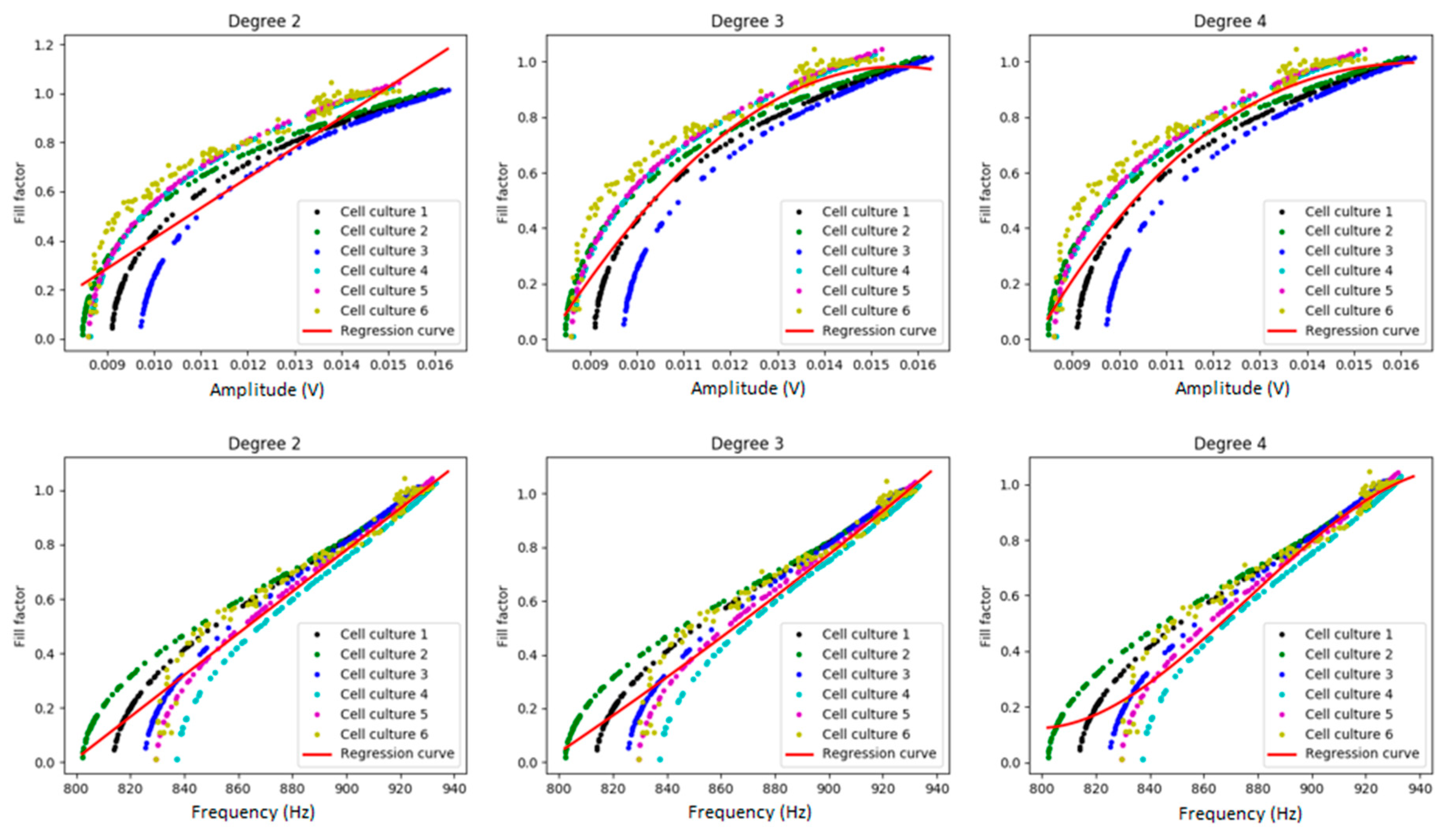
2.3.2. Polynomial Regression Model
2.3.3. Regression Trees
2.3.4. Gaussian Processes
2.4. Experimental Procedure
3. Results
3.1. N2A Cell Culture
3.2. Myoblast Cell Cultures
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Affonso, C.; Sassi, R.J.; Marques, R. Biological image classification using rough-fuzzy artificial neural network. Expert Syst. Appl. 2015, 42, 9482–9488. [Google Scholar] [CrossRef]
- Turki, T.; Taguchi, Y.-H. Machine learning algorithms for predicting drugs–tissues relationships. Expert Syst. Appl. 2019, 127, 167–186. [Google Scholar] [CrossRef]
- Eriksson, L.; McCready, C. Characterizing a bioprocess with advanced data analytics. Bio Pharm Int. 2018, 31, 18–23. [Google Scholar]
- Giaever, I.; Keese, C.R. Micromotion of mammalian cells measured electrically. Proc. Nail. Acad. Sci. USA 1991, 88, 7896–7900. [Google Scholar] [CrossRef] [PubMed]
- Szulcek, R.; Bogaard, H.J.; van Nieuw Amerongen, G.P. Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J. Vis. Exp. 2014, 85, e51300. [Google Scholar] [CrossRef] [PubMed]
- Anchan, A.; Kalogirou-Baldwin, P.; Johnson, R.; Kho, D.T.; Joseph, W.; Hucklesby, J.; Finlay, G.J.; O’Carrol, S.J.; Angel, C.E.; Graham, E.S. Real-time measurement of melanoma cell-mediated human brain endothelial barrier disruption using electric cell-substrate impedance sensing technology. Biosensors 2019, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Gamal, W.; Wu, H.; Underwood, I.; Jia, J.; Smith, S.; Bagnaninchi, P.O. Impedance-based cellular assays for regenerative medicine. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170226. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Nguyen, D.; Greve, D.W.; Domach, M.M. Simulation of microelectrode impedance changes due to cell growth. IEEE Sens. 2004, 4, 576–583. [Google Scholar] [CrossRef]
- Olmo, A.; Yúfera, A. Computer simulation of microelectrode based bio-impedance measurements with COMSOL. In Proceedings of the 3rd International Conference on Biomedical Electronics and Devices (Biodevices), Valencia, Spain, 20–23 January 2010; pp. 178–182. [Google Scholar]
- Hamilton, S.J.; Hauptmann, A. Deep D-Bar: Real-time electrical impedance tomography imaging with deep neural networks. IEEE Trans. Med. Imaging 2018, 37, 2367–2377. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wu, H.; Huang, Y.; Yang, Y.; Jia, J. Accelerated structure-aware sparse Bayesian learning for three-dimensional electrical impedance tomography. IEEE Trans. Ind. Inf. 2019, 15, 5033–5041. [Google Scholar] [CrossRef]
- Serrano, J.A.; Huertas, G.; Maldonado-Jacobi, A.; Olmo, A.; Pérez, P.; Martín, M.E.; Daza, P.; Yúfera, A. An empirical-mathematical approach for calibration and fitting cell-electrode electrical models in bioimpedance tests. Sensors 2018, 18, 2354. [Google Scholar] [CrossRef] [PubMed]
- Huertas, G.; Maldonado, A.; Yufera, A.; Rueda, A.; Huertas, J.L. The bio-oscillator: A circuit for cell-culture assays. IEEE Trans. Circuits Syst. II Express Briefs 2015, 62, 164–168. [Google Scholar] [CrossRef]
- Pérez, P.; Huertas, G.; Olmo, A.; Maldonado, J.A.; Serrano, J.A.; Martin, M.E.; Daza, P.; Yufera, A. Remote cell growth sensing using self-sustained bio-oscillations. Sensors 2018, 18, 2550. [Google Scholar] [CrossRef] [PubMed]
- Applied Biophysics. Available online: https://www.biophysics.com/ (accessed on 25 October 2019).
- Yuste, Y.; Serrano, J.A.; Olmo, A.; Maldonado, A.; Pablo, P.; Huertas, G.; Pereira, S.; de la Portilla, F.; Yúfera, A. Monitoring muscle stem cell cultures with impedance spectroscopy. In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies, Madeira, Portugal, 19–21 January 2018; pp. 96–99. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B. Scikit-learn: Machine Learning in Python. JMLR 2011, 12, 2825–2830. [Google Scholar]
- Breiman, L. Classification and Regression Trees; Routledge: Abingdon-on-Thames, UK, 2017. [Google Scholar]
- Rasmussen, C.E.; Nickisch, H. Gaussian processes for machine learning (GPML) toolbox. J. Mach. Learn. Res. 2010, 11, 3011–3015. [Google Scholar]
- Haisong, L.; Hannaneh, H.; Shuyu, L.; Christopher, Y.; Yichao, Z.; Bo, W.; Meghana, M.; Yibo, W.; Kimber, K.; Wenzhuo, Y.; et al. A wearable electrofluidic actuation system. Lab Chip 2019, 19, 2966–2972. [Google Scholar]
| Variable | Description |
|---|---|
| x1 | Amplitude of the sine wave generated by the OBT circuit and Hz(s) 1 |
| x2 | Frequency of the sine wave generated by the OBT circuit and Hz(s) 1 |
| y | Fill-factor corresponding to the amplitude and frequency values. It’s the parameter to predict, which can be compared with experimental data in [12,16]. |
| Regression Model | MSE and Standard Deviation | ||
|---|---|---|---|
| Bidimensional | Amplitude | Frequency | |
| Linear Regression | 1.41 × 10−3 (1 × 10−3) | 6.55 × 10−3 (3.45 × 10−3) | 1.76 × 10−3 (1.1 × 10−3) |
| Polynomial Regression (d = 2) | 3.3 × 10−4 (3.4 × 10−4) | 1.2 × 10−3 (1 × 10−3) | 1 × 10−3 (7.9 × 10−4) |
| Polynomial Regression (d = 3) | 1.6 × 10−4 (1.6 × 10−4) | 7.9 × 10−4 (6 × 10−4) | 1.1 × 10−3 (7.4 × 10−4) |
| Polynomial Regression (d = 4) | 8.44 × 10−5 (1.1 × 10−4) | 5.7 × 10−4 (3.3 × 10−4) | 9.45 × 10−4 (6.9 × 10−4) |
| Regression Trees | 9.78 × 10−4 (6.9 × 10−4) | 1 × 10−3 (5.7 × 10−4) | 2.2 × 10−3 (1.39 × 10−3) |
| Gaussian Processes | 1.6 × 10−4 (3.4 × 10−4) | 4.34 × 10−4 (3.4 × 10−4) | 1 × 10−3 (7.2 × 10−4) |
| Regression Model | MSE and Standard Deviation | ||
|---|---|---|---|
| Bidimensional | Amplitude | Frequency | |
| Linear Regression | 4.7 × 10−3 (2.5 × 10−3) | 1.3 × 10−2 (4.8 × 10−3) | 5.9 × 10−3 (3.8 × 10−3) |
| Polynomial Regression (d = 2) | 5.1 × 10−3 (3.7 × 10−3) | 8.4 × 10−3 (6.8 × 10−3) | 6 × 10−3 (3.7 × 10−3) |
| Polynomial Regression (d = 3) | 2.4 × 10−3 (1.5 × 10−3) | 9 × 10−3 (8.3 × 10−3) | 4.9 × 10−3 (3 × 10−3) |
| Polynomial Regression (d = 4) | 1.2 × 10−3 (8 × 10−4) | 9.5 × 10−3 (8.8 × 10−3) | 5.1 × 10−3 (3.2 × 10−3) |
| Regression Trees | 7 × 10−3 (3.8 × 10−3) | 1.3 × 10−2 (1 × 10−2) | 6.6 × 10−3 (3.3 × 10−3) |
| Gaussian Processes | 4.1 × 10−3 (5.2 × 10−3) | 1.4 × 10−2 (1 × 10−2) | 6.1 × 10−3 (3.8 × 10−3) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García, E.; Pérez, P.; Olmo, A.; Díaz, R.; Huertas, G.; Yúfera, A. Data-Analytics Modeling of Electrical Impedance Measurements for Cell Culture Monitoring. Sensors 2019, 19, 4639. https://doi.org/10.3390/s19214639
García E, Pérez P, Olmo A, Díaz R, Huertas G, Yúfera A. Data-Analytics Modeling of Electrical Impedance Measurements for Cell Culture Monitoring. Sensors. 2019; 19(21):4639. https://doi.org/10.3390/s19214639
Chicago/Turabian StyleGarcía, Elvira, Pablo Pérez, Alberto Olmo, Roberto Díaz, Gloria Huertas, and Alberto Yúfera. 2019. "Data-Analytics Modeling of Electrical Impedance Measurements for Cell Culture Monitoring" Sensors 19, no. 21: 4639. https://doi.org/10.3390/s19214639
APA StyleGarcía, E., Pérez, P., Olmo, A., Díaz, R., Huertas, G., & Yúfera, A. (2019). Data-Analytics Modeling of Electrical Impedance Measurements for Cell Culture Monitoring. Sensors, 19(21), 4639. https://doi.org/10.3390/s19214639







