The Advent of Salivary Breast Cancer Biomarker Detection Using Affinity Sensors
Abstract
1. Introduction
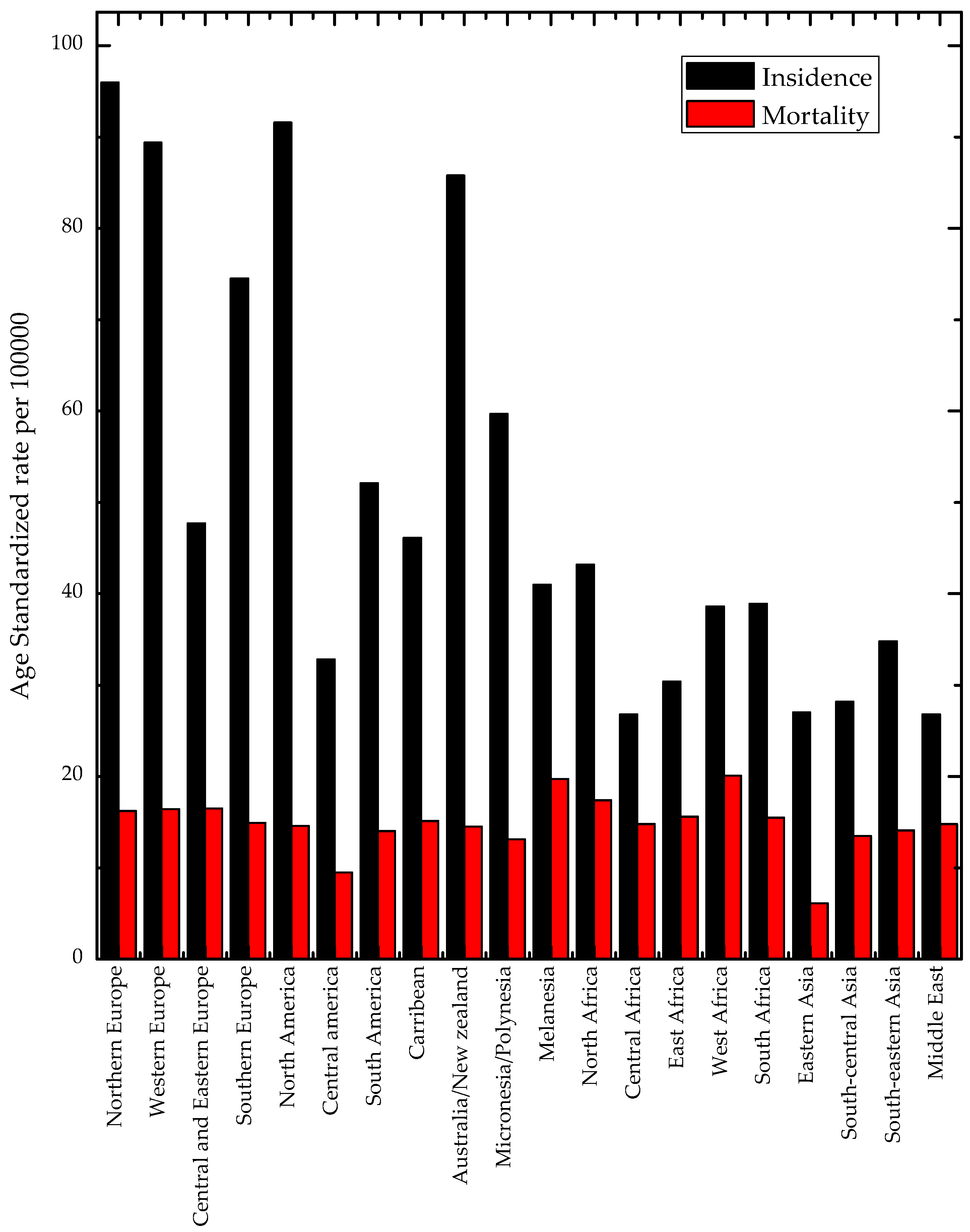
2. Prevalence of Breast Cancer Internationally, Regionally, and Locally
Regional and National Characteristics of Breast Cancer
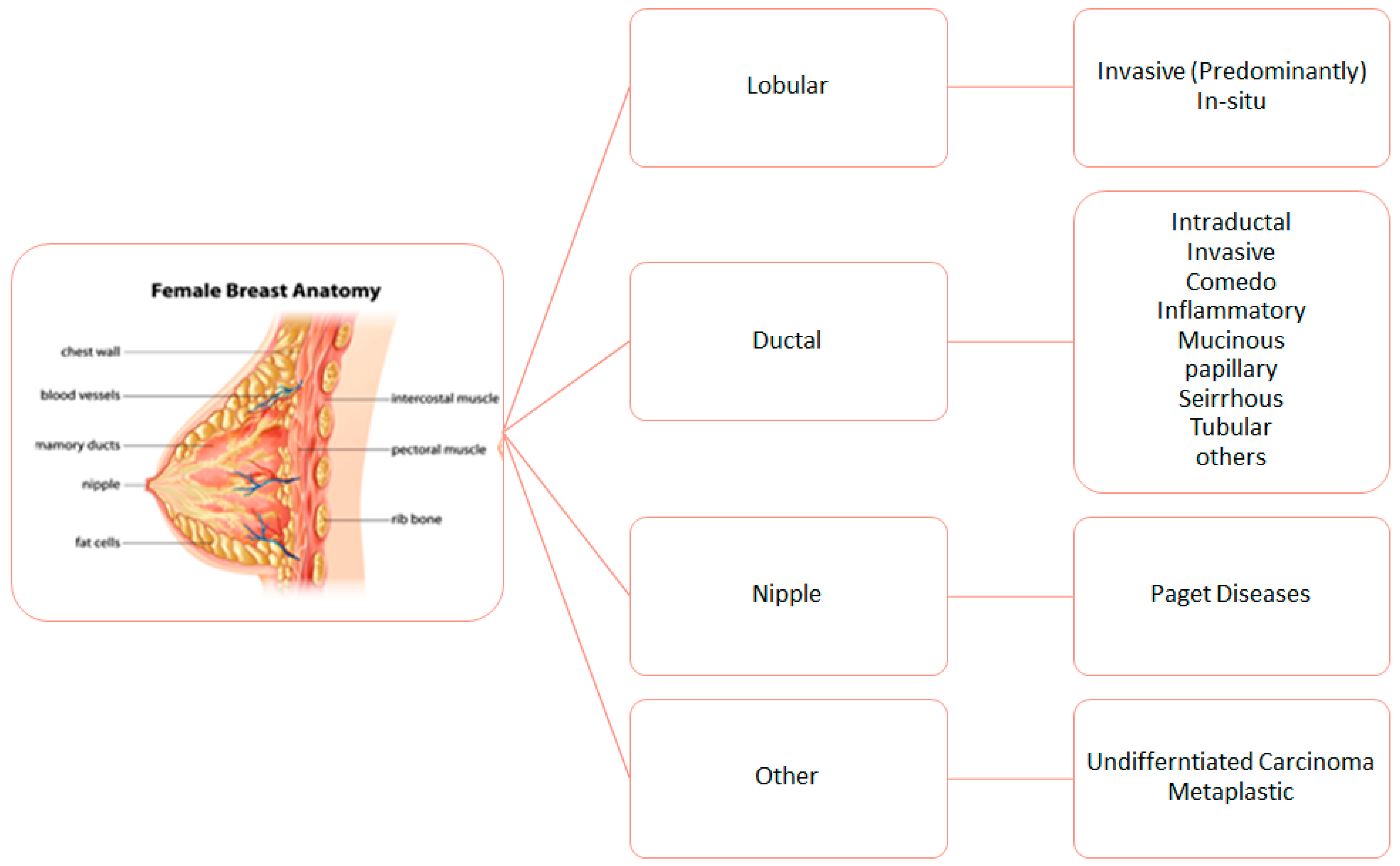
3. Biological Characteristics of Breast Cancer
Regional Immunohistochemical Characteristics of BC
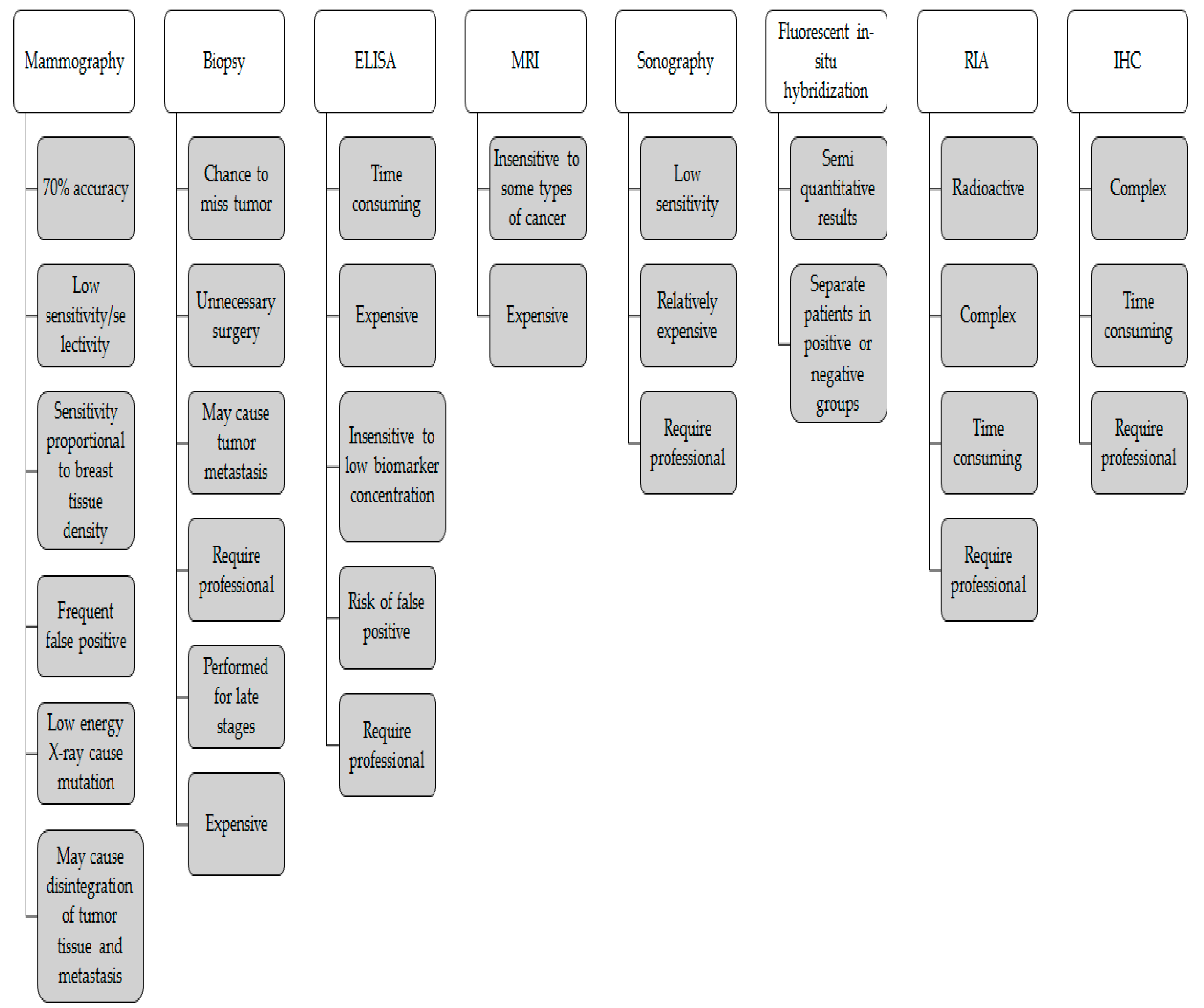
4. Early Detection of Breast Cancer Biomarkers Diagnosis Tests
4.1. Mammography
4.2. Clinical-Breast Examination (CBE) and Breast Self-Examination (BSE)
5. Body Fluids and the BC Biomarkers Found in Them
5.1. Blood
5.2. Saliva
5.3. Others
5.4. BC Biomarkers Found in Biological Fluids
5.4.1. Human Epidermal Receptor (HER) Family
5.4.2. CEA
5.4.3. Mucins 1
5.4.4. Tumor Protein 53 (p53)
5.4.5. VEGF and EGF
5.4.6. Other Proteins
5.4.7. Autoantibodies
5.4.8. MicroRNA
6. Affinity Sensors (AS)
- Direct assay: also known as label free assay, the biomarker is immobilized right on the surface where a detection antibody against the biomarker is used for sensing. Direct assay is the most preferred due to it being low cost and a simple, one step, procedure for the detection of target molecules, its main limitation is the lack of significant change during the recognition of the target [75,76].
- Capture assay ‘‘sandwich’’: A capturing antibody is first immobilized on the surface before the addition of the biomarker. Then the biomarker is sandwiched between it and an intermediate antibody before a third labeled antibody for the detection. This method is used to enhance the signal that is generated by the recognition of the target. Several materials have been used as labels or tags for labeling or even for enhancing the detection. The most notable ones are:
- ○
- Nanoparticles (NP): NPs are particles that range in size from 1 nm to 100 nm, ranging in materials from inorganic to organic in nature. They have been used in AS set-ups mainly due to their optical properties which differ from normal materials through their plasmonic properties which change with any surface change providing the basis of sensing. Also, they have useful fluorescent and electrochemical properties which have been exploited in different set-ups [77].
- ○
- Enzymes and proteins that can generate a fluorescence signal or an electrochemical signal through oxido-reduction reaction [78].
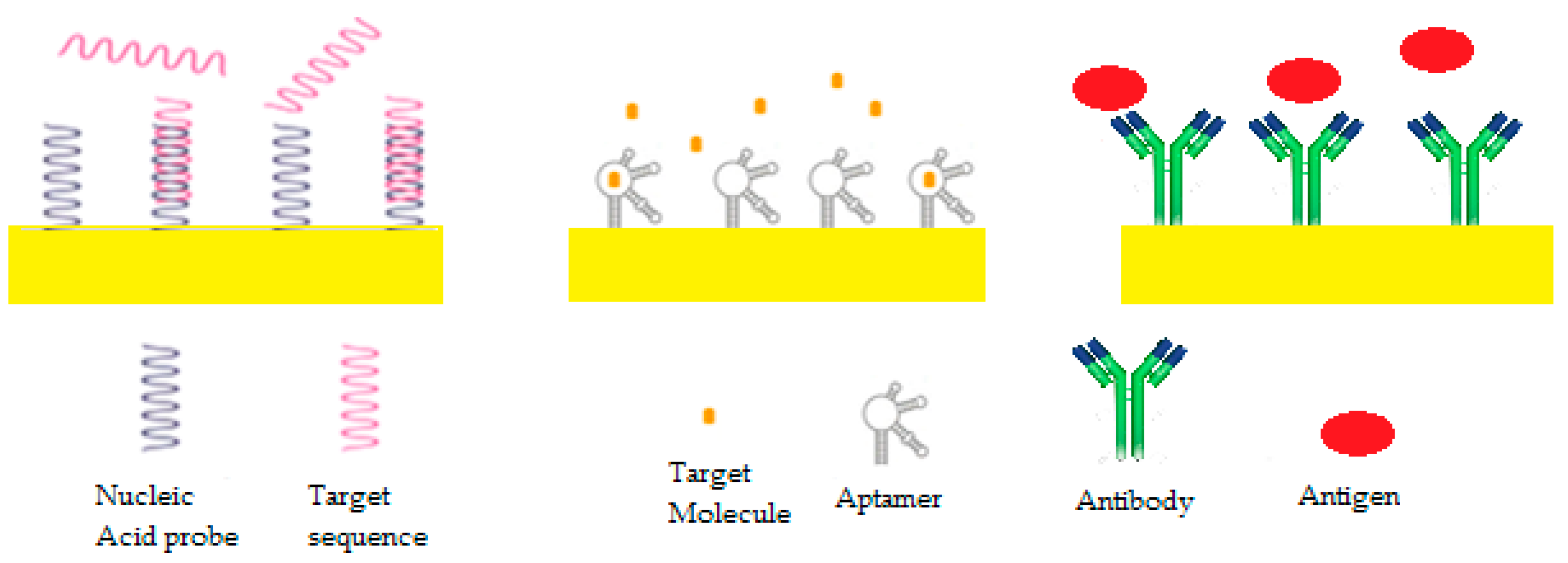
6.1. Bioreceptors
6.1.1. DNA/RNA AS
6.1.2. Peptide Nucleic Acid (PNA) AS
6.1.3. Antigen/Antibody
6.1.4. Enzyme Biosensors
6.1.5. Aptameric Biosensors
6.1.6. Molecularly Imprinted Polymers (MIPs)
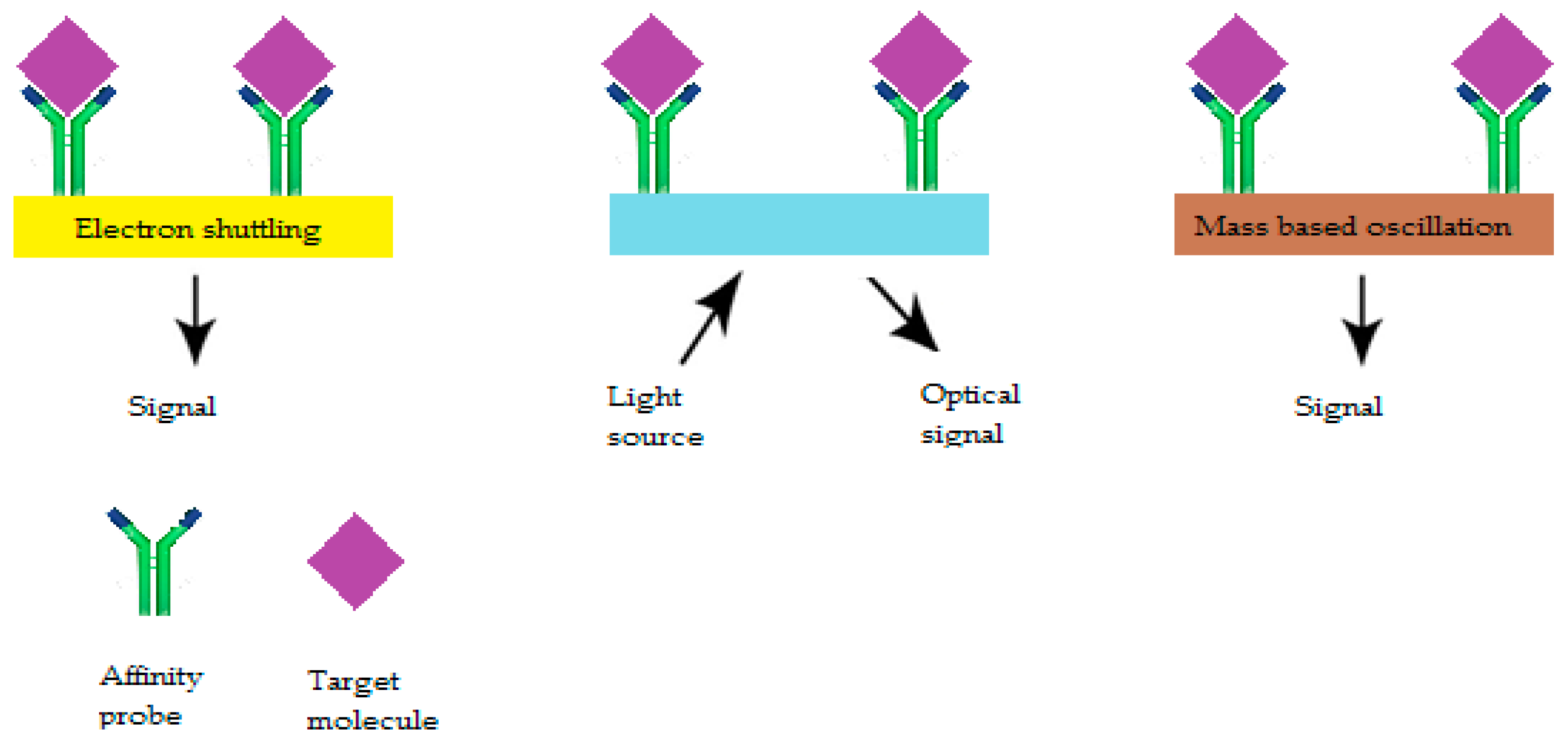
6.2. Biotransducers
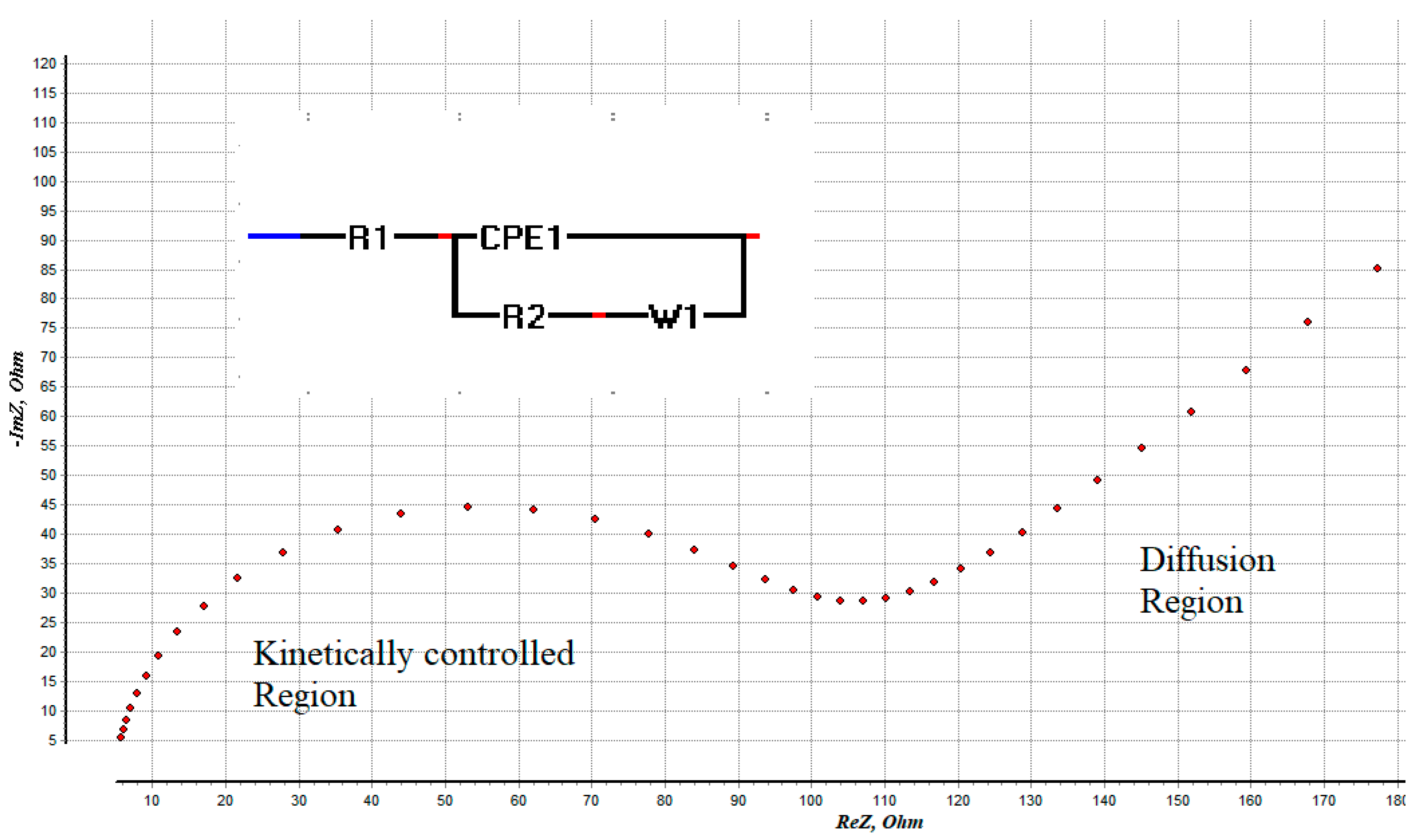
6.2.1. Electrochemical AS
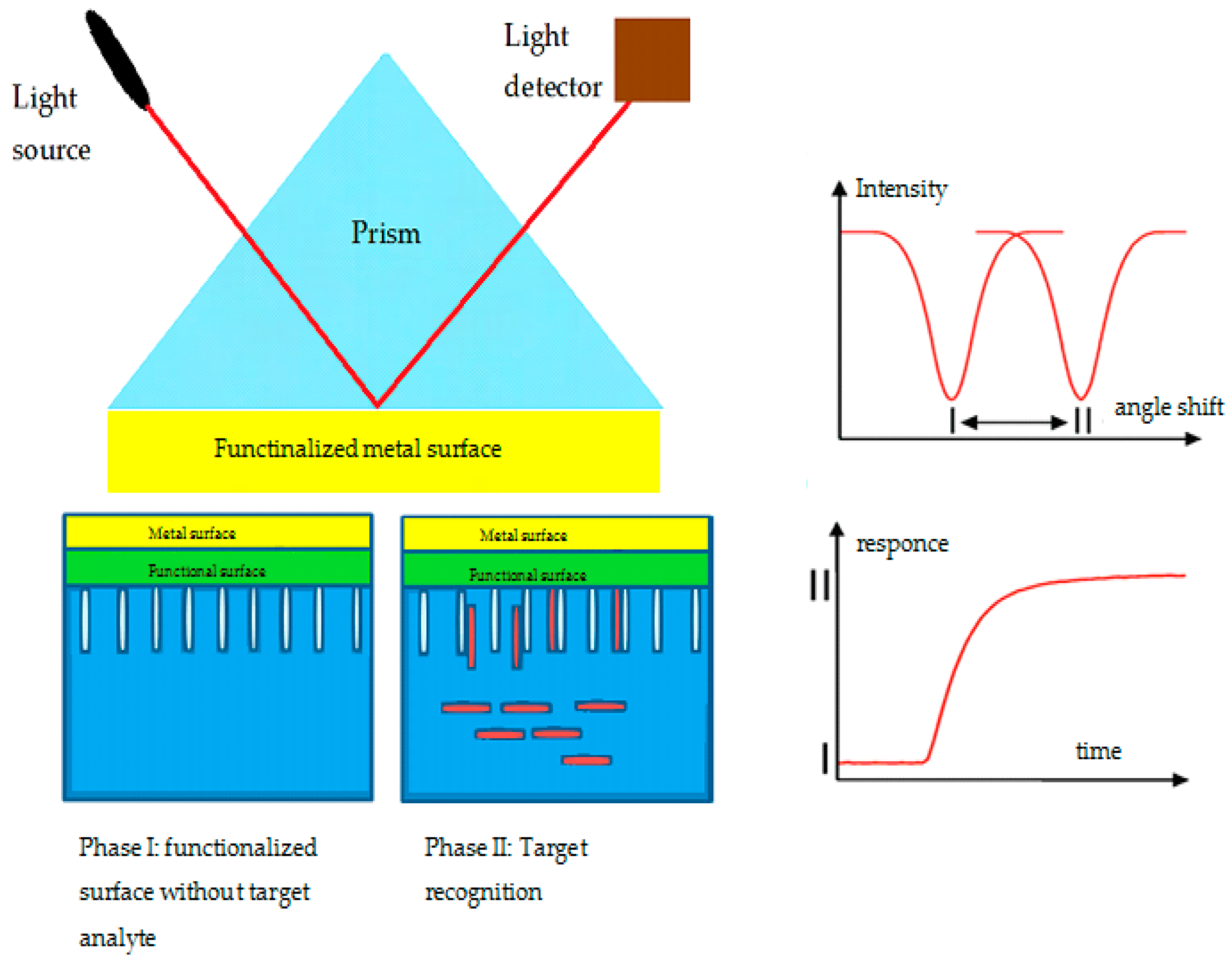
6.2.2. Optical AS
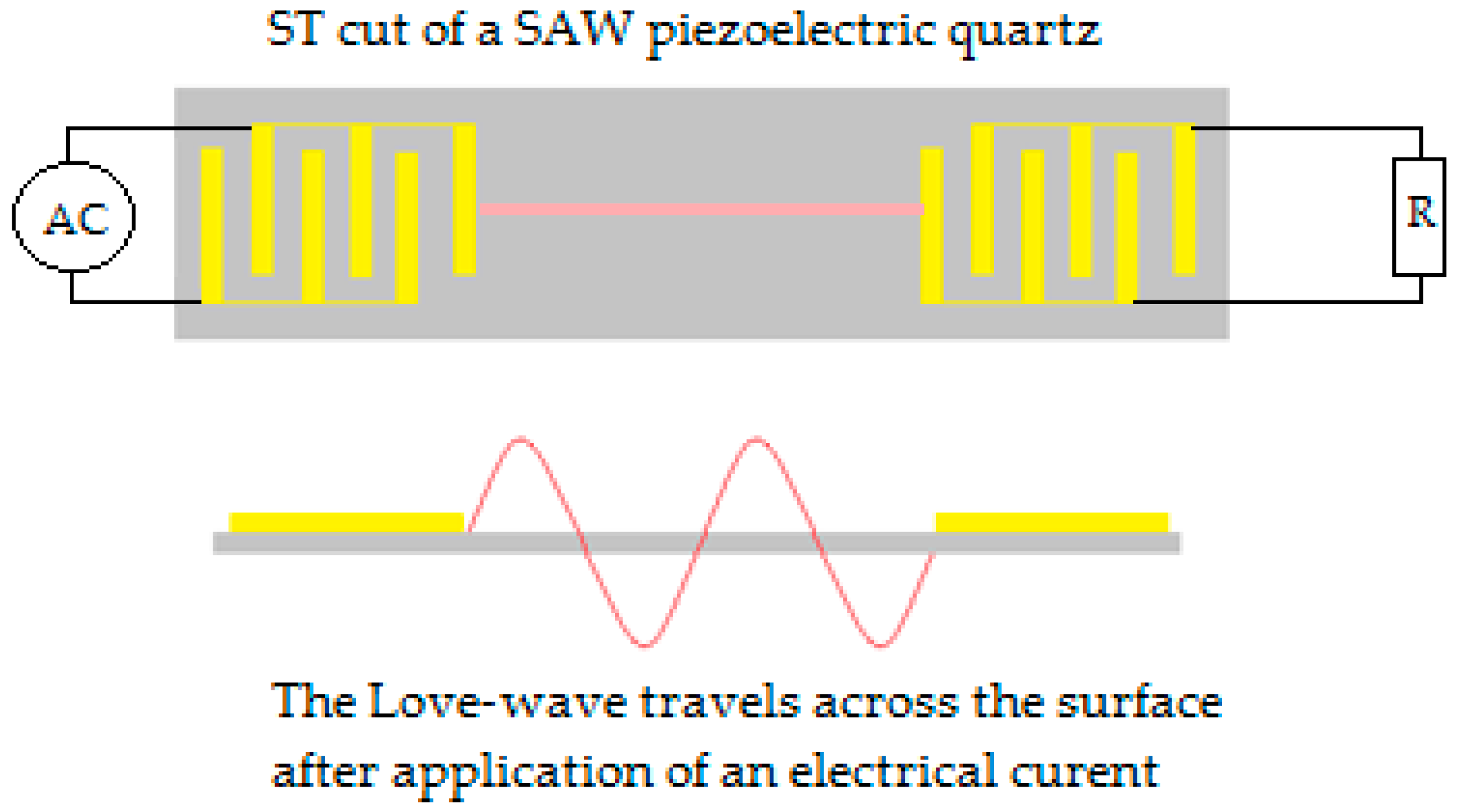
6.2.3. Acoustic Wave Sensor
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- De Vita, V.T. A book review of: “UICC manual of clinical oncology”. Cancer Investig. 2006, 24, 106. [Google Scholar] [CrossRef]
- Løberg, M.; Lousdal, M.L.; Bretthauer, M.; Kalager, M. Benefits and harms of mammography screening. Breast Cancer Res. 2015, 17, 1–12. [Google Scholar] [CrossRef]
- Van Bockstal, M.; Floris, G.; Galant, C.; Lambein, K.; Libbrecht, L. A plea for appraisal and appreciation of immunohistochemistry in the assessment of prognostic and predictive markers in invasive breast cancer. Breast 2018, 37, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Zeidan, B.A.; Townsend, P.A.; Garbis, S.D.; Copson, E.; Cutress, R.I. Clinical proteomics and breast cancer. Surgeon 2015, 13, 271–278. [Google Scholar] [CrossRef]
- Miller, A.B.; Baines, C.J. The role of clinical breast examination and breast self-examination. Prev. Med. 2011, 53, 118–120. [Google Scholar] [CrossRef]
- Croswell, J.M.; Ransohoff, D.F.; Kramer, B.S. Principles of cancer screening: Lessons from history and study design issues. Semin. Oncol. 2010, 37, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2017, 88, 217–231. [Google Scholar] [CrossRef]
- Jayanthi, V.S.P.K.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Greabu, M.; Battino, M.; Mohora, M.; Totan, A.; Didilescu, A.; Spinu, T.; Totan, C.; Miricescu, D.; Radulescu, R. Saliva—A diagnostic window to the body, both in health and in disease. J. Med. Life 2009, 2, 124–132. [Google Scholar] [PubMed]
- Liu, J.; Duan, Y. Saliva: A potential media for disease diagnostics and monitoring. Oral Oncol. 2012, 48, 569–577. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2018, 144, 1941–1953. [Google Scholar] [CrossRef]
- Pilleron, S.; Sarfati, D.; Janssen-Heijnen, M.; Vignat, J.; Ferlay, J.; Bray, F.; Soerjomataram, I. Global cancer incidence in older adults, 2012 and 2035: A population-based study. Int. J. Cancer 2018, 58, 49–58. [Google Scholar] [CrossRef]
- Ly, D.; Forman, D.; Ferlay, J.; Brinton, L.A.; Cook, M.B. An international comparison of male and female breast cancer incidence rates. Int. J. Cancer 2012, 132, 1918–1926. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available online: http://gco.iarc.fr/today (accessed on 8 February 2019).
- Preat, F.; Simon, P.; Noel, J.-C. Differences in breast carcinoma immunohistochemical subtypes between immigrant Arab and European women. Diagn. Pathol. 2014, 9, 26. [Google Scholar] [CrossRef]
- Tao, Z.; Shi, A.; Lu, C.; Song, T.; Zhang, Z.; Zhao, J. Breast cancer: Epidemiology and etiology. Cell Biochem. Biophys. 2014, 72, 333–338. [Google Scholar] [CrossRef]
- Makki, J. Diversity of breast carcinoma: Histological subtypes and clinical relevance. Clin. Med. Insights Pathol. 2015, 8, 23–31. [Google Scholar] [CrossRef]
- Jones, R.L.; Constantinidou, A.; Reis-Filho, J.S. Molecular classification of breast cancer. Surg. Pathol. Clin. 2012, 5, 701–717. [Google Scholar] [CrossRef]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Bertucci, F.; Finetti, P.; Vermeulen, P.; Van Dam, P.; Dirix, L.; Birnbaum, D.; Viens, P.; Van Laere, S. Genomic profiling of inflammatory breast cancer: A review. Breast 2014, 23, 538–545. [Google Scholar] [CrossRef]
- Kamel, D.; Brady, B.; Tabchy, A.; Mills, G.B.; Hennessy, B. Proteomic classification of breast cancer. Curr. Drug Targets 2012, 13, 1495–1509. [Google Scholar] [CrossRef][Green Version]
- Lal, S.; Reed, A.E.M.; de Luca, X.M.; Simpson, P.T. Molecular signatures in breast cancer. Methods 2017, 131, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Hoang, L.N.; Gilks, B.C. Hereditary breast and ovarian cancer syndrome: Moving beyond BRCA1 and BRCA2. Adv. Anat. Pathol. 2018, 25, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan-Haq, A.N. MYC-Driven pathways in breast cancer subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef]
- Marabelli, M.; Cheng, S.C.; Parmigiani, G. Penetrance of ATM gene mutations in breast cancer: A meta-analysis of different measures of risk. Genet. Epidemiol. 2016, 40, 425–431. [Google Scholar] [CrossRef]
- McVeigh, T.P.; Choi, J.K.; Miller, N.M.; Green, A.J.; Kerin, M.J. Lobular breast cancer in a CDH1 splice site mutation carrier: Case report and review of the literature. Clin. Breast Cancer 2014, 14, e47–e51. [Google Scholar] [CrossRef]
- Kriege, M.; Hollestelle, A.; Jager, A.; Huijts, P.E.A.; Berns, E.M.; Sieuwerts, A.M.; Meijer-Van Gelder, M.E.; Collée, J.M.; Devilee, P.; Hooning, M.J.; et al. Survival and contralateral breast cancer in CHEK2 1100delC breast cancer patients: Impact of adjuvant chemotherapy. Br. J. Cancer 2014, 111, 1004–1013. [Google Scholar] [CrossRef]
- Yamamoto, M.; Hosoda, M.; Nakano, K.; Jia, S.; Hatanaka, K.C.; Takakuwa, E.; Hatanaka, Y.; Matsuno, Y.; Yamashita, H. P53 accumulation is a strong predictor of recurrence in estrogen receptor-positive breast cancer patients treated with aromatase inhibitors. Cancer Sci. 2014, 105, 81–88. [Google Scholar] [CrossRef]
- Chouchane, L.; Boussen, H.; Sastry, K.S.R. Breast cancer in Arab populations: Molecular characteristics and disease management implications. Lancet Oncol. 2013, 14, e417–e424. [Google Scholar] [CrossRef]
- Chahine, G.; El Rassy, E.; Khazzaka, A.; Saleh, K.; Rassy, N.; Khalife, N.; Atallah, D. Characteristics of incident female breast cancer in Lebanon, 1990–2013: Descriptive study of 612 cases from a hospital tumor registry. Cancer Epidemiol. 2015, 39, 303–306. [Google Scholar] [CrossRef]
- Sabel, M.S. Principles of breast cancer screening. In Essentials of Breast Surgery; Elsevier: Amsterdam, The Netherlands, 2009; pp. 19–40. [Google Scholar]
- Dabbous, F.M.; Dolecek, T.A.; Berbaum, M.L.; Friedewald, S.M.; Summerfelt, W.T.; Hoskins, K.; Rauscher, G.H. Impact of a false-positive screening mammogram on subsequent screening behavior and stage at breast cancer diagnosis. Cancer Epidemiol. Biomark. Prev. 2017, 26, 397–403. [Google Scholar] [CrossRef]
- Puliti, D.; Duffy, S.W.; Miccinesi, G.; De Koning, H.; Lynge, E.; Zappa, M.; Paci, E. Overdiagnosis in mammographic screening for breast cancer in Europe: A literature review. J. Med. Screen. 2012, 19, 42–56. [Google Scholar] [CrossRef]
- Wender, R.C.; Brawley, O.W.; Fedewa, S.A.; Gansler, T.; Smith, R.A. A blueprint for cancer screening and early detection: Advancing screening’s contribution to cancer control. CA Cancer J. Clin. 2018, 50–79. [Google Scholar] [CrossRef]
- Junod, B.; Zahl, P.-H.; Kaplan, R.M.; Olsen, J.; Greenland, S. An investigation of the apparent breast cancer epidemic in France: Screening and incidence trends in birth cohorts. BMC Cancer 2011, 11, 401. [Google Scholar] [CrossRef][Green Version]
- Haddad, F.G.; Kourie, H.R.; Adib, S.M. Trends in mammography utilization for breast cancer screening in a Middle-Eastern country: Lebanon 2005–2013. Cancer Epidemiol. 2015, 39, 819–824. [Google Scholar] [CrossRef]
- Brennan, M.E. The role of clinical breast examination in cancer screening for women at average risk: A mini review. Maturitas 2016, 92, 61–63. [Google Scholar] [CrossRef]
- Dehn, R.; Asprey, D. Essential Clinical Procedures: Expert Consult-Online and Print; Saunders: Philadelphia, PA, USA, 2013. [Google Scholar]
- Schwab, F.D.; Huang, D.J.; Schmid, S.M.; Schötzau, A.; Güth, U. Self-detection and clinical breast examination: Comparison of the two “classical” physical examination methods for the diagnosis of breast cancer. Breast 2015, 24, 90–92. [Google Scholar] [CrossRef]
- Gold, P.; Freedman, S.O. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 1965, 121, 439–462. [Google Scholar] [CrossRef]
- Murray, R.K. Illustrated Biochemistry; McGraw-Hill: New York, NY, USA, 2009. [Google Scholar]
- Pepe, M.S.; Feng, Z.; Janes, H.; Bossuyt, P.M.; Potter, J.D. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: Standards for study design. J. Natl. Cancer Inst. 2008, 100, 1432–1438. [Google Scholar] [CrossRef]
- Hanash, S.M.; Baik, C.S.; Kallioniemi, O. Emerging molecular biomarkers-blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 2011, 8, 142–150. [Google Scholar] [CrossRef]
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; De Luca Canto, G.; Acevedo, A.C.; Guerra, E.N.S. Salivary biomarkers in the diagnosis of breast cancer: A review. Crit. Rev. Oncol. Hematol. 2017, 110, 62–73. [Google Scholar] [CrossRef]
- Chiu, M.L.; Lawi, W.; Snyder, S.T.; Wong, P.K.; Liao, J.C.; Gau, V. Matrix effects—A challenge toward automation of molecular analysis. JALA J. Assoc. Lab. Autom. 2010, 15, 233–242. [Google Scholar] [CrossRef]
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative diagnostic methods for early prostate cancer detection through urine analysis: A review. Cancers 2018, 10, 123. [Google Scholar] [CrossRef]
- Erbes, T.; Hirschfeld, M.; Rücker, G.; Jaeger, M.; Boas, J.; Iborra, S.; Mayer, S.; Gitsch, G.; Stickeler, E. Feasibility of urinary {microRNA} detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 2015, 15, 193. [Google Scholar] [CrossRef]
- Guo, C.; Li, X.; Ye, M.; Xu, F.; Yu, J.; Xie, C.; Cao, X.; Guo, M.; Yuan, Y.; Zheng, S. Discriminating patients with early-stage breast cancer from benign lesions by detection of oxidative {DNA} damage biomarker in urine. Oncotarget 2017, 8, 53100–53109. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef] [PubMed]
- Grace, P.B.; Taylor, J.I.; Low, Y.L.; Luben, R.N.; Mulligan, A.A.; Botting, N.P.; Dowsett, M.; Welch, A.A.; Khaw, K.T.; Wareham, N.J.; et al. Phytoestrogen concentrations in serum and spot urine as biomarkers for dietary phytoestrogen intake and their relation to breast cancer risk in European Prospective Investigation of Cancer and Nutrition-Norfolk. Cancer Epidemiol. Biomark. Prev. 2004, 13, 698–708. [Google Scholar]
- Thomas, H.V.; Reeves, G.K.; Key, T.J.A. Endogenous estrogen and postmenopausal breast cancer: A quantitative review. Cancer Causes Control 1997, 8, 922–928. [Google Scholar] [CrossRef]
- Zelig, U.; Barlev, E.; Bar, O.; Gross, I.; Flomen, F.; Mordechai, S.; Kapelushnik, J.; Nathan, I.; Kashtan, H.; Wasserberg, N.; et al. Early detection of breast cancer using total biochemical analysis of peripheral blood components: A preliminary study. BMC Cancer 2015, 15, 408. [Google Scholar] [CrossRef]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Levenson, V.V. Biomarkers for early detection of breast cancer: What, when, and where? Biochim. Biophys. Acta Gen. Subj. 2007, 1770, 847–856. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L.; Dellinger, T.; Dai, X.; Kingman, A.; Thigpen, J.T. The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: A preliminary study. Clin. Cancer Res. 2000, 6, 2363–2370. [Google Scholar]
- Streckfus, C.; Bigler, L.; Tucci, M.; Thigpen, J.T. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Investig. 2000, 18, 101–109. [Google Scholar] [CrossRef]
- Herbst, R.S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 21–26. [Google Scholar] [CrossRef]
- Kjaer, I.M.; Bechmann, T.; Brandslund, I.; Madsen, J.S. Prognostic and predictive value of EGFR and EGFR-ligands in blood of breast cancer patients: A systematic review. Clin. Chem. Lab. Med. 2018, 56, 688–701. [Google Scholar] [CrossRef]
- Ludovini, V.; Gori, S.; Colozza, M.; Pistola, L.; Rulli, E.; Floriani, I.; Pacifico, E.; Tofanetti, F.R.; Sidoni, A.; Basurto, C.; et al. Evaluation of serum HER2 extracellular domain in early breast cancer patients: Correlation with clinicopathological parameters and survival. Ann. Oncol. 2008, 19, 883–890. [Google Scholar] [CrossRef]
- Streckfus, C.F.; Arreola, D.; Edwards, C.; Bigler, L. Salivary protein profiles among her2/neu-receptor-positive and -negative breast cancer patients: Support for using salivary protein profiles for modeling breast cancer progression. J. Oncol. 2012, 2012, 413256. [Google Scholar] [CrossRef]
- Kabel, A.M. Tumor markers of breast cancer: New prospectives. J. Oncol. Sci. 2017, 3, 5–11. [Google Scholar] [CrossRef]
- Duffy, M.J.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Gam, L.-H. Breast cancer and protein biomarkers. World J. Exp. Med. 2012, 2, 86–91. [Google Scholar] [CrossRef]
- Duffy, M.J.; Evoy, D.; McDermott, E.W. CA 15-3: Uses and limitation as a biomarker for breast cancer. Clin. Chim. Acta 2010, 411, 1869–1874. [Google Scholar] [CrossRef] [PubMed]
- Gasco, M.; Shami, S.; Crook, T. The p53 pathway in breast cancer. Breast Cancer Res. 2002, 4, 70. [Google Scholar] [CrossRef] [PubMed]
- Brooks, M.N.; Wang, J.; Li, Y.; Zhang, R.; Elashoff, D.; Wong, D.T. Salivary protein factors are elevated in breast cancer patients. Mol. Med. Rep. 2008, 1, 375–378. [Google Scholar] [CrossRef]
- Banys-Paluchowski, M.; Witzel, I.; Riethdorf, S.; Pantel, K.; Rack, B.; Janni, W.; Fasching, P.A.; Aktas, B.; Kasimir-Bauer, S.; Hartkopf, A.; et al. The clinical relevance of serum vascular endothelial growth factor (VEGF) in correlation to circulating tumor cells and other serum biomarkers in patients with metastatic breast cancer. Breast Cancer Res. Treat. 2018, 172, 93–104. [Google Scholar] [CrossRef]
- Chapman, C.; Murray, A.; Chakrabarti, J.; Thorpe, A.; Woolston, C.; Sahin, U.; Barnes, A.; Robertson, J. Autoantibodies in breast cancer: Their use as an aid to early diagnosis. Ann. Oncol. 2007, 18, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Laidi, F.; Bouziane, A.; Errachid, A.; Zaoui, F. Usefulness of salivary and serum auto-antibodies against tumor biomarkers HER2 and MUC1 in breast cancer screening. Asian Pac. J. Cancer Prev. 2016, 17, 335–339. [Google Scholar] [CrossRef]
- Takahashi, R.U.; Miyazaki, H.; Ochiya, T. The roles of microRNAs in breast cancer. Cancers 2015, 7, 598–616. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. Micrornas: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef]
- Karunakaran, C.; Rajkumar, R.; Bhargava, K. Introduction to Biosensors. Biosens. Bioelectron. 2015, 60, 1–68. [Google Scholar]
- Bahadir, E.B.; Sezgintürk, M.K. A review on impedimetric biosensors. Artif. Cells Nanomed. Biotechnol. 2016, 44, 248–262. [Google Scholar] [CrossRef]
- Selvolini, G.; Marrazza, G. MIP-based sensors: Promising new tools for cancer biomarker determination. Sensors 2017, 17, 718. [Google Scholar] [CrossRef]
- Elshafey, R.; Tavares, A.C.; Siaj, M.; Zourob, M. Electrochemical impedance immunosensor based on gold nanoparticles-protein G for the detection of cancer marker epidermal growth factor receptor in human plasma and brain tissue. Biosens. Bioelectron. 2013, 50, 143–149. [Google Scholar] [CrossRef]
- Chamorro-Garcia, A.; Merkoçi, A. Nanobiosensors in diagnostics. Nanobiomedicine 2016, 3, 1–26. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Oligonucleotide and DNA microarrays as versatile tools for rapid diagnostics. In Portable Biosensing of Food Toxicants and Environmental Pollutants; CRC Press: Boca Raton, FL, USA, 2013; pp. 571–604. ISBN 9781466576339. [Google Scholar]
- Marques, R.C.B.; Costa-Rama, E.; Viswanathan, S.; Nouws, H.P.A.; Costa-García, A.; Delerue-Matos, C.; González-García, M.B. Voltammetric immunosensor for the simultaneous analysis of the breast cancer biomarkers CA 15-3 and HER2-ECD. Sens. Actuators B Chem. 2018, 255, 918–925. [Google Scholar] [CrossRef]
- D’Agata, R.; Giuffrida, M.C.; Spoto, G. Peptide nucleic acid-based biosensors for cancer diagnosis. Molecules 2017, 22, 1951. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, G.-Y. Current conjugation methods for immunosensors. Nanomaterials 2018, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, B.C. Machine tool slideways using multirecess hydrostatic thrust bearings. J. Inst. Eng. Mech. Eng. Div. 1973, 54, 91–94. [Google Scholar]
- Felix, F.S.; Angnes, L. Electrochemical immunosensors—A powerful tool for analytical applications. Biosens. Bioelectron. 2018, 102, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xie, Q.; Yang, D.; Xiao, H.; Fu, Y.; Tan, Y.; Yao, S. Recent advances in electrochemical glucose biosensors: A review. RSC Adv. 2013, 3, 4473. [Google Scholar] [CrossRef]
- Rahman, G. Recent trends in the development of electrochemical glucose biosensors. Int. J. Biosens. Bioelectron. 2017, 3, 210–213. [Google Scholar] [CrossRef]
- Liu, K.; Dong, H.; Deng, Y. Recent advances on rapid detection of pesticides based on enzyme biosensor of nanomaterials. J. Nanosci. Nanotechnol. 2016, 16, 6648–6656. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Shadjou, N.; De, M. Trends in analytical chemistry aptamer-based assay of biomolecules: Recent advances in electro-analytical approach. Trends Anal. Chem. 2017, 89, 119–132. [Google Scholar] [CrossRef]
- Liu, M.; Yu, X.; Chen, Z.; Yang, T.; Yang, D.; Liu, Q.; Du, K.; Li, B.; Wang, Z.; Li, S.; et al. Aptamer selection and applications for breast cancer diagnostics and therapy. J. Nanobiotechnol. 2017, 15, 81. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.A.; Nguyen, H.L.; Nguyen, D.T.; Do, Q.P.; Tran, L.D. Electrosynthesized poly(1,5-diaminonaphthalene)/polypyrrole nanowires bilayer as an immunosensor platform for breast cancer biomarker CA 15-3. Curr. Appl. Phys. 2017, 17, 1422–1429. [Google Scholar] [CrossRef]
- Hong, C.; Yuan, R.; Chai, Y.; Zhuo, Y. Ferrocenyl-doped silica nanoparticles as an immobilized affinity support for electrochemical immunoassay of cancer antigen 15-3. Anal. Chim. Acta 2009, 633, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Jiao, X.; Chen, D. Ultrasensitive electrochemical immunosensor for CA 15-3 using thionine-nanoporous gold-graphene as a platform and horseradish peroxidase-encapsulated liposomes as signal amplification. Analyst 2012, 137, 4440–4447. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Li, S.; Turner, A.P.F. Electrochemical immunosensor with N-doped graphene-modified electrode for label-free detection of the breast cancer biomarker CA 15-3. Biosens. Bioelectron. 2013, 43, 25–29. [Google Scholar] [CrossRef]
- Akter, R.; Jeong, B.; Choi, J.S.; Rahman, M.A. Ultrasensitive nanoimmunosensor by coupling non-covalent functionalized graphene oxide platform and numerous ferritin labels on carbon nanotubes. Biosens. Bioelectron. 2016, 80, 123–130. [Google Scholar] [CrossRef]
- Ribeiro, J.A.; Pereira, C.M.; Silva, A.F.; Sales, M.G.F. Disposable electrochemical detection of breast cancer tumour marker CA 15-3 using poly(Toluidine Blue) as imprinted polymer receptor. Biosens. Bioelectron. 2018, 109, 246–254. [Google Scholar] [CrossRef]
- Pacheco, J.G.; Silva, M.S.V.; Freitas, M.; Nouws, H.P.A.; Delerue-Matos, C. Molecularly imprinted electrochemical sensor for the point-of-care detection of a breast cancer biomarker (CA 15-3). Sens. Actuators B Chem. 2018, 256, 905–912. [Google Scholar] [CrossRef]
- Cui, Z.; Wu, D.; Zhang, Y.; Ma, H.; Li, H.; Du, B.; Wei, Q.; Ju, H. Ultrasensitive electrochemical immunosensors for multiplexed determination using mesoporous platinum nanoparticles as nonenzymatic labels. Anal. Chim. Acta 2014, 807, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Chai, Y.; Yuan, R. A super intramolecular self-enhanced electrochemiluminescence immunosensor based on polymer chains grafted on palladium nanocages. Nanoscale 2014, 6, 10316. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Sun, M.; Liu, W.; Li, S.; Wang, X.; Chu, C.; Yan, M.; Yu, J. Disposable electrochemical immunosensor based on peroxidase-like magnetic silica—Graphene oxide composites for detection of cancer antigen 153. Sens. Actuators B Chem. 2014, 192, 317–326. [Google Scholar] [CrossRef]
- Park, Y.M.; Kim, S.J.; Kim, K.; Han, Y.D.; Yang, S.S.; Yoon, H.C. Lectin-based optical sensing for quantitative analysis of cancer antigen CA15-3 as a breast cancer marker. Sens. Actuators B Chem. 2013, 186, 571–579. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, Q.; Zhang, L.; Wang, L.; Zeng, Y.; Huang, H. Combined detection of breast cancer biomarkers based on plasmonic sensor of gold nanorods. Sens. Actuators B Chem. 2015, 221, 1391–1397. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, H.; Wang, H.; Zhuo, Y.; Yuan, R.; Chai, Y. Electrochemiluminescence biosensor based on 3-D DNA nanomachine signal probe powered by protein-aptamer binding complex for ultrasensitive Mucin 1 detection. Anal. Chem. 2017, 89, 4280–4286. [Google Scholar] [CrossRef]
- Tallapragada, S.D.; Layek, K.; Mukherjee, R.; Mistry, K.K.; Ghosh, M. Development of screen-printed electrode based immunosensor for the detection of HER2 antigen in human serum samples. Bioelectrochemistry 2017, 118, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Gohring, J.T.; Dale, P.S.; Fan, X. Detection of HER2 breast cancer biomarker using the opto-fluidic ring resonator biosensor. Sens. Actuators B Chem. 2010, 146, 226–230. [Google Scholar] [CrossRef]
- Ilkhani, H.; Sarparast, M.; Noori, A.; Bathaie, S.Z.; Mousavi, M.F. Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker, using gold nanoparticles as a signaling probe. Biosens. Bioelectron. 2015, 74, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.G.; Rebelo, P.; Freitas, M.; Nouws, H.P.A.; Delerue-Matos, C. Breast cancer biomarker (HER2-ECD) detection using a molecularly imprinted electrochemical sensor. Sens. Actuators B Chem. 2018, 273, 1008–1014. [Google Scholar] [CrossRef]
- Yang, S.; You, M.; Zhang, F.; Wang, Q.; He, P. A sensitive electrochemical aptasensing platform based on exonuclease recycling amplification and host-guest recognition for detection of breast cancer biomarker HER2. Sens. Actuators B Chem. 2018, 258, 796–802. [Google Scholar] [CrossRef]
- Shen, C.; Zeng, K.; Luo, J.; Li, X.; Yang, M.; Rasooly, A. self-assembled DNA generated electric current biosensor for HER2 analysis. Anal. Chem. 2017, 89, 10264–10269. [Google Scholar] [CrossRef]
- Ravalli, A.; da Rocha, C.G.; Yamanaka, H.; Marrazza, G. A label-free electrochemical affisensor for cancer marker detection: The case of HER2. Bioelectrochemistry 2015, 106, 268–275. [Google Scholar] [CrossRef]
- Arya, S.K.; Zhurauski, P.; Jolly, P.; Batistuti, M.R.; Mulato, M.; Estrela, P. Capacitive aptasensor based on interdigitated electrode for breast cancer detection in undiluted human serum. Biosens. Bioelectron. 2018, 102, 106–112. [Google Scholar] [CrossRef]
- Gruhl, F.J.; Rapp, M.; Länge, K. Label-free detection of breast cancer marker HER-2/neu with an acoustic biosensor. Procedia Eng. 2010, 5, 914–917. [Google Scholar] [CrossRef][Green Version]
- Li, R.; Huang, H.; Huang, L.; Lin, Z.; Guo, L.; Qiu, B.; Chen, G. Electrochemical biosensor for epidermal growth factor receptordetection with peptide ligand. Electrochim. Acta 2013, 109, 233–237. [Google Scholar] [CrossRef]
- Omidfar, K.; Darzianiazizi, M.; Ahmadi, A.; Daneshpour, M.; Shirazi, H. A high sensitive electrochemical nanoimmunosensor based on Fe3O4/TMC/Au nanocomposite and PT-modified electrode for the detection of cancer biomarker epidermal growth factor receptor. Sens. Actuators B Chem. 2015, 220, 1311–1319. [Google Scholar] [CrossRef]
- Lu, Y.; Ivanoff-Reyes, P.; Lee, K.; Solanki, A.; Ku, C.-J. Zinc Oxide-Based Thin Film Transistor Biosensors with High Sensitivity and Selectivity. U.S. Patent Application No. 2013/0,221,346 A1, 29 August 2013. [Google Scholar]
- Myung, S.; Solanki, A.; Kim, C.; Park, J.; Kim, K.S.; Lee, K.B. Graphene-encapsulated nanoparticle-based biosensor for the selective detection of cancer biomarkers. Adv. Mater. 2011, 23, 2221–2225. [Google Scholar] [CrossRef]
- Vasudev, A.; Kaushik, A.; Bhansali, S. Electrochemical immunosensor for label free epidermal growth factor receptor (EGFR) detection. Biosens. Bioelectron. 2013, 39, 300–305. [Google Scholar] [CrossRef]
- Zhao, L.; Li, C.; Qi, H.; Gao, Q.; Zhang, C. Electrochemical lectin-based biosensor array for detection and discrimination of carcinoembryonic antigen using dual amplification of gold nanoparticles and horseradish peroxidase. Sens. Actuators B Chem. 2016, 235, 575–582. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, Z. Fabrication of an ultrasensitive electrochemical immunosensor for CEA based on conducting long-chain polythiols. Biosens. Bioelectron. 2013, 46, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; She, Z.; Ma, T.; Tian, S.; Kraatz, H.-B. Electrochemical detection of carcinoembryonic antigen. Biosens. Bioelectron. 2018, 102, 610–616. [Google Scholar] [CrossRef]
- Viswanathan, S.; Rani, C.; Anand, A.V.; Ho, J.-A.A. Disposable electrochemical immunosensor for carcinoembryonic antigen using ferrocene liposomes and MWCNT screen-printed electrode. Biosens. Bioelectron. 2009, 24, 1984–1989. [Google Scholar] [CrossRef]
- Wen, W.; Huang, J.-Y.; Bao, T.; Zhou, J.; Xia, H.-X.; Zhang, X.-H.; Wang, S.-F.; Zhao, Y.-D. Increased electrocatalyzed performance through hairpin oligonucleotide aptamer-functionalized gold nanorods labels and graphene-streptavidin nanomatrix: Highly selective and sensitive electrochemical biosensor of carcinoembryonic antigen. Biosens. Bioelectron. 2016, 83, 142–148. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Deng, D.; Luo, L.; He, H.; Wang, Z. Water-dispersible graphene/amphiphilic pyrene derivative nanocomposite: High AuNPs loading capacity for CEA electrochemical immunosensing. Sens. Actuators B Chem. 2017, 248, 966–972. [Google Scholar] [CrossRef]
- Wang, K.; He, M.-Q.; Zhai, F.-H.; He, R.-H.; Yu, Y.-L. A label-free and enzyme-free ratiometric fluorescence biosensor for sensitive detection of carcinoembryonic antigen based on target-aptamer complex recycling amplification. Sens. Actuators B Chem. 2017, 253, 893–899. [Google Scholar] [CrossRef]
- Li, H.; Shi, L.; Sun, D.-e.; Li, P.; Liu, Z. Fluorescence resonance energy transfer biosensor between upconverting nanoparticles and palladium nanoparticles for ultrasensitive CEA detection. Biosens. Bioelectron. 2016, 86, 791–798. [Google Scholar] [CrossRef]
- Li, S.; Wan, Y.; Su, Y.; Fan, C.; Bhethanabotla, V.R. Gold nanoparticle-based low limit of detection Love wave biosensor for carcinoembryonic antigens. Biosens. Bioelectron. 2017, 95, 48–54. [Google Scholar] [CrossRef]
- Crulhas, B.P.; Karpik, A.E.; Delella, F.K.; Castro, G.R.; Pedrosa, V.A. Electrochemical aptamer-based biosensor developed to monitor PSA and VEGF released by prostate cancer cells. Anal. Bioanal. Chem. 2017, 409, 6771–6780. [Google Scholar] [CrossRef]
- Pan, L.H.; Kuo, S.H.; Lin, T.Y.; Lin, C.W.; Fang, P.Y.; Yang, H.W. An electrochemical biosensor to simultaneously detect VEGF and PSA for early prostate cancer diagnosis based on graphene oxide/ssDNA/PLLA nanoparticles. Biosens. Bioelectron. 2017, 89, 598–605. [Google Scholar] [CrossRef]
- Lin, C.-W.; Wei, K.-C.; Liao, S.; Huang, C.-Y.; Sun, C.-L.; Wu, P.-J.; Lu, Y.-J.; Yang, H.-W.; Ma, C.-C.M. A reusable magnetic graphene oxide-modified biosensor for vascular endothelial growth factor detection in cancer diagnosis. Biosens. Bioelectron. 2015, 67, 431–437. [Google Scholar] [CrossRef]
- Fu, X.M.; Liu, Z.J.; Cai, S.X.; Zhao, Y.P.; Wu, D.Z.; Li, C.Y.; Chen, J.H. Electrochemical aptasensor for the detection of vascular endothelial growth factor (VEGF) based on DNA-templated Ag/Pt bimetallic nanoclusters. Chin. Chem. Lett. 2016, 27, 920–926. [Google Scholar] [CrossRef]
- Qureshi, A.; Gurbuz, Y.; Niazi, J.H. Capacitive aptamer-antibody based sandwich assay for the detection of VEGF cancer biomarker in serum. Sens. Actuators B Chem. 2015, 209, 645–651. [Google Scholar] [CrossRef]
- Du, D.; Wang, J.; Lu, D.; Dohnalkova, A.; Lin, Y. Multiplexed electrochemical immunoassay of phosphorylated proteins based on enzyme-functionalized gold nanorod labels and electric field-driven acceleration. Anal. Chem. 2011, 83, 6580–6585. [Google Scholar] [CrossRef]
- Afsharan, H.; Navaeipour, F.; Khalilzadeh, B.; Tajalli, H.; Mollabashi, M.; Ahar, M.J.; Rashidi, M.R. Highly sensitive electrochemiluminescence detection of p53 protein using functionalized Ru-silica nanoporous@gold nanocomposite. Biosens. Bioelectron. 2016, 80, 146–153. [Google Scholar] [CrossRef]
- Adley, C.C.; Ryan, M.P. Conductometric biosensors for high throughput screening of pathogens in food. In High Throughput Screening for Food Safety Assessment; Elsevier: Amsterdam, The Netherlands, 2015; Volume 47, pp. 315–326. ISBN 9780857098078. [Google Scholar]
- De Moraes, A.; Kubota, L. Recent trends in field-effect transistors-based immunosensors. Chemosensors 2016, 4, 20. [Google Scholar] [CrossRef]
- Randviir, E.P.; Banks, C.E. Electrochemical impedance spectroscopy: An overview of bioanalytical applications. Anal. Methods 2013, 5, 1098–1115. [Google Scholar] [CrossRef]
- Ciucci, F. Modeling electrochemical impedance spectroscopy. Curr. Opin. Electrochem. 2018, 13, 132–139. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef]
- Ligler, F.; Taitt, C. Optical biosensors. Opt. Biosens. 2008, 60, 91–100. [Google Scholar]
- Mejía-Salazar, J.R.; Oliveira, O.N. Plasmonic biosensing focus review. Chem. Rev. 2018, 118, 10617–10625. [Google Scholar] [CrossRef]
- Tainaka, K.; Sakaguchi, R.; Hayashi, H.; Nakano, S.; Liew, F.F.; Morii, T. Design strategies of fluorescent biosensors based on biological macromolecular receptors. Sensors 2010, 10, 1355–1376. [Google Scholar] [CrossRef]
- González-Vera, J.A.; Morris, M.C. Fluorescent reporters and biosensors for probing the dynamic behavior of protein kinases. Proteomes 2015, 3, 369–410. [Google Scholar] [CrossRef]
- Muzyka, K. Current trends in the development of the electrochemiluminescent immunosensors. Biosens. Bioelectron. 2014, 54, 393–407. [Google Scholar] [CrossRef]
- Wohltjen, H.; Dessy, R. Surface acoustic wave probe for chemical analysis. I. Introduction and instrument description. Anal. Chem. 1979, 51, 1458–1464. [Google Scholar] [CrossRef]
- Länge, K.; Rapp, B.E.; Rapp, M. Surface acoustic wave biosensors: A review. Anal. Bioanal. Chem. 2008, 391, 1509–1519. [Google Scholar] [CrossRef]
- Fogel, R.; Limson, J.; Seshia, A.A. Acoustic biosensors. Essays Biochem. 2016, 60, 101–110. [Google Scholar] [CrossRef]


| Molecular Subtypes | ER * | PR ** | HER2 *** | Histological Grade | Basal Marker | Proliferation Cluster | P53 Mutation | Outcome |
|---|---|---|---|---|---|---|---|---|
| Luminal A | + | + | − | Low | − | Low | Low | Good |
| Luminal B | ± | ± | −/+ | High | − | High | Intermediate | Intermediate |
| Basal like | − | − | − | High | + | High | High | Poor |
| HER+/ER− | − | −/+ | −/+ | High | ± | High | High | Poor |
| Molecular apocrine | − | − | ± | Intermediate/High | ± | High | Intermediate/High | Poor |
| Claudine-low | − | − | − | High | High | High | High | Intermediate |
| c-erbB-2 | CA 15-3 | p53 | EGFR *** | |||||
|---|---|---|---|---|---|---|---|---|
| Subject Status | C * U/mg | TSP ** mg/mL | C U/mg | TSP mg/mL | C pmol/mg | TSP mg/mL | C fmol/mg | TSP mg/mL |
| Control | 71.13 ± 66.07 | 1.48 ± 60.09 | 2.27 ± 1.54 | 1.25 ± 0.82 | 177.1 ± 61.3 | 1.25 ± 0.82 | 1.03 ± 0.69 | 1.25 ± 0.82 |
| Benign | 63.75 ± 67.00 | 1.58 ± 0.08 | 2.22 ± 1.95 | 1.44 ± 0.92 | 180.7 ± 70.7 | 1.44 ± 0.92 | 0.37 ± 0.31 | 1.44 ± 0.92 |
| Malignant | 143.58 ± 611.53 | 1.67 ± 60.10 | 5.26 ± 4.12 | 1.71 ± 0.79 | 134.6 ± 63.8 | 1.71 ± 0.79 | 0.92 ± 0.80 | 1.71 ± 0.79 |
| Protein | Electrode Type | Essay Type | Amplification | Detection Methods | Limit of Detection | Medium of Detection | Reference |
|---|---|---|---|---|---|---|---|
| CA15-3 9 | Screen-printed carbon electrode, functionalyzed with polypyrrole | Sandwich assay | Magnetic beads | Amperometric | 0.02 U/mL | PBS 1 | [90] |
| CA15-3 | Gold electrode | label free assay | Ferrocenecarboxylic doped nanoparticles | Amperometric | 0.64 U/mL | PBS, human serum | [91] |
| CA15-3 | Gold/Graphene glassy carbon electrode | Sandwich assay | HRP 2-encapsulated liposomes | Differential pulse voltammetry | 5 µU/mL | PBS, human serum | [92] |
| CA15-3 | N-doped graphene sheets on glassy carbon electrode | label free assay | - | Differential pulse voltammetry | 0.012 U/mL | PBS, human serum | [93] |
| CA15-3 | Graphene oxide modified gold electrode | Sandwich assay | Ab 3/ferritin/multiwall carbon nanotube | Differential pulse voltammetry | 0.009 U/mL | PBS, human serum | [94] |
| CA15-3 | MIPs 4/SPE 5 | Direct assay | Toluidine blue | Differential pulse voltammetry | 0.1 U/mL | PBS/artificial human serum | [95] |
| CA15-3 | MIPs/gold SPE | Direct assay | Hexacyanoferrate (II/III) | Differential pulse voltammetry/ EIS 6 | 1.5 U/mL | PBS/human serum | [96] |
| CA15-3, CA125 10, CEA 11 | screen-printed carbon electrode | Sandwich assay | platinum nanoparticles | Conductometry | 0.002 U/mL, 0.001 U/mL and 7.0 pg/mL for CA125, CA153 and CEA, respectively | PBS, human serum | [97] |
| HER2 12+ CA15-3 | Screen printed carbon electrode | label free assay | gold nanoparticles | Linear Sweep voltammetry | 5.0 U/mL for CA 15-3 and 2.9 ng/mLfor HER2 | PBS | [80] |
| CA15-3 | Glassy carbon electrode modified with gold nanoparticles | Sandwich assay | Palladium nanocage captured Ru(II) luminophore | ECL 7 | 0.003 U/mL | PBS, Human serum | [98] |
| CA15-3 | graphene oxide modified screen printed carbon electrode | Sandwich assay | Modified magnetic nanoparticles | ECL | 2.8 × 10−4 U/mL | PBS, Human serum | [99] |
| CA15-3 | Gold electrode | Sandwich assay | 2 types of Lectin modified fluoromicrobeads, Sambucus nigra agglutinin and peanut agglutinin lectins | Fluorescence | 1.2 U/mL for PNA and 0.4 U/mL for SNA | PBS | [100] |
| CA15-3 | Gold nanorods | Direct assay | Plasmonic | 0.1 nM | Human serum | [101] | |
| CA15-3 | Glassy carbon electrode | Direct assay | 3D DNA nanomachine probes using Protein-Aptamer binding complex, a mimic peroxidase | ECL | 0.62 fg/mL | PBS, Human serum | [102] |
| HER2 | screen printed electrode | Sandwich assay | HRP linked Ab | Cyclic Voltammetry | 4 ng/mL | PBS, human plasma | [103] |
| HER2 | screen printed electrode | Sandwich assay | Magnetic beads modified with enzymes, affibody were used instead of antibody | Differential pulse voltammetry | 1.8 ng/mL | PBS, human serum | [104] |
| HER2 | Gold electrode | Sandwich assay | Aptamer, DNA primer | Amperometric | 1 pg/mL | PBS, human plasma | [105] |
| HER2 | MIPs/gold SPE | Label-free assay | Ferro-ferricyanide | Differential pulse voltammetry | 1.6 ng/L | PBS, human serum | [106] |
| HER2 | Aptamer hybridized on ferrocene labeled DNA gold nanoparticles | Sandwich assay | horse radish peroxidase labeled DNA gold nanoparticles | Conductometry | 4.9 ng/mL | PBS, human serum | [107] |
| HER2 | Gold electrode | Sandwich assay | Aptamer, DNA primer | Conductometry | - | PBS, human serum | [108] |
| HER2 | screen printed graphite | label free assay | gold nanoparticles | EIS | 6.0 μg/L | PBS, human serum | [109] |
| HER2 | Gold electrode | label free assay | DNA aptamer | EIS | 1 pM | PBS, Diluted human serum | [110] |
| HER2 | 36°YX-LiTaO 3 device with gold transducers | label free assay | Neutravidin and Protein A | Surface acoustic wave | 2 ng/mL | PBS | [111] |
| EGFR 13 | Gold electrode | label free assay | ferrocene bead coupled with peptide | EIS | 0.37 ng/mL | PBS, Diluted human serum | [112] |
| EGFR | Gold electrode | label free assay | gold nanoparticles | EIS | 0.34 pg/mL in PBS and 0.88 pg/mL in human plasma | PBS, human plasma | [77] |
| EGFR | screen printed electrode | Sandwich assay | Ferro oxide/Chitosan/Gold nanoparticles | differential pulse voltammetry | - | PBS, human plasma | [113] |
| EGFR | Zinc-Oxide | Direct assay | - | FET-based sensor | 10 fM | Goat serum | [114] |
| EGFR + HER2 | Graphene encapsulated nanoparticles | Direct assay | - | FET 8-based sensor | 1 pM for HER2 and 100 pM for EGFR | PBS | [115] |
| EGFR | Gold electrode | label free assay | - | Cyclic Voltammetry | 1 pg/mL | PBS | [116] |
| CEA | screen printed electrode | label free assay | Lectin | Chronoamperometry | 0.03 ng/mL | PBS, human plasma | [117] |
| CEA | Gold electrode | label free assay | gold nanoparticles | Differential pulse voltammetry | 0.015 fg/mL | PBS, human plasma | [118] |
| CEA | gold electrode | Sandwich assay | gold nanoparticles | Square wave Voltammetry | 0.2 ng/mL | PBS, human plasma | [119] |
| CEA | screen printed carbon electrode | Sandwich assay | ferrocene carboxylic acid liposome | Square wave Voltammetry | 1 pg/mL | PBS, human plasma, human saliva | [120] |
| CEA | Graphene electrode | Sandwich assay | Gold nanorods modified with HRP and hairpin-oligonucleotide | Conductometry | 1.5 pg/mL | PBS, human serum | [121] |
| CEA | Graphene electrode | label free assay | gold nanoparticles | EIS | 0.06 ng/mL | PBS, human plasma | [122] |
| CEA | Aptamer nanocluster pair | Direct assay | - | Fluorescence | 0.1 ng/mL | 3-(N-Morpholino) propanesulfonic acid buffer, diluted human serum | [123] |
| CEA | Palladium-converting nanoparticles | Direct assay | - | resonance energy transfer | 2 pg/mL | Tris-HCl buffer, diluted human serum | [124] |
| CEA | ST 90°-X quartz | Sandwich assay | Gold nanoparticles | Love wave | 30 pg/mL | PBS | [125] |
| VEGF 14 +PSA 15 | Gold electrode | label free assay | aptamer | Square wave Voltammetry | 1.1 ng/mL | PBS, cell lysate | [126] |
| VEGF +PSA | Gold electrode modified with graphene oxide/ssDNA | label free assay | Poly-L-lactide nanoparticles | Differential pulse voltammetry | 50 pg/mL | PBS, human plasma | [127] |
| VEGF | Gold electrode | label free assay | Magnetic graphene oxide | Differential pulse voltammetry | 31.25 pg/mL | PBS, Human plasma | [128] |
| VEGF | Glass carbon electrode modified aptamer | Sandwich assay | Gold platinum nanocluster | Amperometric | 4.6 pmol/L | PBS | [129] |
| VEGF | Gold electrode | Sandwich assay | magnetic beads | EIS | 401 pg/mL | Diluted human serum | [130] |
| p53 16 | Sandwich assay for multiple detection | Gold nanorod, enzyme label | Square wave Voltammetry | 5 pM | PBS | [131] | |
| p53 | Glassy carbon electrode | Sandwich assay | Streptavidin modified gold nanoparticles | ECL | 22.8 fM | PBS, cell lysate | [132] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abrao Nemeir, I.; Saab, J.; Hleihel, W.; Errachid, A.; Jafferzic-Renault, N.; Zine, N. The Advent of Salivary Breast Cancer Biomarker Detection Using Affinity Sensors. Sensors 2019, 19, 2373. https://doi.org/10.3390/s19102373
Abrao Nemeir I, Saab J, Hleihel W, Errachid A, Jafferzic-Renault N, Zine N. The Advent of Salivary Breast Cancer Biomarker Detection Using Affinity Sensors. Sensors. 2019; 19(10):2373. https://doi.org/10.3390/s19102373
Chicago/Turabian StyleAbrao Nemeir, Imad, Joseph Saab, Walid Hleihel, Abdelhamid Errachid, Nicole Jafferzic-Renault, and Nadia Zine. 2019. "The Advent of Salivary Breast Cancer Biomarker Detection Using Affinity Sensors" Sensors 19, no. 10: 2373. https://doi.org/10.3390/s19102373
APA StyleAbrao Nemeir, I., Saab, J., Hleihel, W., Errachid, A., Jafferzic-Renault, N., & Zine, N. (2019). The Advent of Salivary Breast Cancer Biomarker Detection Using Affinity Sensors. Sensors, 19(10), 2373. https://doi.org/10.3390/s19102373








